Abstract
Explaining the structure of terrestrial and aquatic food webs remains one of the most important challenges of ecological theory. Most existing models use emergent properties of food webs, such as diversity and connectance as parameters, to determine other food-web descriptors. Lower-level processes, in particular adaptation (whether by behavioral, developmental, or evolutionary mechanisms), are usually not considered. Here, we show that complex, realistic food webs may emerge by evolution from a single ancestor based on very simple ecological and evolutionary rules. In our model, adaptation acts on body size, whose impact on the metabolism and interactions of organisms is well established. Based on parameters defined at the organism scale, the model predicts emergent properties at the food-web scale. Variations of two key parameters (width of consumption niche and competition intensity) allow very different food-web structures and functionings to emerge, which are similar to those observed in some of the best-documented food webs.
Keywords: complex adaptive system, evolutionary assembly, macroevolution, food-web structure
Although prevailing food-web models, such as the cascade model (1, 2), the niche model (3), and the more recent nested-hierarchy model (4), are able to describe food-web structure satisfactorily, they fail to provide clear mechanisms explaining how this structure emerges. There are two reasons for this shortcoming. First, these models are parameterized by using some emergent properties of observed food webs (usually diversity and connectance), although these properties result from lower-level processes. Second, they consider only binary food webs in which species and trophic links are either present or absent, but are not quantified. The dynamical aspects of food webs, linked to population dynamics and adaptation processes, although ubiquitous in ecosystems (5), are absent from these theoretical studies.
Consideration of adaptive processes may provide new approaches to understand the structure and functioning of food webs. A few recent pioneering studies (6–8) have suggested that complex food webs may emerge from evolutionary assembly processes. These models consider variations of a large set of traits that determine the strength of predator–prey interactions. Considering a large number of traits is appealing because many characteristics are likely to have a role in trophic interactions. However, because these traits are numerous and are not explicitly identified, it is not possible to understand the evolutionary dynamics in terms of selective pressure or to test model predictions by using empirical data. Here, we propose a simpler approach in which body size is the single biological trait subject to evolution. This simple approach allows us to make testable predictions on how evolution shapes ecosystem structure and functioning.
Model and Methods
The reasons for using body size as a key trait are numerous. The trophic cascade model (1, 2) is based on a hierarchy among species, and body size is a good candidate to explain this hierarchy (9, 10). Body mass, M, is also tightly linked to individual metabolism, B, by the allometric relation B ≅ Ma. The exponent a in this relation is usually 0.75 (11–13), or –0.25 if metabolism is measured per unit mass (mass-specific metabolic rate) (13). Because of this allometric relation, it is possible to correlate body size and a number of life-history traits of organisms, thereby making a link between organismic and community scales (13, 14). We modeled the population dynamics of species i with biomass Ni and body size xi by
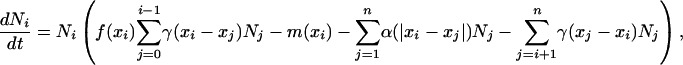 |
[1] |
where the xi are ranked by increasing values, f(xi) is the production efficiency of species i, and m(xi) is its mass-specific mortality rate. Because these two parameters are related directly to mass-specific metabolic rate, they are assumed to depend on body size (13): f(xi) = f0xi–0.25 and m(xi) = m0xi–0.25.
The function γ(xi – xj) describes the consumption rate exerted by predator i on prey j. It is assumed to be a Gaussian function with standard deviation s and a maximum value when the body sizes of the predator and the prey are separated by a distance d as follows:
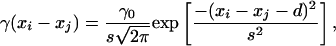 |
with xi > xj (Fig. 1). The choice of this type of function is based on the idea that, for a predator of a given size, energy gains should increase with its prey body size, whereas the probability of such successful attacks should decrease with the prey body size. As a result, body size should then be optimum at an intermediate value. This type of relationship between interaction strength and body size is supported by empirical data (13, 15, 16).
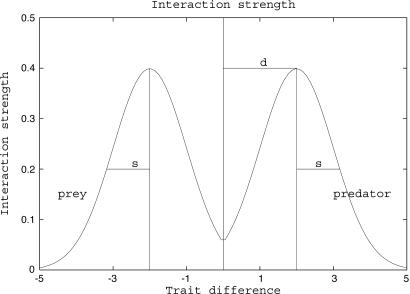
Fig. 1.
Trophic interactions. Interaction strength, as measured by function γ, between two species as a function of the difference between their body sizes is shown. The vertical line indicates a target species. Species whose traits are larger are its potential predators, and species whose traits are smaller are its potential prey. Mutations acting on the body-size trait of the target species will give birth to types whose trophic interactions are slightly different.
Exploitation competition among individuals of similar size is implicit in the γ function because they consume the same kind of resources. Also, these individuals of similar body size may also hamper, interfere physically, or even harm each other while competing for the resource. This interaction is called “interference competition.” The function α(|xi – xj|) corresponds to the interference-competition rate and is defined as follows:
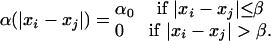 |
The value of β was chosen to be small (0.25), which means that competition is assumed to occur mainly within species or with closely related types. Interference competition among species with similar body sizes has been shown in some groups (17); the mechanisms involved may be interference (18, 19). Habitat partitioning leads to similar effects (20). Our qualitative results do not hinge on this assumption of interference competition among similar-sized types because we also considered the case in which this competition is absent (α0 = 0).
Last, N0 is the amount of inorganic nutrient whose trait x0 = 0 is expressed for mathematical convenience but does not evolve. Its dynamics is defined as follows:
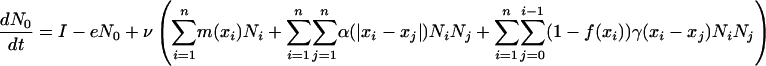 |
[2] |
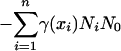 |
where I is the input of inorganic nutrient, e is its output rate, and v is the percentage of remaining nutrient within the system during the recycling process.
Numerical simulations were performed by using the Runge–Kutta method in fortran 90. Food webs emerge progressively from a single ancestor by mutation–selection processes. For each population, the mutation rate was 10–6 per unit mass at each time step. If a mutation occurs in a population x, a new population is created whose trait is drawn randomly in the interval [0.8x, 1.2x]. The initial biomass of the mutant is 10–20, which is also the threshold biomass below which any population is assumed to go extinct.
The initial population has a trait x = d and consumes inorganic nutrient. The simulations were run during 108 time steps. Their computation time varied from a few hours to several weeks depending on the total diversity reached (varying from 1 to >200 species). The total time for the 201 simulations exceeded 1 year. Of these simulations, we focus here on the results of the 36 simulations that varied the two parameters that proved to be critical in determining food-web structure [i.e., niche width (nw = 0.5, 1, 2, 3, 4, and 5) and competition intensity (α0 = 0, 0.1, 0.2, 0.3, 0.4, and 0.5)]. Niche width is defined as nw = s2/d, a relative measure of the variance of body sizes consumed by each species.
Trophic position is determined recursively from the bottom to the top of the food web. The trophic position of a target species is defined as the average trophic position of the species it consumes weighted by the proportion of nutrient these represent in the diet of the target species, plus one. For details about the method, see ref. 21.
Results
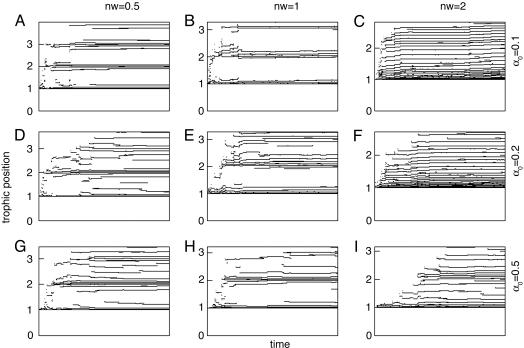
As shown in Fig. 2, the structure of the food web gradually stabilizes after an initial period of strong diversification. It is mainly determined by niche width and, to a lesser extent, by competition intensity. If niche width and competition intensity are small, species are packed in distinct trophic levels. If niche width or competition intensity are large, the structure of the food web is somewhat blurred, with no distinct trophic levels. The community then appears as a continuum of species homogeneously spaced along an axis of body size or trophic position. Given the link between body size and predation (Fig. 1), trophic position is strongly correlated with body size. Thus, the patterns displayed by Fig. 2 would be identical if body size, instead of trophic position, were plotted against time.
Fig. 2.
Food-web structure. The evolution of simulated food webs during 108 time steps for three values of niche width (nw) and competition intensity (α0) is shown. Trophic position is computed as explained in Model and Methods. Because this measure is strongly correlated with body size, similar patterns would be obtained by using body size. The following parameter values were used in these simulations: I = 10, e = 0.1, v = 0.5, γ0 = 1, d = 2, f0 = 0.3, m0 = 0.1, and β = 0.25.
The observed patterns (Fig. 2) can be explained by the way that the food web structures itself in a bottom-up manner. If niches are narrow, then only those species whose size is close to d are able to consume the basal resource efficiently enough to survive. Smaller or larger species appear transiently but are not favored in the long run. Similarly, only species that are packed at size ≈2d can consume species with size d efficiently. Therefore, they have a selective advantage over species with intermediate body size. More generally, species from trophic level i evolve to a body size close to id. Thus, when niche width is small, the food web is strongly structured in groups of different sizes corresponding to distinct trophic levels (Fig. 2 A and B). If niches are broad, the advantage (in terms of resource consumption) of species with sizes that are multiples of d is less important compared with species with intermediate sizes. This advantage may then be offset by other processes, such as the dependence of mean generation time [which is the inverse of mortality rate m(xi)] on body size and interference competition.
Although predation tends to pack species in distinct trophic levels, interference competition tends to have the opposite effect of homogenizing the body-size distribution. This homogenization occurs because species compete only if their body sizes are similar enough. When competition is intense, it has a more important role than predation does, and the structure of the food web is less distinct. This effect is particularly visible for nw = 0.5 or 1 (Fig. 2 G and H). When niche width is large (nw = 2), the trophic structure is blurred in all three cases.
Our results show that complex food webs may emerge from simple ecological and evolutionary rules in a system that initially contains a single species consuming an inorganic nutrient. The variety of emerging structures is comparable with empirical observations of real ecosystems. Food webs with distinct trophic levels are commonly found in freshwater ecosystems (22–24), whereas food webs with a more continuous trophic structure may be more common in soil terrestrial or marine ecosystems (25).
Although interaction strength is a continuous property that emerges from the coevolutionary dynamics, most empirical data and food-web models (1, 3) are binary (presence or absence of species and trophic links). Therefore, we transformed the food webs obtained here into binary food webs to make comparisons with these previous studies. Each type was then regarded as a species, and a consumption link was considered to be present between any two species if their interaction strength γ exceeded the threshold value of 0.15. The food web was then comparable with the classical binary webs studied extensively in the ecological literature. Variations in the threshold value may affect estimates of connectance, the proportion of omnivores, food-chain length, and the distribution among bottom, intermediate, and top species. This variability underlines the limits of the binary approach to describing food webs because the threshold value depends on the reliability of the determination of trophic interactions in the field. Although the threshold value may have an impact on the absolute value of the aggregated food-web parameters measured, it is unlikely to have an impact on their qualitative variations with niche width and competition intensity.
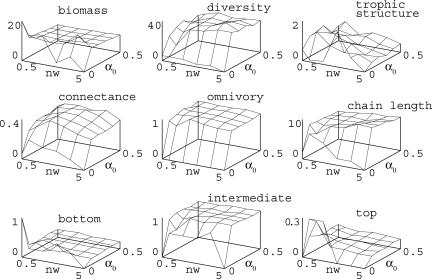
The structure of the resulting binary food webs was described by using classical parameters. Quantitative changes in the emergent properties of our model food webs with niche width and competition intensity are displayed in Fig. 3. Connectance is measured as 2L/S(S–1), where L is the total number of links and S is the number of species. Chain length corresponds to the mean chain length in the food web. Omnivory is measured by the proportion of species that are able to eat species positioned at different levels in the food chain. “Bottom” corresponds to the proportion of species feeding on the inorganic nutrient, “top” corresponds to the proportion of species that are not consumed by any other species, and “intermediate” corresponds to the remaining species in the food web. Fig. 3 also shows variations of a trophic structure index. It corresponds to the ratio between the variance of the difference between two adjacent body sizes and the expected variance of this variance for a random distribution of these traits (26). This index is expected to be high if species are packed in distinct trophic levels and low if species are homogeneously distributed along the body-size axis.
Fig. 3.
Emergent properties of simulated food webs as functions of niche width (nw) and competition intensity (α0). Properties were measured at the end of simulations (i.e., after 108 time steps). See Model and Methods for computation of the various properties. Parameter values are as in Fig. 2.
However, for many food-web properties, competition intensity appears to be more important than niche width (Fig. 3). In particular, substantial variations in these properties occur between the cases in which interference competition is absent (α0 = 0) and present, even at a low level (α0 = 0.1). Webs without interference competition contain very few species (usually three or four). Because evolution of the community is then only driven by consumption, their structure is chain-like (high trophic structure index, low connectance, and low omnivory). Populations then tend also to have unstable dynamics.
Interference competition favors the emergence of diversity (Fig. 3). The trophic structure index confirms the important role of the niche width shown in Fig. 2. As explained above, competition tends to homogenize the body-size distribution and, therefore, to increase the connectance as well as the omnivory. As the connections between species become more numerous, the mean chain length increases. If the competition intensity is fixed, connectance is maximum at intermediate value of the niche width. This intermediate optimum may be explained by the fact that increasing niche width blurs the trophic structure (Fig. 2), making it more connected, but it decreases the interaction strength (the γ function being normalized), thus decreasing the probability of recording this interaction. This decrease in the mean interaction strength as nw increases also explains why the omnivory index decreases. Last, note that variations of the total biomass with the parameters are weak, which means that the evolved communities are similar in their efficiency to exploit the resource.
Based on these results, we selected the niche width and competition intensity giving the best match between the properties of our model ecosystems and those of seven of the best-documented food webs. By using the data shown in Fig. 3, we interpolated the values for connectance, omnivory, chain length, and distribution among bottom, intermediate, and top species in our model food webs. Least-square fitting of the interpolated functions to the observed properties of the seven documented food webs allowed us to determine the niche width and competition intensity that would best match model predictions and observations. Biomass and diversity were not included because these two properties are determined mainly by nutrient input I and production efficiency f(xi) in our model. Also, we did not consider the trophic structure index in the comparison because this property is not documented in empirical data. Although we are aware that a lot of other properties could have been included, we limited our study to the six properties mentioned above, for two main reasons. First, some properties (such as number of loops) are not relevant given the assumptions of the model. Second, our model cannot be fully predictive because data on niche width and competition intensity are not available for the observed food webs. We performed the comparison between predicted and observed webs to show simply that the variety of structures found in the model is representative of the variety of structures observed in nature. Reliable data on interaction strength in food webs would make the comparison with the results of our model much more interesting.
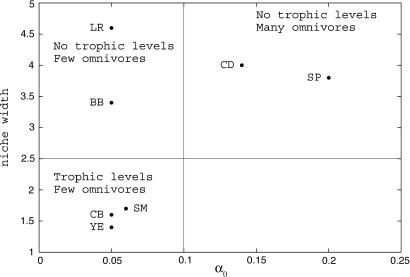
Observed food webs fall into three groups (Fig. 4). One group includes the food webs of the Ythan Estuary, the Chesapeake Bay, and St. Martin Island. These food webs have very similar properties and are comparable with evolved food webs in which competition is weak and niches are narrow (well differentiated trophic levels and little omnivory). Another group includes Little Rock Lake and Bridge Brook Lake, which have properties that are comparable with evolved food webs in which competition is weak and niches are broad (poorly defined trophic structure). The third group consists of Coachella Desert and Skipwith Pond. Their properties are comparable with evolved food webs in which competition is strong and niches broad (blurred trophic levels and large proportion of omnivores). The characteristics of these empirical food webs and their model counterparts are summarized in Table 1. For comparison, the corresponding predictions from the niche model are also included. The relative errors of the two models show that our evolutionary model is usually better at fitting the emergent properties than the niche model is. Comparison between the niche model and ours is limited by the fact that the niche model has been used to fit more properties and considers connectance as a parameter. However, note that the evolutionary model is unable to reproduce the high proportion of basal species (Skiptwith Pond) or the very low connectance (Chesapeake Bay and Ythan Estuary) that are sometimes observed in food webs.
Fig. 4.
Comparisons with documented food webs. Niche width and competition intensity that lead to food-web properties that best fit those of seven well documented food webs (BB, Bridge Brook Lake; CB, Chesapeake Bay; CD, Coachella Desert; LR, Little Rock Lake; SM, St. Martin Island; SP, Skipwith Pond; and YE, Ythan Estuary). We use the emergent properties given in Table 1. The main characteristics of the corresponding model food webs (level of omnivory and trophic structure) are indicated to give a better idea of the structure of the evolved communities. Empirical data are from Cattin et al. (4). Parameters are the same as in Fig. 2.
Table 1.
Properties of empirical food webs (D) and corresponding properties predicted by our evolutionary model (E) and the niche model (N) (3)
| Mean error
|
St. Martin Island
|
Coachella Desert
|
Ythan Estuary
|
Chesapeake Bay
|
Bridge Brook Lake
|
Little Rock Lake
|
Skipwith Pond
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value | E | N | E | N | D | E | N | D | E | N | D | E | N | D | E | N | D | E | N | D | E | N | D |
| Connectance, % | 99 | Parameter | 0.26 | * | 0.12 | 0.26 | * | 0.31 | 0.23 | * | 0.061 | 0.24 | * | 0.072 | 0.2 | * | 0.17 | 0.15 | * | 0.12 | 0.27 | * | 0.32 |
| (116) | (16) | (277) | (233) | (15) | (25) | (15) | |||||||||||||||||
| Chain length, % | 26 | 35 | 4.15 | 5.76 | 4.2 | 6.2 | 6.95 | 7.18 | 3.79 | 6.39 | 4.89 | 4 | 3.28 | 2.77 | 4.14 | 4.86 | 2.55 | 4.6 | ** | ** | 5.44 | 6.23 | 4.81 |
| (1) | (37) | (14) | (3) | (22) | (31) | (44) | (18) | (62) | (91) | (13) | (30) | ||||||||||||
| Omnivore, % | 19 | 22 | 54 | 63 | 60 | 76 | 75 | 79 | 42 | 61 | 53 | 46 | 42 | 38 | 48 | 61 | 36 | 47 | ** | ** | 74 | 77 | 60 |
| (10) | (5) | (4) | (5) | (22) | (15) | (21) | (11) | (33) | (69) | (23) | (28) | ||||||||||||
| Top, % | 22 | 52 | 17 | 9 | 17 | 5 | 6 | 0 | 24 | 8 | 38 | 21 | 14 | 28 | 4 | 10 | 0 | 4 | 5 | 0 | 5 | 6 | 4 |
| (0) | (47) | (37) | (79) | (25) | (50) | (25) | (33) | ||||||||||||||||
| Intermediate, % | 7 | 10 | 63 | 75 | 69 | 80 | 86 | 90 | 55 | 74 | 53 | 60 | 60 | 62 | 68 | 73 | 68 | 82 | 85 | 87 | 79 | 85 | 92 |
| (9) | (9) | (11) | (4) | (4) | (40) | (3) | (3) | (0) | (7) | (6) | (2) | (14) | (8) | ||||||||||
| Bottom, % | 75 | 60 | 20 | 17 | 14 | 15 | 9 | 10 | 21 | 18 | 9 | 19 | 26 | 10 | 28 | 17 | 32 | 14 | 10 | 13 | 16 | 9 | 4 |
| (43) | (21) | (50) | (10) | (22) | (33) | (90) | (160) | (13) | (47) | (8) | (23) | (300) | (125) | ||||||||||
For comparison, the relative errors of the two models are indicated between parentheses. See Model and Methods for computation of these properties. Empirical data are from Cattin et al. (4).
Connectance is a parameter of the niche model, not a result.
Not available.
Discussion
The qualitative results presented here are robust to variations in other parameters. In particular, we varied nutrient input I (1, 10, and 100), basal production efficiency f0 (0.1, 0.3, and 0.5), and the threshold parameter β below which two species compete (0.1, 0.25, and 0.5). Other simulations were performed at very low values of the competition rate α0 (0.001, 0.01, 0.025, 0.05, and 0.075) because several emergent food-web properties are particularly sensitive to changes in this range (Fig. 3). We also replaced the linear functional response of the predator by a Holling type II function. In total, 201 simulations were made to check the robustness of the results. The production efficiency f(xi) and inorganic nutrient input I have large, positive effects on phenotypic diversity, total biomass, and maximum trophic position, in agreement with many ecological data and models (23, 27–29). The production efficiency f(xi) and mortality rate m(xi) also affect the distribution of biomass within the food web (top species having a lower biomass than bottom species), as expected for most food webs (30). The tendencies exhibited by the emergent food-web properties shown in Fig. 3, as well as the conditions for the emergence of the trophic structure displayed by Fig. 2, are qualitatively robust to changes in these parameters and in changing a Holling type I into a Holling type II functional response. Although other types of functional responses are possible to model predator prey interactions (Holling type III and ratio-dependent responses), it is not clear a priori how considering these functions should affect the results, although we did not test this possibility. Last, our measure of connectance inevitably depends on the particular threshold value of interaction strength above which a trophic interaction was chosen to be present in the binary food webs. All quantitative data and models of interaction strength will be confronted with this threshold problem when assessing this descriptor. This problem shows the limit of connectance when dealing with quantitative data.
Compared with static models (1, 3, 4), our approach has the advantage of dealing with quantitative data and dynamical properties of food webs. Evolution acting on individual traits provides a mechanistic explanation for the emergence of food-web structure, whereas static models use emergent properties as parameters, making it impossible to identify lower-level mechanisms. Fundamental features such as the joint emergence of the structure and functioning of the food web are interesting common points with other evolutionary models (6–8). We also find, as these models did, that after a sufficiently long time, the structure of the system no longer changes even though changes in species composition may occur. Other similarities include a rapid emergence and then a stabilization of diversity and the fact that large avalanches of extinctions are rare. However, two aspects distinguish our work from other studies. First, our model is able to reproduce the emergence of distinct trophic levels (Fig. 2). Second, our model incorporates a single, clearly identified trait as well as important components of evolution such as trade-offs among life-history parameters. Only two parameters are needed to obtain the variety of results shown in Figs. 2 and 3. The trait is unique and measurable, so that the link between evolutionary dynamics and food-web emergence is more direct, and the mechanisms and results can be readily tested.
Although it was built from an evolutionary perspective, our model may also be related to community-assembly models (31–34). Under certain conditions, the assumptions of our evolutionary model should be a good approximation to community assembly by colonization processes. It should be true in particular if species migrate from a neighboring community and there is a spatial correlation among communities in terms of species traits. Because the mutation rate constrains the difference between invaders and residents, the results of this model may not hold for invasion by species that are very different from the resident species. However, even in this case, the results of our model may be quite robust given the fairly large range of mutation (20%) that was considered here. Therefore, it is not surprising that some of the results presented here are similar to results found in colonization models. For example, we also observe a decrease in community invasibility through time (31–34), and the robustness of the final state of the food webs emerging in our model echoes the small number of endpoints in community-assembly models (33–34).
We do not claim that body size or any other single trait is sufficient for understanding all aspects of food-web structure and functioning. For example, our model does not account for functional differences between autotrophs and heterotrophs. As a consequence, it might be best suited to describing the evolution of the “animal” part of the food web. It is less likely to apply to organisms that have an extracellular consumption, such as decomposers, and to plant–herbivore interactions (which may explain why it has difficulties in predicting high proportions of basal species). Our model could give a good description of aquatic or soil food webs, because they seem to be largely size-structured. It is also likely to be applicable to ecosystems that are relatively closed, ensuring that local adaptation is important, or in open systems in which invaders have traits that are not too different from resident populations. In contrast, it should not apply to systems invaded by exotic species introduced by human activities. If it were considered as a macroevolutionary model sensu stricto, we expect that the process of the evolutionary assembly would require long periods of time and, hence, take place on large spatial scales. Last, if rules applied here to link size and consumption were reversed, the model might be used to portray parasite–host interactions.
We think that the main strengths of our model lie in the fact that it provides (i) an approach to investigate the links between adaptive dynamics, community ecology, and ecosystem functioning, and (ii) testable predictions on the evolutionary consequences of body size for food-web structure. Although a single trait was considered here, the variety of structures obtained is impressive, and the extension of our evolutionary framework to include a few other well chosen traits should enlighten our understanding of natural food webs.
Acknowledgments
We thank Peter Abrams, Blandine Descamps-Julien, Régis Ferrière, Florence Hulot, Mathew Leibold, Isabelle Olivieri, and Elisa Thébault for helpful comments on an early version of the manuscript.
Author contributions: N.L. and M.L. designed research; N.L. performed research and analyzed data; and N.L. and M.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cohen, J. E., Briand, F. & Newman, C. M. (1990) Community Food-Webs: Data and Theory (Springer, Berlin), Vol. 20.
- 2.Solow, A. R. & Beet, A. R. (1998) Ecology 79, 2013–2018. [Google Scholar]
- 3.Williams, R. J. & Martinez, N. D. (2000) Nature 404, 180–183. [DOI] [PubMed] [Google Scholar]
- 4.Cattin, M. F., Bersier, L. F., Banasek-Richter, C., Baltensperger, R. & Gabriel, J. P. (2004) Nature 427, 835–839. [DOI] [PubMed] [Google Scholar]
- 5.Paine, R. T. (1988) Ecology 69, 1648–1654. [Google Scholar]
- 6.Caldarelli, G., Higgs, P. & McKane, A. J. (1998) J. Theor. Biol. 193, 345–358. [DOI] [PubMed] [Google Scholar]
- 7.Drossel, B., Higgs, P. G. & McKane, A. J. (2001) J. Theor. Biol. 208, 91–107. [DOI] [PubMed] [Google Scholar]
- 8.McKane, A. J. (2004) Eur. Phys. J. B 38, 287–295. [Google Scholar]
- 9.Warren, P. H. & Lawton, J. H. (1987) Oecologia 74, 231–235. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. E., Pimm, S. L., Yodzis, P. & Saldana, J. (1993) J. Anim. Ecol. 62, 67–78. [Google Scholar]
- 11.Hemmingsen, A. M. (1960) Rep. Steno. Mem. Hosp. 9, 1–110. [Google Scholar]
- 12.Kleiber, M. (1961) The Fire of Life: An Introduction to Animal Energetics (Wiley, New York).
- 13.Peters, R. H. (1983) The Ecological Implications of Body Size (Cambridge Univ. Press, Cambridge, U.K.).
- 14.Enquist, B. J., Economo, E. P., Huxman, T. E., Allen, A. P., Ignace, D. D. & Gillooly, J. F. (2003) Nature 423, 639–642. [DOI] [PubMed] [Google Scholar]
- 15.Berlow, E. L., Neutel, A. M., Cohen, J. E., De Ruiter, P. C. & Ebenman, B. (2004) J. Anim. Ecol. 73, 585–598. [Google Scholar]
- 16.Emmerson, M. C. & Raffaelli, D. (2004) J. Anim. Ecol. 73, 399–409. [Google Scholar]
- 17.Bowers, M. A. & Brown, J. H. (1982) Ecology 63, 391–400. [Google Scholar]
- 18.Rosenzweig, M. L. & Sterner, P. (1970) Ecology 51, 217–224. [Google Scholar]
- 19.Hafner, M. S. (1977) J. Anim. Ecol. 46, 925–938. [Google Scholar]
- 20.Price, M. V. (1978) Ecology 59, 910–921. [Google Scholar]
- 21.Vander Zanden, M. J. & Rasmussen, J. B. (1996) Ecol. Monogr. 66, 451–477. [Google Scholar]
- 22.Strong, D. R. (1992) Ecology 73, 747–754. [Google Scholar]
- 23.Persson, L., Diehl, S., Johansson, L., Andersson, G. & Hamrin, S. F. (1992) Am. Nat. 140, 59–84. [Google Scholar]
- 24.Bowlby, J. N. & Roff, J. C. (1986) Ecology 67, 1670–1679. [Google Scholar]
- 25.Polis, G. A. (1991) Am. Nat. 138, 123–155. [Google Scholar]
- 26.Poole, R. W., Ratchke, B. J. & Stiles, F. G. (1979) Science 203, 470–471. [DOI] [PubMed] [Google Scholar]
- 27.Oksanen, L., Fretwell, S. D., Arruda, J. & Niemela P. (1981) Am. Nat. 118, 240–261. [Google Scholar]
- 28.Abrams, P. A. (1993) Am. Nat. 141, 351–371. [Google Scholar]
- 29.Loeuille, N. & Loreau, M. (2004) Theor. Popul. Biol. 65, 285–298. [DOI] [PubMed] [Google Scholar]
- 30.Elton, C. (1927) Animal Ecology (Macmillan, New York).
- 31.Post, W. M. & Pimm, S. L. (1983) Math. Biosci. 64, 169–192. [Google Scholar]
- 32.Drake, J. A. (1990) J. Theor. Biol. 147, 213–233. [Google Scholar]
- 33.Law, R. & Morton, D. R. (1996) Ecology 77, 762–775. [Google Scholar]
- 34.Morton, D. R. & Law, R. (1997) J. Theor. Biol. 187, 321–331. [Google Scholar]