Abstract
The gut microbiota is altered in liver diseases, and several probiotics have been shown to reduce the degree of liver damage. We hypothesized that oral administration of specific Bifidobacterium strains isolated from healthy guts could attenuate liver injury. Five strains were tested in this study. Acute liver injury was induced by D-galactosamine after pretreating Sprague-Dawley rats with the Bifidobacterium strains, and liver function, liver and ileum histology, plasma cytokines, bacterial translocation and the gut microbiome were assessed. Two strains, Bifidobacterium pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10, conferred liver protection, as well as alleviated the increase in plasma M-CSF, MIP-1α and MCP-1 and bacterial translocation. They also ameliorated ileal mucosal injury and gut flora dysbiosis, especially the enrichment of the opportunistic pathogen Parasutterella and the depletion of the SCFA-producing bacteria Anaerostipes, Coprococcus and Clostridium XI. Negative correlations were found between MIP-1α / MCP-1 and Odoribacter (LI09 group) and MIP-1α / M-CSF and Flavonifractor (LI10 group). Our results indicate that the liver protection effects might be mediated through gut microbiota modification, which thus affect the host immune profile. The desirable characteristics of these two strains may enable them to serve as potential probiotics for the prevention or adjuvant treatment of liver injury.
Introduction
Accumulating evidence shows that alterations in the intestinal microbiota play an important role in the progression of liver injury1–3. The liver interacts directly with the intestine through the portal system and the bile secretion system4. Under normal circumstances, a series of local and systemic protective mechanisms, including intestinal colonization resistance, can prevent the passage of potentially pathogenic bacteria or bacterial components, such as lipopolysaccharide, beyond the intestinal lumen5, 6. When the normal liver physiology is disrupted, followed by the weakening of the gut barrier, the movement of these microbes and their products from the intestinal lumen to the liver is likely to aggravate certain liver diseases by enhancing the propagation of inflammation and tissue damage, and spontaneous bacterial peritonitis could even occur7. Thus, a therapy aimed at preserving the intestinal colonization resistance by modulating the gut flora, may aid in the prevention or adjuvant treatment of hepatic injury.
A previous study showed that the Bifidobacterium/Enterobacteriaceae (B/E) ratio, which may indicate the microbial colonization resistance of the bowel, decreased significantly in patients with liver diseases8. Bifidobacterium species are generally used as probiotics because of their associated health-promoting benefits and “GRAS” (generally recognized as safe) status9. Bifidobacterium longum, a major ingredient of VSL#3 (VSL#3 is a high potency probiotic medical food designated for ulcerative colitis, irritable bowel syndrome, etc. and it contains 8 diverse strains of bacteria), and Bifidobacterium pseudocatenulatum can potentially attenuate cirrhosis10–14, and Bifidobacterium catenulatum has been proven to be an effective treatment for acute liver injury15, 16. However, the definitive mechanism by which Bifidobacterium strains protect against liver injury and their influence on modulation of gut microbiota composition have not been completely explored in previous studies.
In this study, we examined the effects of Bifidobacterium longum LI06 (CGMCC 10385), Bifidobacterium longum LI07 (CGMCC 10386), Bifidobacterium pseudocatenulatum LI08 (CGMCC 10387), Bifidobacterium pseudocatenulatum LI09 (CGMCC 10388) and Bifidobacterium catenulatum LI10 (CGMCC 10389) on acute liver injury induced by D-galactosamine (D-GalN). Furthermore, we explored the relevant mechanism by which these potential probiotic strains protect against liver injury by examining plasma inflammatory cytokines, terminal ileum histology, bacterial translocation and the gut flora composition. Our results showed that B. pseudocatenulatum LI09 and B. catenulatum LI10 could attenuate D-GalN induced liver damage, as well as systemic inflammatory responses, bacterial translocation, ileal mucosal injury and gut flora dysbiosis, with gut microbiota modification.
Results
Bifidobacterium pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10 alleviated the D-GalN-induced liver injury
Twenty-four hours after the induction of liver damage, none of the rats had died. To examine the effects of the Bifidobacterium strains on D-GalN-induced acute liver injury, we first compared the liver function of Bifidobacterium-treated rats with that of positive acute liver injury control (PC) rats. Among the five Bifidobacterium strains used in this study, oral administration with B. longum LI06, B. pseudocatenulatum LI08, B. pseudocatenulatum LI09 and B. catenulatum LI10 significantly reduced the D-GalN-induced increase in alanine aminotransferase (ALT) and total bile acid, but B. longum LI07 did not (Table 1). The increase in glutamyltransferase induced by D-GalN was significantly reduced by B. longum LI06, B. pseudocatenulatum LI08 and B. catenulatum LI10. In addition, only administration with B. pseudocatenulatum LI09 and B. catenulatum LI10 relieved the increase in aspartate aminotransferase (AST) and glycylproline dipeptidyl aminopeptidase. Therefore, B. pseudocatenulatum LI09 and B. catenulatum LI10 seemed to more efficiently ameliorate the destruction of liver function induced by D-GalN.
Table 1.
Effects of pretreatment with five Bifidobacterium strains on liver function during D-GalN-induced acute liver injury.
| ALT (U/L) | AST (U/L) | GGT (U/L) | GPDA (U/L) | TBA (µmol/L) | TB (µmol/L) | ALB (g/L) | |
|---|---|---|---|---|---|---|---|
| LI06 (n = 9) | 5005.0 ± 4147.8** | 7505.0 ± 6705.4 | 10.9 ± 8.4* | 303.6 ± 161.9 | 261.6 ± 167.0* | 12.5 ± 10.3 | 37.2 ± 3.1 |
| LI07 (n = 9) | 6340.0 ± 5733.0 | 9135.0 ± 7725.8 | 14.6 ± 13.1 | 318.8 ± 203.9 | 284.6 ± 192.6 | 43.6 ± 83.7 | 36.7 ± 2.9 |
| LI08 (n = 9) | 6431.1 ± 2933.8* | 8200.0 ± 4038.7 | 13.2 ± 6.6* | 356.3 ± 135.3 | 326.0 ± 116.4* | 23.7 ± 12.5 | 36.8 ± 1.5 |
| LI09 (n = 9) | 4191.1 ± 2777.8*** | 5800.0 ± 3601.0** | 13.8 ± 11.4 | 272.4 ± 137.0** | 285.2 ± 136.9* | 17.4 ± 12.8 | 36.8 ± 1.7 |
| LI10 (n = 9) | 3888.9 ± 3190.7*** | 5471.1 ± 4247.0** | 10.8 ± 8.2* | 250.2 ± 151.0** | 274.6 ± 177.0* | 16.3 ± 12.5 | 37.1 ± 2.1 |
| PC (n = 8) | 10000.0 ± 2058.1 | 12050.0 ± 3342.2 | 21.5 ± 9.0 | 427.4 ± 56.3 | 440.1 ± 60.6 | 68.6 ± 88.0 | 34.5 ± 3.7 |
| NC (n = 6) | 49.5 ± 7.8*** | 101.2 ± 12.2*** | NA | 65.7 ± 14.1*** | 54.2 ± 24.3*** | 1.8 ± 0.4 | 37.3 ± 2.4 |
Values are expressed as the mean ± SD. The serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), glutamyltransferase (GGT), glycylproline dipeptidyl aminopeptidase (GPDA), total bile acid (TBA), total bilirubin (TB) and albumin (ALB) were determined after 24 h of D-galactosamine (D-GalN) administration. Compared with the PC group, *p < 0.05, **p < 0.01, ***p < 0.001.
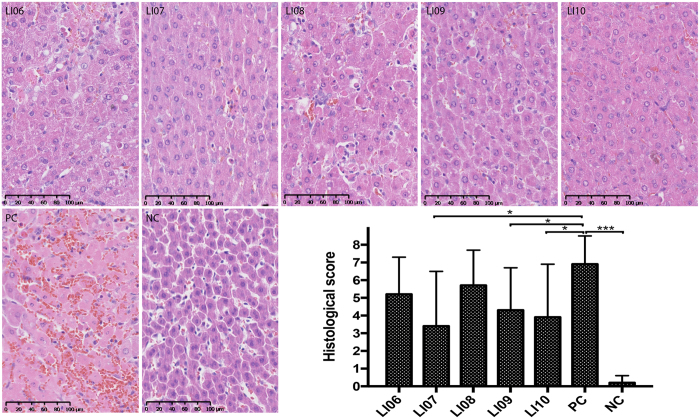
To further confirm the effects of these Bifidobacterium strains on liver injury, we examined the liver morphology of rats in different groups. Twenty-four hours after the induction of liver injury, all groups, except the negative acute liver injury control (NC) group, demonstrated hepatic histological abnormalities (Fig. 1). Administration of B. longum LI07, B. pseudocatenulatum LI09 and B. catenulatum LI10 significantly ameliorated D-GalN-induced hepatic degeneration or necrosis and inflammatory cell infiltration. The results from two aspects, liver function and liver histology, both indicate the protective effects of B. pseudocatenulatum LI09 and B. catenulatum LI10 against hepatic injury.
Figure 1.
Effects of pretreatment with five Bifidobacterium strains on hepatic histological abnormalities during D-galactosamine (D-GalN)-induced acute liver injury. Representative images of hepatic haematoxylin and eosin (H&E) staining and histological scores of livers based on these images. Values are expressed as the mean ± SD. *p < 0.05, ***p < 0.001 compared with the positive acute liver injury control (PC) group.
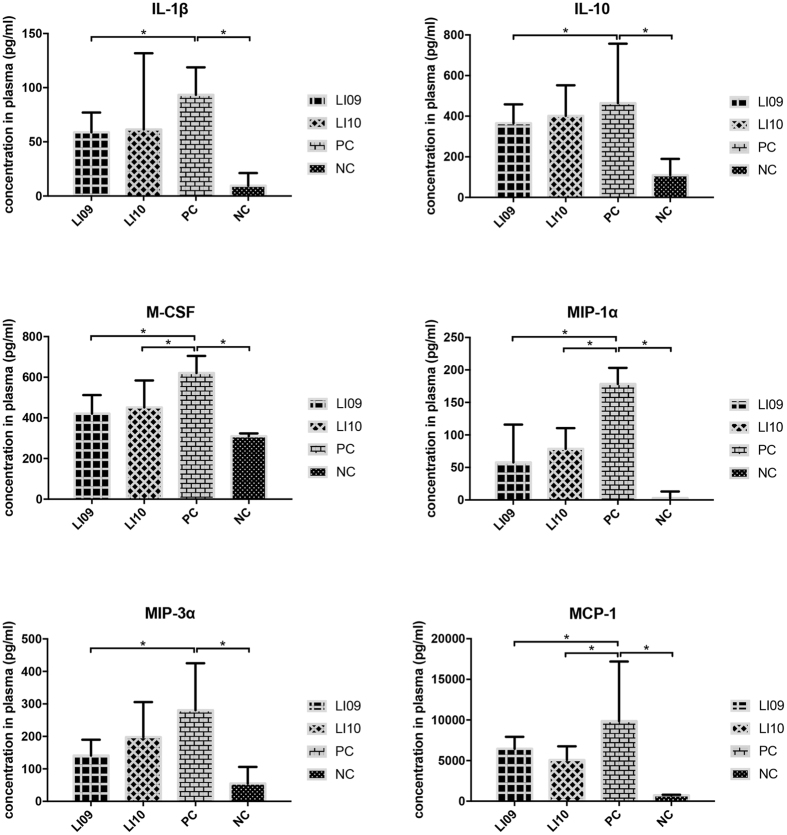
B. pseudocatenulatum LI09 and B. catenulatum LI10 reduced the D-GalN-induced increase in plasma inflammatory cytokines
Since liver injury is aggravated by inflammatory molecules, we next examined whether these two strains could relieve inflammation by measuring a total of 24 different plasma cytokines. Twenty-four hours after D-GalN treatment, the plasma levels of all detected cytokines increased in the PC group compared with those in the NC group. Administration of B. pseudocatenulatum LI09 significantly alleviated the D-GalN-induced increase in the proinflammatory cytokine interleukin (IL)-1β and the immunomodulatory cytokine IL-10 (Fig. 2). Among the chemokines, the increase in macrophage inflammatory protein 1 alpha (MIP-1α) and monocyte chemoattractant protein 1 (MCP-1) was attenuated when the rats were treated with either B. pseudocatenulatum LI09 or B. catenulatum LI10. However, the increase in another chemokine, MIP-3α, was only ameliorated in B. pseudocatenulatum LI09-treated rats. In addition, the increase in macrophage colony-stimulating factor (M-CSF) was also alleviated in the groups treated with these two strains. These results suggest that the reduction of chemokine levels may play a pivotal role in the process by which Bifidobacterium improves liver injury. By comparison, Administration of none of the B. longum LI06, B. longum LI07 or B. pseudocatenulatum LI08 could alleviate the increase of all these chemokines simultaneously (see Supplementary Fig. S1).
Figure 2.
Effects of pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 on hypercytokinemia during D-GalN-induced acute liver injury. Values are expressed as the median with interquartile range. *p < 0.05 compared with the PC group.
B. pseudocatenulatum LI09 and B. catenulatum LI10 ameliorated the D-GalN-induced bacterial translocation
To determine how the inflammatory reaction could be attenuated, we investigated the gut barrier function, since oral administration of Bifidobacterium strains first and directly impacts gut. We examined the bacterial translocation to evaluate the protective effects of these two strains on D-GalN-induced destruction of the gut barrier. As shown in Table 2, administration of either B. pseudocatenulatum LI09 or B. catenulatum LI10 reduced the population of bacteria that translocated to mesenteric lymph nodes (MLNs), indicating the restorative effects of these two strains on gut barrier function. By contrast, administration of none of the B. longum LI06, B. longum LI07 or B. pseudocatenulatum LI08 could cut down the population of bacteria which translocated to MLNs (see Supplementary Table S1).
Table 2.
Effects of pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 on bacterial translocation during D-GalN-induced acute liver injury.
| MLN (log10 CFU/g) | |
|---|---|
| LI09 (n = 9) | 2.0 ± 1.4* |
| LI10 (n = 9) | 2.3 ± 1.2* |
| PC (n = 8) | 3.7 ± 0.5 |
| NC (n = 6) | 2.8 ± 0.3* |
Values are expressed as the mean ± SD. *p < 0.05 compared with the PC group.
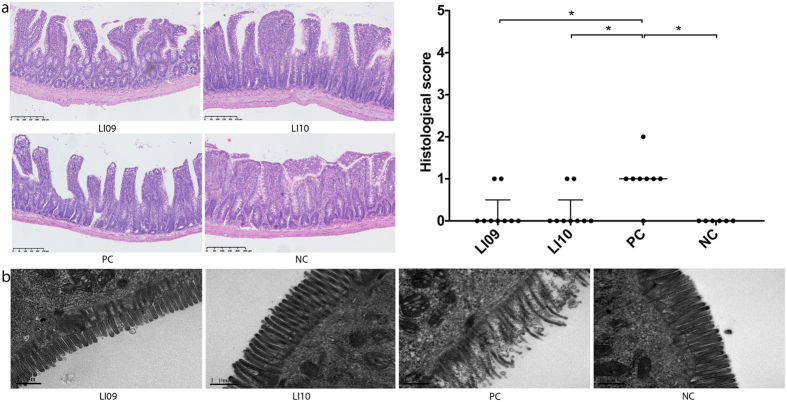
B. pseudocatenulatum LI09 and B. catenulatum LI10 attenuated the D-GalN-induced terminal ileum injury
Since the integrity of the intestinal mucosa is an important structure that supports the gut barrier function, we explored whether the intestinal mucosa integrity was damaged during D-GalN-induced liver injury and whether administering these strains improved the condition. As evaluated by haematoxylin and eosin (H&E) staining17, pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 alleviated the D-GalN-induced histological abnormalities of the terminal ileum by decreasing the incidence of subepithelial Gruenhagen’s space and epithelial lifting (Fig. 3a). In comparison, pretreatment with none of the B. longum LI06, B. longum LI07 or B. pseudocatenulatum LI08 could alleviate the histological abnormalities significantly (see Supplementary Fig. S2). We further observed the intestinal mucosal ultrastructure by transmission electron microscopy. Pretreatment with these two strains improved the ruptured, sparse, and stunted phenotypes of the intestinal epithelial cell microvilli (Fig. 3b).
Figure 3.
Effects of pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 on ileal histological abnormalities during D-GalN-induced acute liver injury. (a) Representative images of distal ileal H&E staining and histological scores of distal ileums based on these images. Values are expressed as the median with interquartile range. *p < 0.05 compared with the PC group. (b) Representative electron microscopy images of the distal ileum. Microvilli of the intestinal epithelial cells were examined.
B. pseudocatenulatum LI09 and B. catenulatum LI10 alleviated the D-GalN-induced alterations of the gut microbiome
Since the normal flora acts as another form of intestinal barrier, and the ecological balance of the gut flora is disrupted in D-GalN-induced hepatic injury, we next explored how the gut microbiota changed upon the administration of these strains during D-GalN-induced liver injury by metagenomic sequencing of the bacterial 16 S rRNA V3–V4 regions. From 59 rat caecum content samples, 1 489 572 qualified reads were filtered for downstream analysis. A total of 5000 reads were randomly chosen from each sample to equilibrate the sequencing depth. Based on ≥97% sequence identity, 497 qualified operational taxonomic units (OTUs) were clustered. Four OTUs were discarded because of their low coverage (fewer than 5 samples).
The species diversity calculated by the Simpson diversity indices and the community richness determined by the Chao1 indices were similar between the PC group and the LI09 (p = 0.161, p = 0.332, respectively) or LI10 group (p = 0.161, p = 0.180, respectively) (Fig. 4a). Actually, the Simpson indices and Chao1 indices were also similar between the PC group and the LI06 or LI07 group. However, the Simpson index of LI08 group was higher than that of PC group, although the Chao1 indices were similar between these two groups (see Supplementary Fig. S3). An unweighted UniFrac principal coordinate analysis (PCoA) was used to analyse the overall structural changes of microbial communities (Fig. 4b). PC samples were roughly separated from NC samples (p = 0.013) along PC2, which explained 8.7% of the total variations. When the rats were administered with either B. pseudocatenulatum LI09 or B. catenulatum LI10 before D-GalN treatment, the gut microbiome was distinctly separated from that of the PC samples (p < 0.001, p = 0.015, respectively) and did not differ from that of the NC samples (p = 0.050, p = 0.607, respectively) along PC2, demonstrating that oral administration of B. pseudocatenulatum LI09 or B. catenulatum LI10 ameliorates the D-GalN-induced alterations of the gut microbiome.
Figure 4.
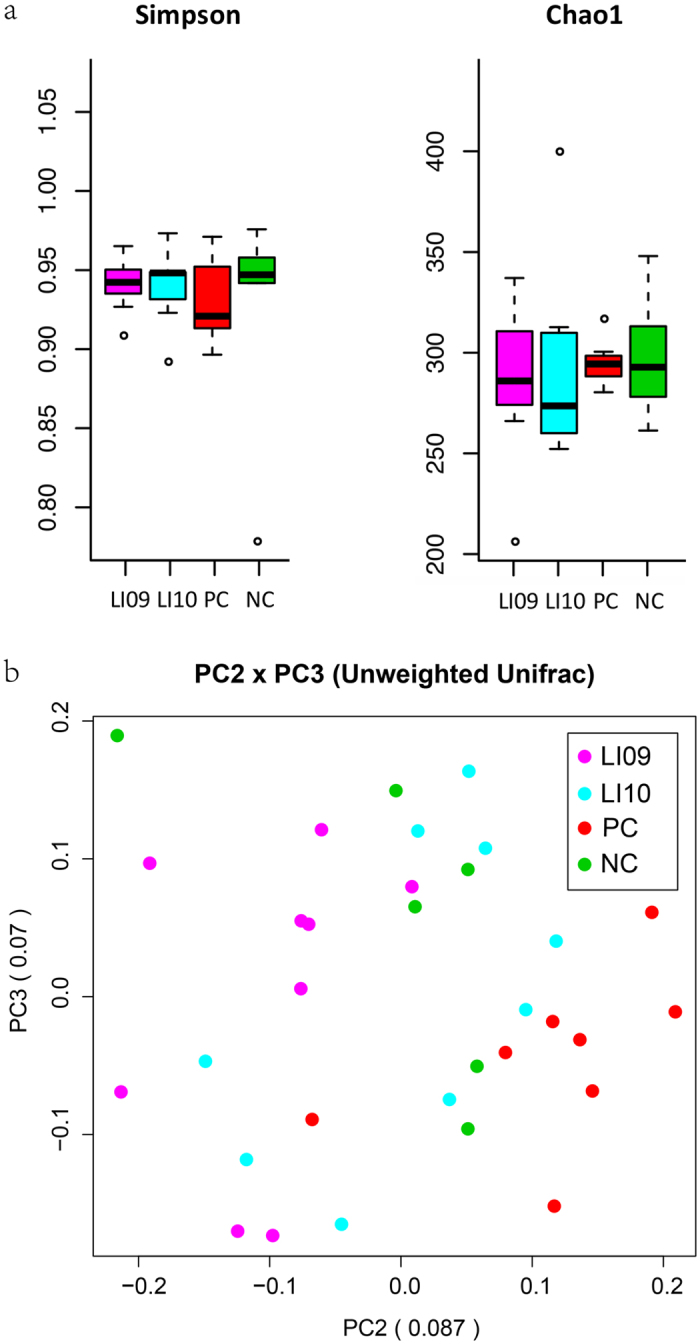
Effects of pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 on the overall structural changes of gut microbial communities during D-GalN-induced acute liver injury. (a) The alpha diversity of the gut microbiome, determined by Simpson index and Chao1 index, in the LI09 or LI10 group was compared with that in the PC group. Values are expressed as the median with interquartile range. (b) The principal coordinate analysis (PCoA) plot shows the beta diversity of the gut microbiome with unweighted UniFrac distance derived from 16S-based sequencing data. Composition profiles of the caecal flora in the LI09 or LI10 group was compared with those in the PC and negative acute liver injury control (NC) groups.
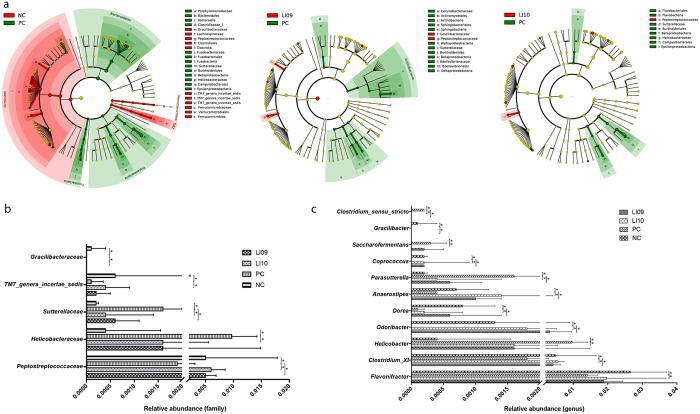
D-GalN-induced acute hepatic injury clearly affected the intestinal microbial structure at the phylum level (Fig. 5a). Compared with the NC group, the PC group demonstrated a marked depletion in Firmicutes, Verrucomicrobia and TM7 and a significant enrichment in Bacteroidetes, Proteobacteria and Fusobacteria. Oral B. pseudocatenulatum LI09 administration ameliorated the D-GalN-induced Proteobacteria enrichment, and administration of B. catenulatum LI10 alleviated the D-GalN-induced TM7 depletion.
Figure 5.
Effects of pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 on the alterations of gut bacterial taxonomic abundance during D-GalN-induced acute liver injury. (a) Bacterial taxa identified as differentially abundant between two groups analysed by linear discriminant analysis effect size (LEfSe). The bacterial taxa of the NC, LI09 and LI10 groups were compared with those of the PC group at different levels. Green indicates those bacterial taxa whose abundance were higher in the PC group, and red indicates those bacterial taxa whose abundance were higher in the other group. In addition, yellow indicates those bacterial taxa whose abundance show non-significant differences between the two groups. (b) The relative abundance at the bacterial family level in the LI09 or LI10 group was compared with that in the PC group. (c) The relative abundance at the bacterial genus level in the LI09 or LI10 group was compared with that in the PC group. Values are expressed as the median with interquartile range. *p < 0.05 compared with the PC group.
Furthermore, acute liver injury had a much wider impact on the gut microbiome at the family and genus levels (Fig. 5). Odoribacter, which belongs to family Porphyromonadaceae of phylum Bacteroidetes, was depleted in the PC group, and administration of either B. pseudocatenulatum LI09 or B. catenulatum LI10 attenuated the D-GalN-induced Odoribacter depletion.
Administration of B. pseudocatenulatum LI09 or B. catenulatum LI10 contributed considerably to preventing the D-GalN-induced alterations in the abundance of Clostridiales of phylum Firmicutes. Specifically, genus Clostridium_sensu_stricto, the main component of family Clostridiaceae_1, was enriched in the PC group compared with that in the NC group, and administration of either B. pseudocatenulatum LI09 or B. catenulatum LI10 ameliorated its enrichment. Conversely, genus Clostridium XI, the main member of Peptostreptococcaceae, was depleted in the PC group, and this depletion was alleviated by pretreatment with either of these two strains. Only B. pseudocatenulatum LI09 attenuated the D-GalN-induced depletion of genus Gracilibacter. In addition, pretreatment with B. pseudocatenulatum LI09 or B. catenulatum LI10 also ameliorated the depletion of genera Dorea, Anaerostipes and Coprococcus, which all belong to family Lachnospiraceae. Although there was no difference in the abundance of family Ruminococcaceae between the PC and NC groups, genus Saccharofermentans, within this family, was enriched in the PC group; this enrichment was alleviated when the rats were pretreated with B. catenulatum LI10. Genus Flavonifractor, another Ruminococcaceae member, was depleted in the PC group, and treatment with B. catenulatum LI10 attenuated its depletion.
B. pseudocatenulatum LI09 or B. catenulatum LI10 administration significantly prevented the D-GalN-induced enrichment of opportunistic pathogens belonging to phylum Proteobacteria. Parasutterella, which was the primary contributor to the enrichment of class Betaproteobacteria, was enriched in the PC group, and administration of either B. pseudocatenulatum LI09 or B. catenulatum LI10 ameliorated its enrichment. In addition, the D-GalN-induced enrichment of another Proteobacteria member, genus Helicobacter, was attenuated in the group treated with B. catenulatum LI10.
By comparison, administration of none of the B. longum LI06, B. longum LI07 or B. pseudocatenulatum LI08 could alleviate the depletion of Odoribacter, Clostridium XI, Dorea, Anaerostipes and Coprococcus and the enrichment of Clostridium_sensu_stricto and Parasutterella simultaneously (see Supplementary Table S2).
Correlations between involved inflammatory cytokines and gut bacterial genera were found in the groups treated with these two strains
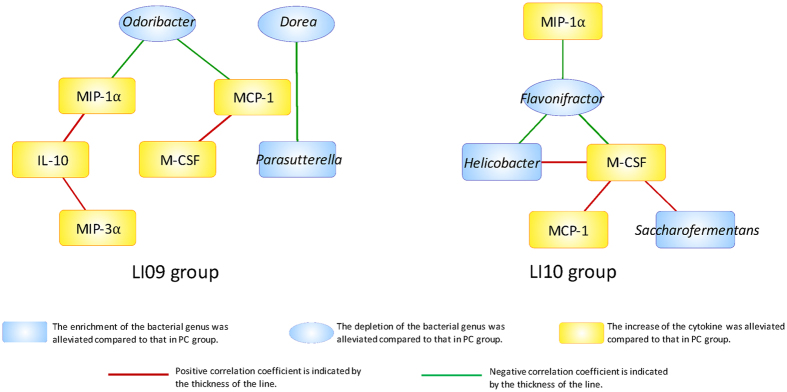
To verify that alleviating the alterations in the gut microbial composition plays an important role in reducing the increase in inflammatory cytokines, a systems biology approach (correlation-network analysis) was used to identify key linkages between the involved inflammatory cytokines and gut bacterial genera in acute liver injury (Fig. 6). In the B. pseudocatenulatum LI09-treated group, the chemokines MIP-1α and MCP-1 were negatively correlated with Odoribacter, while in the B. catenulatum LI10-treated group, the chemokine MIP-1α, along with M-CSF, was negatively correlated with Flavonifractor. Moreover, in both groups, a positive association was found between MCP-1 and M-CSF.
Figure 6.
Correlation-network analysis of plasma cytokines and gut bacterial genera of D-GalN-sensitized rats who received B. pseudocatenulatum LI09 or B. catenulatum LI10. A Spearman correlation analysis was performed, and only correlations with p < 0.05 are displayed. Blue nodes represent bacterial genera, and yellow nodes represent cytokines. The ellipses indicate that the depletion of the bacterial genera was alleviated compared to that in the PC group, and the round rectangles indicate that the enrichment of the bacterial genera was alleviated or the increase of the cytokines was alleviated compared to that in the PC group. A red line connecting nodes represents positive correlation, and a green line represents negative correlation. Values of the corresponding correlation coefficients are indicated by the thickness of the line; the thicker the line, the greater the coefficient.
Discussion
Acute liver failure, a severe form of liver damage caused by various factors, is an infrequent but life-threatening disease, with death occurring in up to 50% of the cases18, 19. The standard therapy for liver failure, liver transplantation, has improved the survival rate by approximately 40%; however, this option is strongly limited because of the shortage of donor livers20, 21. Thus, an artificial liver system (ALS), with the primary aim of detoxifying blood, is widely used in clinics until the patient’s own liver regenerates or a donor liver becomes available; nevertheless, the cost and safety of ALS remain a concern22. In this study, oral administration of B. pseudocatenulatum LI09 or B. catenulatum LI10 had extensive beneficial effects on induced liver damage in experimental rats, including reducing ALT, AST, glycylproline dipeptidyl aminopeptidase and total bile acid levels, as well as hepatic inflammation and necrosis, and attenuating systemic inflammatory responses, which could be of great importance in the prevention or adjuvant treatment of clinical acute liver failure in the future.
By contrast, administration of B. longum LI06 or B. pseudocatenulatum LI08 did not demonstrate significant protective effects on induced liver damage. And administration of B. longum LI07 showed beneficial effects only in the aspect of liver histology, which is a subjective indicator. Therefore, we didn’t consider B. longum LI07 as a candidate probiotics either.
Two strains from the same species, B. pseudocatenulatum LI08 and B. pseudocatenulatum LI09, demonstrated different effects. A similar situation was encountered by LX Lv et al.23, who reported that Lactobacillus salivarius LI01 were beneficial in the prevention of acute liver injury and Lactobacillus salivarius LI02 did not. Lactobacillus rhamnosus is another example. There are more than twenty kinds of strains of this species stored at American type culture collection (ATCC), but only the strain Lactobacillus rhamnosus GG (ATCC 53103) has been studied extensively on its various health benefits and become the world’s most studied probiotic bacterium. These conflicting findings may be explained by different sources of the strains which carry different genes and therefore have differences in their phenotypes.
There is increasing evidence suggesting that systemic inflammatory response syndrome (SIRS) occurs in acute liver failure through the release of damage-associated molecular patterns (DAMPs) and proinflammatory cytokines as a result of massive hepatic cell necrosis, which in turn plays a key role in the clinical course and outcome in acute liver failure patients24. In our study, strong inflammatory responses, as evidenced by significant increase in the levels of both proinflammatory cytokines (TNF- α, IL-1α, IL-1β, IL-2, and IL-6) and anti-inflammatory cytokine (IL-10), as well as chemokines (MCP-1, MIP-1α and MIP-3α) and colony-stimulating factor (M-CSF), were observed during acute liver injury progression. Lower levels of MCP-1, MIP-1α and M-CSF were seen at 24 h in both the LI09 and LI10 groups than in the PC group. MCP-1 and MIP-1α, both important members of the CC-chemokine family, recruit and activate monocytes/macrophages to the injured tissue area and regulate proinflammatory cytokines and adhesion molecules. They have been reported to play an important role in the early inflammatory response. M-CSF affects cells of the mononuclear phagocytic lineage in several ways, including regulating their survival, proliferation, and differentiation and as a major chemoattractant for these cells25. In the absence of M-CSF, monocytes could not be recruited to the sites of injured tissue due to the lack of MCP-1 synthesis26. Here, our data showed that administration of either B. pseudocatenulatum LI09 or B. catenulatum LI10 reduced the increased levels of MCP-1, MIP-1α and M-CSF (positively correlated with MCP-1), which may have led to the protection against liver injury, as suggested by the alleviation of the increase in serum ALT & AST and the amelioration of hepatic inflammation and necrosis.
The innate immune response is triggered by the recognition of translocated bacteria/bacterial products, followed by the release of chemokines and cytokines, leading eventually to bacterial killing7. In our study, bacterial translocation in the groups treated with these two strains was evaluated. MLNs are the first line of defence once the gut barrier has been breached. We found that the population of translocated bacteria to MLNs did decrease when rats were treated with either B. pseudocatenulatum LI09 or B. catenulatum LI10, which is consistent with the lower inflammatory response in these two groups.
Generally, only traces of gut-derived bacteria and their components can traffic in the portal vein to the liver due to the gut barrier, which is weakened upon the occurrence of liver disease. A disrupted intestinal barrier facilitates this translocation.
The integrity of the intestinal mucosa is one of the two important pillars that support the gut barrier function. In a liver disease situation, intestinal venous congestion will happen, followed by enterocyte necrosis or apoptosis, which manifest as damaged intestinal mucosal integrity. In this study, the incidence of subepithelial Gruenhagen’s space and epithelial lifting determined by H&E staining and the ruptured, sparse, and stunted microvilli of the intestinal epithelial cells determined by transmission electron microscopy indeed dropped in the LI09 and LI10 groups.
The intestinal colonization resistance, or the control of the growth of opportunistic microorganisms, is the other pillar. The normal flora acts as a barrier against colonization of potentially pathogenic microorganisms and against overgrowth of already present opportunistic microorganisms (mainly aerobic gram-negative bacteria)27, 28. The ecological balance of the gut flora is disrupted in hepatic injury, since bile is secreted abnormally and intestinal peristalsis slows down. Probiotic strains or their metabolites can modify the gut microbiological ecology. In our study, oral administration of B. pseudocatenulatum LI09 or B. catenulatum LI10 alleviated the D-GalN-induced alterations in the gut microbiome, as evidenced by the observation that the gut microbiome in samples pretreated with either B. pseudocatenulatum LI09 or B. catenulatum LI10 was significantly separated from that in PC samples and did not differ from that in NC samples in the PCoA analysis.
Bacterial phylotypes whose abundance was enriched in rats with acute liver damage were mostly associated with phylum Proteobacteria, including Betaproteobacteria and Epsilonproteobacteria. Genus Parasutterella, belonging to class Betaproteobacteria, was significantly enriched in PC samples. A high prevalence of these potentially pathogenic phylotypes has also been demonstrated in patients with Crohn’s disease29. In this study, administration of either B. pseudocatenulatum LI09 or B. catenulatum LI10 ameliorated the enrichment of genus Parasutterella, indicating the beneficial effects of these two strains. In addition, genus Helicobacter, belonging to class Epsilonproteobacteria, was also found to be significantly enriched in PC samples. The most widely known species of this genus is Helicobacter pylori, and some strains of this species, as well as several strains of non-H. pylori Helicobacter, are pathogenic to humans, as they are strongly associated with peptic ulcers, chronic gastritis and gastric cancer. The enrichment of genus Helicobacter was alleviated in the LI10 group.
Bacterial phylotypes with reduced abundance in rats with acute liver damage were strongly associated with class Clostridia (phylum Firmicutes), including Lachnospiraceae, Peptostreptococcaceae and Gracilibacteraceae, but not Clostridiaceae_1. Lachnospiraceae is known to participate in fibre fermentation in the human intestine30. The by-products of carbohydrate fermentation, such as butyrate, nourish cells lining the colon. In addition, these short-chain fatty acids (SCFA) seem to have anti-inflammatory effects by inducing regulatory T cells (Tregs) and, as a result, calibrate our immune system. The absence of these fermentation-related bacteria causes a decline in SCFA production and often correlates with diseases, such as asthma and inflammatory bowel disease31. Genera Dorea, Anaerostipes and Coprococcus, within family Lachnospiraceae, were all found to be depleted in the PC group. The latter two are both butyrate-producing bacteria. A decline of Coprococcus has been demonstrated in patients with intestinal32, 33, neuropsychological34, 35, infectious36, and atopic diseases37, as well as liver diseases38–40. A depletion of Dorea has been found in patients with intestinal41 and infectious diseases36, along with liver disease42, and a suppression of Anaerostipes has been shown in patients with paratuberculosis infection43. In addition, genus Clostridium XI, a SCFA-producing microorganism, within the family Peptostreptococcaceae, was also found to be significantly depleted in the PC group. The depletion of Dorea, Anaerostipes, Coprococcus and Clostridium XI in D-GalN-sensitized rats was attenuated by both the B. pseudocatenulatum LI09 and B. catenulatum LI10 strains, which further demonstrated the regulatory capacity of these two strains. Although there was no difference in the abundance of family Ruminococcaceae between the PC and NC groups, genus Flavonifractor, capable of cleaving the flavonoid C-ring44, within this family, was depleted in the PC group. A decline of a species belonging to Flavonifractor has also been demonstrated in obese individuals45. Treatment with B. catenulatum LI10 attenuated this depletion. Conversely, genus Clostridium_sensu_stricto (family Clostridiaceae_1), another member of class Clostridia, was found to be enriched in the PC group, which is consistent with some infant illnesses such as food allergies46–48 and necrotizing enterocolitis49. Its enrichment was ameliorated by either B. pseudocatenulatum LI09 or B. catenulatum LI10.
In addition, genus Odoribacter, another butyrate producer50, belonging to family Porphyromonadaceae (phylum Bacteroidetes), was also depleted in PC samples. The results of this study suggested that the abundance change of this genus in rats with acute liver injury is mostly accordance with metabolic syndrome50, 51. Its depletion was alleviated in both the LI09 and LI10 groups.
In short, these two strains both played a role in attenuating the depletion of health-promoting bacteria and ameliorating the enrichment of pathogenic bacteria, therefore partly maintaining the intestinal colonization resistance, which contributes to protecting the gut barrier function.
The regulation of the levels of MIP-1α and MCP-1 in the LI09 group and the levels of MIP-1α and M-CSF in the LI10 group might be associated with increased Odoribacter and Flavonifractor respectively, as evidenced by the significantly robust correlations, which are likely due to these two strains. As discussed above, the butyrate-producing genus Odoribacter and flavonoid-cleaving genus Flavonifractor, were both depleted in metabolic syndrome, which is closely associated with chronic low-grade inflammation. Seeing that MCP-1, MIP-1α and M-CSF are important chemoattractants for mononuclear phagocytic cells in the inflammatory response, it is indeed possible that B. pseudocatenulatum LI09 and B. catenulatum LI10 ultimately down-regulate the cytokines MCP-1, MIP-1α and M-CSF by modulating Odoribacter and Flavonifractor respectively. However, the underlying mechanisms will be possibly investigated in the future in view of the limited isolates and undeveloped culture techniques of Odoribacter and Flavonifractor at this stage.
In conclusion, our study shows the beneficial effects of B. pseudocatenulatum LI09 and B. catenulatum LI10 against liver injury induced by D-GalN, which might be mediated through the protection of the gut barrier function, specifically by alleviating intestinal flora dysbiosis and preventing intestinal epithelial cell damage. The restored gut barrier function may contribute to reducing bacterial translocation and finally down-regulating the overactive immune response. We will next evaluate their roles in attenuating liver damage in other experimental models of liver injury to facilitate the development of probiotic products in the future.
Methods
Strains and culture conditions
Five strains were used in this study: B. longum LI06 (CGMCC 10385), B. longum LI07 (CGMCC 10386), B. pseudocatenulatum LI08 (CGMCC 10387), B. pseudocatenulatum LI09 (CGMCC 10388), and B. catenulatum LI10 (CGMCC 10389). All strains were isolated from healthy volunteers, whose fecal samples obtained were immediately placed in an anaerobic environment before homogenized and plated on trypticase-phytone-yeast (QingDao RiShui, Ltd., Qingdao, China) agar. Five isolates were screened out through identifying with 16 s rDNA sequencing, stored at −80 °C and deposited in the China General Microbiological Culture Collection Center (CGMCC). The bacterial strains were revived following a standard approach (https://www.atcc.org/How_to_Revive_Cultures.aspx#bacteria2) and anaerobically cultured for 36 h at 37 °C. The cells were harvested by centrifugation at 6,000 g for 10 min. Subsequently, these cells were washed twice with normal saline and resuspended to a concentration of 3 × 109 colony-forming units/ml in normal saline for further use.
Animals and experimental design
Fifty-nine male Sprague-Dawley rats (from the Experimental Animal Center of Zhejiang Province, China), with an initial weight of 250–350 g, were randomized into seven groups with different treatments as follows: PC group (8 rats) and NC group (6 rats) both received 1 ml of physiologic saline by daily gavage; while LI06 group (9 rats), LI07 group (9 rats), LI08 group (9 rats), LI09 group (9 rats) and LI10 group (9 rats) received 1 ml (3 × 109 colony-forming units/ml) of B. longum LI06, B. longum LI07, B. pseudocatenulatum LI08, B. pseudocatenulatum LI09 and B. catenulatum LI10 strains by daily gavage respectively. All animals were fed normal food (standard rat chow), kept at room temperature (22 °C) with a controlled 12-h light/dark cycle, and orally administered saline or bacteria through an oro-gastric tube once daily for 7 days. Acute liver injury was induced on the 8th day by an intraperitoneal injection of D-galactosamine (G0500, Sigma, Saint Louis, MO, USA) at a dose of 1.1 g/kg body weight in all groups except the NC group. All experimental procedures were performed in accordance with the 2011 National Institutes of Health Guide for the care and use of laboratory animals. The study protocol was approved by the Animal Care committee of Zhejiang University, China.
Sample collections
Twenty-four hours after the induction of liver damage, the animals were anaesthetized and subjected to laparotomy through a large midline incision under aseptic conditions. Blood samples were collected from the inferior vena cava for liver function tests and to measure plasma cytokine levels. MLNs of the ileocaecal area were picked for bacterial translocation assay and caecal contents were obtained for gut bacterial microflora analysis. Tissue samples from the left lobe of the liver and ileum biopsied from a site approximately 2 cm away from the ileocaecal valve were harvested for histologic evaluation. All of the fifty-nine rats were analyzed for each following assay.
Liver function tests
Blood samples were centrifuged at 3,000 g for 10 min to separate the serum for analysis. Concentrations of ALT, AST, glutamyltransferase, glycylproline dipeptidyl aminopeptidase, total bile acid, total bilirubin and albumin in the serum were quantified using a Hitachi 7600 automatic analyser according to the manufacturer’s instructions (Hitachi, Tokyo, Japan)52.
Plasma cytokine analysis
Blood samples were centrifuged at 3,000 g for 10 min to separate the plasma and stored at −80 °C until analysis. The plasma (20 μl) was analysed using magnetic bead suspension arrays with the Bio-Plex Pro Rat Cytokine 24-Plex Panel (Bio-Rad) according to the manufacturer’s instructions. The 24-plex contains antibodies specific for erythropoietin, granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, growth-regulated oncogene-keratinocyte chemoattractant, IFN-gamma (γ), interleukin (IL)-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12(p70), IL-13, IL-17A, IL-18, monocyte chemoattractant protein 1 (MCP-1), macrophage colony-stimulating factor (M-CSF), macrophage inflammatory protein 1 alpha (MIP-1α), MIP-3α, Regulated on Activation, Normal T Cell Expressed and Secreted, tumour necrosis factor alpha (TNF-α), and vascular endothelial growth factor. The samples were examined using a Bio-Plex 200 analyser, and the results were calculated using the Bio-Plex manager 6.0 software53. Values for granulocyte colony-stimulating factor, granulocyte-macrophage colony-stimulating factor and IFN-γ from most samples were below the detection limit, and these cytokines were excluded from the data analysis. Additionally, a few values for IL-1α, IL-2, IL-4, IL-6, IL-13, IL-17A, IL-18, erythropoietin, Regulated on Activation, Normal T Cell Expressed and Secreted and TNF-α were below the detectable limit. Since the standard curve lower limits of all these cytokines were above 0.1 pg/ml, and the statistical analysis was performed by rank sum test, those undetectable values were assumed to be 0.1 pg/ml for statistical purposes54.
Bacterial translocation
A one-fold amount of sample from the MLNs was weighed and milled in a sterile glass homogenizer containing a nine-fold amount of physiological saline. Then, 50 μl of this homogenate (1/10 dilution) was plated on brain-heart infusion agar (BHI, Oxoid, Thermo Fisher Biochemicals Ltd., Beijing, China) in duplicate within 30 min of the sample collection and incubated for 72 h at 37 °C under either aerobic or anaerobic condition. Bacterial translocation was thought to occur if bacteria grew in the culture medium with the MLN homogenate. The number of colony-forming units from each plate was counted, and the number of translocated bacteria was expressed as CFU per gram of tissue.
Histological evaluation
Tissue samples from the left lobe of the liver and the terminal ileum were immediately fixed in 10% neutral formalin when taken from the anesthetized rats and then embedded in paraffin, cut into 2-μm sections, stained with haematoxylin and eosin (H&E), and analysed by a pathologist who was blind to the groups. At least three slides were studied from each specimen.
The tissue damage of the liver was semiquantitatively assess by a histological score consisting of two categories from the Histological Activity Index55, intralobular degeneration and focal necrosis (score, 0–1 or 3–4) and portal inflammation (score, 0–1 or 3–4), which reflect the acute injury of livers. Intestinal mucosal lesions were classified as described by Chiu et al.17. Score 0 is defined as normal mucosa and score 1 is the development of subepithelial Gruenhagen’s space at the tip of the villus. This space is more extended in score 2. In score 3, there is a massive epithelial lifting down the side of the villus. And in score 4, the villus is denuded of epithelium. It is characterized by a loss of the villus itself in score 5.
Electron microscopy
Ileal mucosal specimens fixed in 2.5% glutaraldehyde were post-fixed, dehydrated and embedded in epoxy resin. Ultrathin sections were made and post-stained in uranyl acetate and lead citrate56. The ileal mucosal ultrastructure was analysed on a Philips Tecnai 10 electron microscope (Philips, Eindhoven, the Netherlands). The length, diameter, linear density of microvilli was observed to learn the situation of microvilli damage57.
16 S rRNA sequencing
Caecal content samples (200 mg/aliquot) were immediately frozen upon collection from euthanized rats and stored at −80 °C before analysis. DNA extraction was performed using a QIAamp® Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions.
The bacterial 16 S rRNA V3–V4 regions were amplified by PCR using Phusion® High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA). The universal primer pairs were 319 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′). The reactions were hot started at 98 °C for 30 s, followed by 40 cycles of 98 °C for 10 s, 52 °C for 30 s, and 72 °C for 45 s, with a final extension step at 72 °C for 10 min. The amplified products from different samples were purified using the QIAquick Gel Extraction kit (QIAGEN, Hilden, Germany) and then mixed accordingly to achieve equal amounts in the final mixture. An amplicon library was constructed with a DNA sample preparation kit (Illumina, San Diego, CA).
Sequencing was conducted on an Illumina MiSeq platform (Illumina, San Diego, CA) according to the manufacturer′s instructions. Raw sequence reads were assigned to each sample according to their unique barcode pairs. Overlapping paired-end reads were merged to form tags using FLASH (version 1.2.8)58. The following quality control criteria were used: (1) an exact match to at least one end of barcodes and primers; (2) no undetermined bases in the tags; and (3) the number of mismatches in overlap region was no more than 3.
Operational taxonomic units (OTUs) were clustered with a 97% similarity cut-off using Usearch59. Alpha diversity was determined using the R program60 “rich” and “diversity” packages. Principal coordinate analysis (PCoA) as a standard multivariate statistical technique was performed with the R program “ade4” package to explain differences among microbial communities. The longest sequence in each OTU was chosen as the representative sequence for identification using the RDP (Ribosomal Database Project) database v.11.361. Taxonomic ranks were assigned to each tag using RDP classifier v.2.10.162, using 0.8 as the confidence coefficient.
Statistical Analysis
A t-test (for those with normal distribution) or Mann–Whitney U test (for those with skewed distribution) was used to determine differences between two groups. The Spearman rank correlation coefficient was used for linear correlation analysis. The data were analysed using SPSS 20.0 and GraphPad Prism 7 and presented as the mean ± SD or median with interquartile range. A p-value < 0.05 derived from a two-tailed test was considered statistically significant. The network was presented after the correlation analysis using Cytoscape63.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Science Fund for Creative Research Groups of the National Natural Science Foundation of China (No. 81121002) and the Key Program of the National Natural Science Foundation of China (No. 81330011).
Author Contributions
L.L. designed the experiments; D.F., D.S., W.W., F.G., J.Y. and Y.L. conducted the animal experiments; J.Y. and Y.L. measured the liver function and plasma cytokines; W.W. and F.G. assessed the liver and ileal histology and bacterial translocation; D.F. and D.S. evaluated the gut microbiota; L.L and S.G. analysed the data; Y.C., J.G., A.L. and X.H. provided technical support; L.L., D.F., D.S., L.L., S.G. and W.W. wrote the manuscript; all authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Daiqiong Fang, Ding Shi, Longxian Lv, Silan Gu and Wenrui Wu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-09395-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Celaj S, et al. The microbiota regulates susceptibility to Fas-mediated acute hepatic injury. Lab. Invest. 2014;94:938–949. doi: 10.1038/labinvest.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Possamai, L. A. et al. The role of intestinal microbiota in murine models of acetaminophen-induced hepatotoxicity. Liver Int. 35, 764–773, doi:0.1111/liv.12689 (2015). [DOI] [PMC free article] [PubMed]
- 3.M CCC, et al. Comparing the effects of acute alcohol consumption in germ-free and conventional mice: the role of the gut microbiota. BMC Microbiol. 2014;14 doi: 10.1186/s12866-014-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesaro C, et al. Gut microbiota and probiotics in chronic liver diseases. Dig. Liver Dis. 2011;43:431–438. doi: 10.1016/j.dld.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Kasravi FB, Adawi D, Molin G, Bengmark S, Jeppsson B. Effect of oral supplementation of lactobacilli on bacterial translocation in acute liver injury induced by D-galactosamine. J. Hepatol. 1997;26:417–424. doi: 10.1016/S0168-8278(97)80060-8. [DOI] [PubMed] [Google Scholar]
- 6.Yang YY, et al. Long-term cannabinoid type 2 receptor agonist therapy decreases bacterial translocation in rats with cirrhosis and ascites. J. Hepatol. 2014;61:1004–1013. doi: 10.1016/j.jhep.2014.05.049. [DOI] [PubMed] [Google Scholar]
- 7.Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology. 2005;41:422–433. doi: 10.1002/hep.20632. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, et al. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb. Ecol. 2011;61:693–703. doi: 10.1007/s00248-010-9801-8. [DOI] [PubMed] [Google Scholar]
- 9.Picard C, et al. Review article: bifidobacteria as probiotic agents–physiological effects and clinical benefits. Aliment Pharmacol. Ther. 2005;22:495–512. doi: 10.1111/j.1365-2036.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 10.Velayudham A, et al. VSL#3 probiotic treatment attenuates fibrosis without changes in steatohepatitis in a diet-induced nonalcoholic steatohepatitis model in mice. Hepatology. 2009;49:989–997. doi: 10.1002/hep.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhiman, R. K. et al. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: a randomized, controlled trial. Gastroenterology147, 1327–1337, doi:0.1053/j.gastro.2014.08.031 (2014). [DOI] [PubMed]
- 12.Moratalla A, et al. Protective effect of Bifidobacterium pseudocatenulatum CECT7765 against induced bacterial antigen translocation in experimental cirrhosis. Liver Int. 2014;34:850–858. doi: 10.1111/liv.12380. [DOI] [PubMed] [Google Scholar]
- 13.Moratalla A, et al. Bifidobacterium pseudocatenulatum CECT7765 promotes a TLR2-dependent anti-inflammatory response in intestinal lymphocytes from mice with cirrhosis. Eur. J. Nutr. 2016;55:197–206. doi: 10.1007/s00394-015-0837-x. [DOI] [PubMed] [Google Scholar]
- 14.Moratalla A, et al. Bifidobacterium pseudocatenulatum CECT7765 induces an M2 anti-inflammatory transition in macrophages from patients with cirrhosis. J. Hepatol. 2016;64:135–145. doi: 10.1016/j.jhep.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Xing, H. C. et al. Protective role of supplement with foreign Bifidobacterium and Lactobacillus in experimental hepatic ischemia-reperfusion injury. J. Gastroenterol. Hepatol. 21, 647–656, doi:0.1111/j.1440-1746.2006.04306.x (2006). [DOI] [PubMed]
- 16.Li YT, et al. Effects of gut microflora on hepatic damage after acute liver injury in rats. J Trauma. 2010;68:76–83. doi: 10.1097/TA.0b013e31818ba467. [DOI] [PubMed] [Google Scholar]
- 17.Chiu CJ, McArdle AH, Brown R, Scott HJ, N GF. Intestinal mucosal lesion in low-flow states. I. A morphological, hemodynamic, and metabolic reappraisal. Arch. Surg. 1970;101:478–483. doi: 10.1001/archsurg.1970.01340280030009. [DOI] [PubMed] [Google Scholar]
- 18.Bernal, W. & Wendon, J. Acute liver failure. N. Engl. J. Med. 369, 2525–2534, doi:0.1056/NEJMra1208937 (2013). [DOI] [PubMed]
- 19.Davies NA, Banares R. A new horizon for liver support in acute liver failure. J. Hepatol. 2015;63:303–305. doi: 10.1016/j.jhep.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Bernal W, et al. Lessons from look-back in acute liver failure? A single centre experience of 3300 patients. J. Hepatol. 2013;59:74–80. doi: 10.1016/j.jhep.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 21.O’Grady J. Timing and benefit of liver transplantation in acute liver failure. J. Hepatol. 2014;60:663–670. doi: 10.1016/j.jhep.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Krisper P, et al. Efficacy and safety of anticoagulation with heparin versus heparin plus epoprostenol in patients undergoing extracorporeal liver support with Prometheus. Artif. Organs. 2010;34:84–88. doi: 10.1111/j.1525-1594.2009.00793.x. [DOI] [PubMed] [Google Scholar]
- 23.Lv LX, et al. Administration of Lactobacillus salivarius LI01 or Pediococcus pentosaceus LI05 improves acute liver injury induced by D-galactosamine in rats. Appl Microbiol Biotechnol. 2014;98:5619–5632. doi: 10.1007/s00253-014-5638-2. [DOI] [PubMed] [Google Scholar]
- 24.Antoniades CG, Berry PA, Wendon JA, Vergani D. The importance of immune dysfunction in determining outcome in acute liver failure. J. Hepatol. 2008;49:845–861. doi: 10.1016/j.jhep.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Stanley ER, Guilbert LJ, Tushinski RJ, Bartelmez SH. CSF-1–a mononuclear phagocyte lineage-specific hemopoietic growth factor. J. Cell. Biochem. 1983;21:151–159. doi: 10.1002/jcb.240210206. [DOI] [PubMed] [Google Scholar]
- 26.Guleria I, Pollard JW. Aberrant macrophage and neutrophil population dynamics and impaired Th1 response to Listeria monocytogenes in colony-stimulating factor 1-deficient mice. Infect. Immun. 2001;69:1795–1807. doi: 10.1128/IAI.69.3.1795-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albillos A, de la Hera A. Multifactorial gut barrier failure in cirrhosis and bacterial translocation: working out the role of probiotics and antioxidants. J. Hepatol. 2002;37:523–526. doi: 10.1016/S0168-8278(02)00265-9. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis. 2001;1:101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 29.Chiodini RJ, et al. Microbial Population Differentials between Mucosal and Submucosal Intestinal Tissues in Advanced Crohn’s Disease of the Ileum. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan SH, Louis P, Flint HJ. Cultivable bacterial diversity from the human colon. Lett. Appl. Microbiol. 2007;44:343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- 31.Velasquez-Manoff M. Gut microbiome: the peacekeepers. Nature. 2015;518:S3–11. doi: 10.1038/518S3a. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore) 2014;93 doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malinen, E. et al. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 16, 4532–4540, doi:0.3748/wjg.v16.i36.4532 (2010). [DOI] [PMC free article] [PubMed]
- 34.Kang DW, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keshavarzian A, et al. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 2015;30:1351–1360. doi: 10.1002/mds.26307. [DOI] [PubMed] [Google Scholar]
- 36.Kampmann, C., Dicksved, J., Engstrand, L. & Rautelin, H. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin. Microbiol. Infect. 22, 61 e61–68, 10.1016/j.cmi.2015.09.004 (2016). [DOI] [PubMed]
- 37.Nylund L, et al. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70:241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 38.Ruhlemann, M. C. et al. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut, 10.1136/gutjnl-2016-312180 (2016). [DOI] [PubMed]
- 39.Chen, Y. et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology54, 562–572, doi:0.1002/hep.24423 (2011). [DOI] [PubMed]
- 40.Qin N, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 41.Mondot S, et al. Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut. 2016;65:954–962. doi: 10.1136/gutjnl-2015-309184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajaj JS, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G675–685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrazuria R, Elguezabal N, Juste RA, Derakhshani H, Khafipour E. Mycobacterium avium subspecies paratuberculosis infection modifies gut microbiota under different dietary conditions in a rabbit model. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter J, Moore LH, Dowell VRJ, Bokkenheuser VD. C-Ring Cleavage of Flavonoids by Human Intestinal Bacteria. Appl Environ Microbiol. 1989;55:1203–1208. doi: 10.1128/aem.55.5.1203-1208.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasai C, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15 doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling Z, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr. Allergy Immunol. 2016;27:254–262. doi: 10.1111/pai.12522. [DOI] [PubMed] [Google Scholar]
- 48.Olivares M, et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut. 2015;64:406–417. doi: 10.1136/gutjnl-2014-306931. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y, et al. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: a case-control study. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomez-Arango LF, et al. Increased Systolic and Diastolic Blood Pressure Is Associated With Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension. 2016;68:974–981. doi: 10.1161/HYPERTENSIONAHA.116.07910. [DOI] [PubMed] [Google Scholar]
- 51.Lim, M. Y. et al. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut, 10.1136/gutjnl-2015-311326 (2016). [DOI] [PubMed]
- 52.Cho SM, Lee SG, Kim HS, Kim JH. Establishing pediatric reference intervals for 13 biochemical analytes derived from normal subjects in a pediatric endocrinology clinic in Korea. Clin Biochem. 2014;47:268–271. doi: 10.1016/j.clinbiochem.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Nieminen A, et al. Circulating cytokines in predicting development of severe acute pancreatitis. Crit. Care. 2014;18 doi: 10.1186/cc13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo J, et al. The Serum Profile of Hypercytokinemia Factors Identified in H7N9-Infected Patients can Predict Fatal Outcomes. Sci Rep. 2015;5 doi: 10.1038/srep10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knodell RG, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1:431–435. doi: 10.1002/hep.1840010511. [DOI] [PubMed] [Google Scholar]
- 56.Dias G, et al. First record of gregarines (Apicomplexa) in seminal vesicle of insect. Sci Rep. 2017;7 doi: 10.1038/s41598-017-00289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.BROWN AL. Microvilli of the human jejunal epithelial cell. J Cell Biol. 1962;12:623–627. doi: 10.1083/jcb.12.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 60.Kabacoff, R. I. R in action:data analysis and graphics with R. Manning; Pearson Education, 1–474 (2011).
- 61.Cole JR, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.