Abstract
Several mutations in PTEN-induced putative kinase 1 (PINK1) gene have been reported to be associated with recessive parkinsonism. The encoded protein is predicted to be a Ser/Thr protein kinase targeted to mitochondria. In this study, we have investigated the effects of mutations on PINK1 kinase activity in vitro and on expression levels and localization in mammalian cells. We chose to examine two point mutations: G309D, which was originally reported to be stable and properly localized in cells and L347P, which is of interest because it is present at an appreciable carrier frequency in the Philippines. We were able to confirm kinase activity and produce artificial “kinase-dead” mutants that are stable but lack activity. The L347P mutation grossly destabilizes PINK1 and drastically reduces kinase activity, whereas G309D has much more modest effects on these parameters in vitro. This finding is in line with predictions based on homology modeling. We also examined the localization of PINK1 in transfected mammalian cells by using constructs that were tagged with myc or GFP at either end of the protein. These results show that PINK1 is processed at the N terminus in a manner consistent with mitochondrial import, but the mature protein also exists in the cytosol. The physiological relevance of this observation is not yet clear, but it implies that a portion of PINK1 may be exported after processing in the mitochondria.
Keywords: mitochondria, PARK6, Parkinson's disease
Several genes have now been identified that are causally associated with recessive parkinsonism. Mutations in parkin (1) are a relatively frequent cause of parkinsonism with onset before the age of 50, having a varied but generally mild phenotype. DJ-1 mutations are less common than parkin mutations, but the phenotype is broadly similar (2). Given that recessive mutations in either of these two genes cause parkinsonism, it is likely that mutations in either parkin or DJ-1 lead to loss of dopaminergic neurons in the substantia nigra that project to the striatum. Positron-emission tomography demonstrates a loss of dopaminergic function in parkin (e.g., 3–6) and DJ-1 (7, 8) patients, supporting this idea. Therefore, although detailed pathology of these two genetic forms of parkinsonism is not available, there are clear phenotypic overlaps. This conclusion argues that there are relationships between parkin and DJ-1, but the normal functions of the two wild-type proteins are not obviously linked. Parkin is an E3 protein-ubiquitin ligase (9), whereas DJ-1 may have a number of roles (10) but can affect the ability of neurons to survive oxidative stress generated as a result of mitochondrial damage (11).
Recently, mutations in the PTEN-induced putative kinase 1 (PINK1) gene have been described that are also associated with recessive parkinsonism. Initially, three pedigrees were described with identified mutations: a G309D point substitution in one family and a truncation mutation (W437X) in two additional families (12). Subsequently, several studies have described convincing point mutations or truncations (13–16), indicating that the frequency of mutations is less than that of parkin but greater than DJ-1. There are some populations with an appreciable frequency of mutations: in the Philippines, the L347P mutation is present in 8% of controls, although always in the heterozygous state (15). Detailed phenotypic descriptions of cases linked to this locus have been reported, including positron-emission tomography data (17, 18), and show a pattern of disease broadly similar to parkin or DJ-1.
The PINK1 protein had previously been cloned by two groups examining gene regulation by the tumor suppressor PTEN (19) and differential gene expression in mouse cell lines that vary in their metastatic potential (20). Both studies showed that there was strong prediction of a serine/threonine-directed kinase domain, and Unoki et al. (19) demonstrated that recombinant GST-fusion proteins had autophosphorylation activity. Natural kinase substrates for PINK1 have not yet been identified. In their paper describing PINK1 mutations, Valente and colleagues (12) also noted the presence of a strong mitochondrial targeting peptide at the N terminus of the protein and showed that N-myc-tagged proteins expressed in mammalian cells accumulated in mitochondria.
The identification of PINK1 mutations associated with recessive parkinsonism suggests two important hypotheses. First, as we (21) and others (22) have noted, the mitochondrial localization might support the hypothesis that this organelle is critically involved in the pathogenesis of nigral cell loss. Second, the recessive nature of the PINK1 mutations suggests that loss of function is associated with disease, further implying that the PINK1 kinase activity is important in protecting nigral neurons. Valente et al. (12) have shown that wild-type PINK1, but not the G309D mutation, protects cells against the loss of mitochondrial function resulting from exposure to proteasome inhibitors.
In this study, we examined whether mutations in PINK1 affect kinase activity by using the autophosphorylation assay previously shown to be useful for this protein in vitro (19). We also examined protein processing, localization, and steady-state levels for two mutations: the G309D mutation originally reported by Valente et al. (12) and the L347P mutation prevalent in the Philippines. We show that, like other leucine-to-proline substitutions in α-helixes (23), L347P has a dramatic effect on protein stability in mammalian cells.
Materials and Methods
Homology Modeling of the Kinase Domain of PINK1. Sequence analysis indicated that amino acids 235–554 of human PINK1 were similar to the family of eukaryotic serine/threonine protein kinases. A model of this domain was constructed from a set of homologous kinase domain crystal structures by using techniques implemented in the whatif suite of programs (24).
Plasmids. A cDNA encoding the full-length of human PINK1 (residues 1–581) cloned into the Gateway entry clone was purchased from Genecopoeia (Germantown, MD). Full-length PINK1 was transferred into destination vectors by using Gateway recombination technology (Invitrogen) according to the manufacturer's instructions. We used pcDNA-DEST47 to the fuse GFP to the C terminus of PINK1 and pcDNA-DEST53 to fuse GFP to the N terminus of PINK1. Additionally, we cloned PINK1 into pCM-VTnT (Promega) and introduced a myc-tag on the C or N terminus of PINK1. Two primers for the N-myc tag or two primers for C-myc tag were ligated together and cloned into pCMVTnT digested with XhoI and XbaI to produce pCMVTnT-N-myc or SalI and NotI for pCMVTnT-C-myc. All primer sequences are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The PINK1 gateway entry vector was digested with EcoRI/NotI and EcoRI/SalI to release the PINK1 coding region then ligated to pCMVTnT-N-myc and pCMVTnT-C-myc respectively.
Recessive mutants L347P and G309D were introduced by site-directed mutagenesis by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. To test kinase activities, we made K219A, D362A, and D384A variants as well as a triple kinase-dead mutant (K219A/D362A/D384A) by using the QuikChange multi kit (Stratagene).
For transfections, COS7 or M17 (human neuroblastoma) cells were grown in Opti-MEM supplemented with 10% (vol/vol) FBS (Invitrogen) and were transiently transfected by using FuGENE (Roche Applied Science, Indianapolis) as described in ref. 23.
Protein Purification and in Vitro Kinase Assays. The kinase domain (amino acids 112–496) of wild-type or mutant human PINK1 cDNAs was amplified by using Pfu DNA polymerase (Stratagene) with forward (5′-ACAGAATTCGAGGAAAAACAGGCGGAG-3′) and reverse (5′-TTTCTCGAGCTTGCTGGCCTCTCG-3′) primers, resulting in the creation of EcoRI and XhoI sites. The PCR products were digested with EcoRI and XhoI, and the resulting DNA fragments were ligated into pGEX-5X-1 (Amersham Pharmacia Biosciences) for fusion with the N terminus of GST. PINK1 fusion proteins were expressed in Escherichia coli BL21 (DE3) (Novagen). Cells were grown at 37°C until the OD600 reached 0.7–0.8 and then were induced with isopropyl β-d-thiogalactopyranoside to a final concentration of 0.2 mM at 37°C for 3 h. Cells were harvested and lysed by sonication on ice in lysis buffer containing 50 mM Tris·HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1%, Triton X-100, and protease inhibitors. After centrifugation for 20 min at 12,000 × g, the supernatant was incubated with Glutathione Sepharose 4B beads (Amersham Pharmacia Biosciences) overnight at 4°C. The beads were washed extensively with lysis buffer and prepared for SDS/PAGE by addition of 2× SDS sample buffer and then heating at 95°C for 5 min.
Protein autophosphorylation assays were carried out as described in ref. 20. Samples were fractionated on 10% SDS/PAGE gels, which were stained in Coomassie blue solution overnight to confirm equal loading of protein. For autoradiography, gels were dried and imaged on a Storm phosphorimager (Amersham Pharmacia Biosciences).
Kinase activity was quantified by measuring [32P]ATP incorporation into GST-PINK1 by using a Storm phosphorimager running imagequant software (Amersham Pharmacia Biosciences). These data were normalized by protein expression levels with the total amount of protein on scanned Coomassie-stained gels, also using imagequant. Differences in autophosphorylation activity between different recombinant proteins (n = 3) were analyzed by using ANOVA with Fisher's PLSD post hoc test (Statview, SAS Institute, Cary, NC).
Immunoblotting. Proteins resolved on SDS/PAGE were blotted as described in refs. 11 and 23. Antibodies used were monoclonal anti-GFP (clones 7.1 and 13.1, Roche Applied Science), anti-myc (clone 9E10, Roche Applied Science), and anti-β-actin (clone AC15, Sigma). Blots were imaged by using a Storm phosphorimager, and the relative intensity of immunoreactive bands was measured by using imagequant 5.2 and normalized to β-actin. All data represent the means ± SEM from at least triplicate determinations.
Pulse–Chase Labeling. Cos-7 cells were transiently transfected with GFP-tagged PINK1. Forty-eight hours after transfection, cells were starved in methionine/cysteine-free DMEM (Gibco-BRL) plus 1% l-glutamine for 1 h. Cells were then exposed to 500 μCi (1 Ci = 37 GBq) of [35S]Met/[35S]Cys (Amersham Pharmacia Biosciences) per well for 3 h, rinsed three times with warm PBS, and chased in complete medium containing 1% l-glutamine, 2 mM l-methionine, 2 mM l-cysteine, and 10% FCS for various times up to 8 h. Cells were lysed in buffer containing 150 mM NaCl, 5 mM EDTA, 50 mM Tris (pH 7.6), 0.25% Nonidet P-40, 1 mM PMSF, and protease inhibitors. Transfected proteins were immunoprecipitated by using anti-GFP conjugated to agarose (Vector Laboratories), and products were eluted with sample buffer and separated by SDS/PAGE. After electrophoresis, the gel was fixed (isopropyl alcohol/water/acetic acid, 25:65:10), enhanced with Amplify (Amersham Pharmacia Biosciences) for 30 min, and dried for autoradiography on a Storm PhosphorImager.
Quantitative RT-PCR. Total RNA was isolated from cells 48 h after transfection by using TRIzol (Invitrogen). The isolated RNA was treated with a DNase I (Invitrogen), and 1-μg quantities of total RNA were reverse transcribed separately with SuperScript III reverse transcriptase (Invitrogen) by using oligo(dT) primers. cDNA templates were diluted 10-fold before use in RT-PCR. Primers for PINK1 were 5′-AGACGCTTGCAGGGCTTTC-3′ (forward) and 5′-GGCAATGTAGGCATGGTGG-3′ (reverse). Primers for β-actin were 5′-AGAAGGATTCCTATGTGGGCG-3′ (forward) and 5′-CATGTCGTCCCAGTTGGTGAC-3′ (reverse). Real-time quantitative PCR was performed and analyzed by using the Applied Biosystems 7900 system as described in ref. 25.
Subcellular Fractionation. Subcellular fractions were purified by using a mitochondria isolation kit according the manufacturer's instructions (Pierce). The purity of each fraction was determined by using anti-VDAC1 (1:4,000 monoclonal 31HL, Calbiochem) and anti-Hsp70 (1:400 monoclonal W27, Neomarkers, Fremont, CA).
Immunofluorescence Staining. Cells were stained with MitoTracker (Molecular Probes) and transfected PINK1 as described in refs. 11 and 23 for DJ-1, except that primary antibodies were either anti-GFP or anti-myc (both at 1:200 dilution).
Results
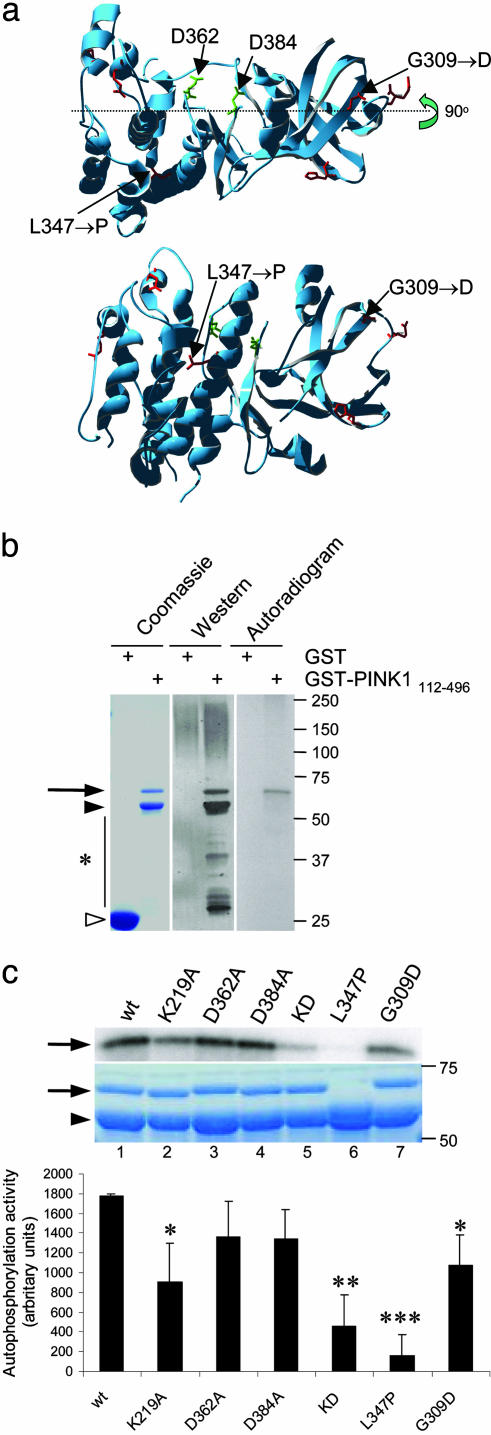
Effects of Mutations on PINK1 Kinase Activity in Vitro. The primary sequence of PINK1 includes three motifs consistent with the protein having kinase activity. For the human protein, these motifs are LAIK (amino acids 216–219), HRD (amino acids 360–362), and DFG (amino acids 384–386), which are predicted to form an ATP orientation site, an active catalytic base, and a chelation site for divalent metal cations, respectively. We modeled the kinase portion of human PINK1 that includes two of these motifs from amino acids 235–554 by using a family of homologous serine/threonine protein kinase domain crystal structures to create a consensus template, which was then modified with the sequence of PINK1. The homology model was subjected to constrained empirical energy minimization to remove any steric clashes and optimize local interactions without allowing the structure to depart significantly from that of the consensus template (Fig. 1a). This model predicts that the putative catalytic aspartate, D362, and the metal-binding site (D384) are relatively close to each other and are likely to form an active site. The G309 and L347 sites where recessive point mutations have been found are not predicted to be within the active site but in areas that may be important for protein folding. This model places G309 in a loop near the PSTAIRE helix and the ATP binding site. A G309D substitution is unlikely to grossly destabilize the fold but could interfere electrostatically with ATP binding or hydrolysis. L347 is predicted to be in the middle of a PSTAIRE helix, a helical segment that forms part of the cyclin binding surface in the cyclin-dependent protein kinases. From this model, we made the following predictions. First, mutating D362 and D384 (in conjunction with K219) would inactivate kinase activity. Second, given the propensity of proline to act as a helix breaker, L347P is likely to be unstable, but G309D would be relatively stable.
Fig. 1.
Kinase activity of PINK1 and the effects of mutations in vitro.(a) Model of kinase domain of PINK1 showing positions of critical aspartates and the two recessive mutations investigated in this study. Lower is rotated by 90° at the indicated axis. (b) Expression and autophosphorylation activity of wild-type PINK1 GST-fusion proteins showing, from left to right, Coomassie-stained SDS/PAGE gel, Western blot, and autoradiogram. Alternate lanes contain GST alone, as indicated. Arrow indicates GST-PINK1 fusion proteins, filled arrowhead shows prominent breakdown product, and open arrowhead shows GST alone. Markers on the right of all gels or blots are in kilodaltons. (c) Kinase activity of PINK1 mutants showing autoradiogram (Top), Coomassie staining (Middle), and quantitation of incorporated radioactivity, corrected for protein loading. Wild-type PINK1 (lane 1) is shown for comparison with artificial kinase mutants (K219A, lane 2; D362A, lane 3; D384A, lane 4; triple kinase-dead mutant, lane 5). Two recessive mutants associated with human parkinsonism, L347P and G309D, are shown in lanes 6 and 7. (Bottom) Bars show the average from three independent experiments, with error bars showing SEM. Differences in activity between variants were assessed by ANOVA with Fisher's PLSD post hoc test; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test these ideas, we generated recombinant wild-type and mutant PINK1 proteins. Full-length PINK1 was toxic to E. coli and poorly soluble (data not shown). Therefore, we made constructs lacking the mitochondrial targeting peptide and part of the C terminus of PINK1, fused to GST as described by Unoki et al. (19). GST-PINK1112–496 constructs were purified from the soluble fraction of bacterial lysates as an intact band of the predicted size of the fusion protein (apparent molecular mass of 70 kDa), although degradation products including a major band at 60 kDa were seen (Fig. 1b). Some PINK1 was deposited into insoluble fractions (data not shown). PINK1 has previously been shown to be capable of autophosphorylation in vitro (19), which we used as a convenient assay for active enzyme. Autophosphorylation was limited to the intact GST-PINK1 protein, not degradation products (Fig. 1b).
We next measured autophosphorylation of PINK1 variants in the same assay (Fig. 1c). There was a significant overall difference between the constructs (P = 0.0002 by one-way ANOVA; n = 3). Of the constructs designed to reduce kinase activity, K219A had a significant effect alone, decreasing activity to 50.6 ± 12.8% of wild type (P = 0.003), whereas both D362A and D384A decreased activity to ≈75% of wild type, neither effect was statistically significant. However, the effect of all three substitutions was stronger than K219A alone, decreasing activity to 25.5 ± 10.5% of wild type (P < 0.0001). We tested two recessive mutations and found that although G309D had somewhat decreased activity (60.5% of wild type; P = 0.01), L347P had drastically decreased activity (<10% of wild type). Recombinant L347P GST-PINK1 fusions were unstable. This result suggests that the L347P mutation destabilizes the protein, supporting our prediction above (Fig. 1c).
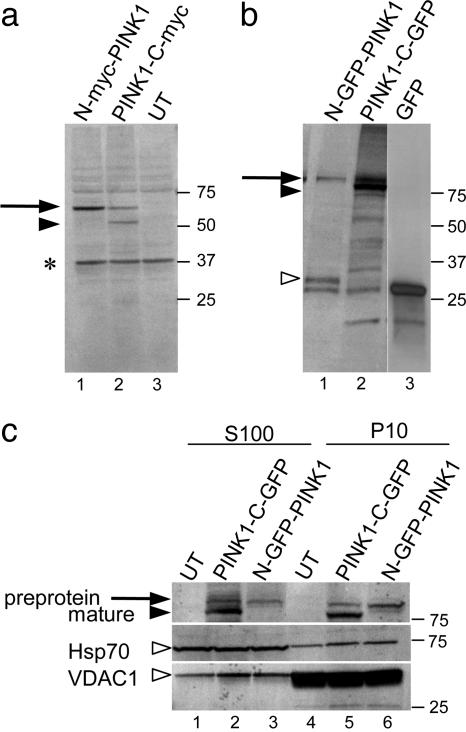
Processing and Mitochondrial Localization of PINK1 in Mammalian Cells. The targeting sequence at the N terminus of PINK1 is predicted to direct the protein toward mitochondria, consistent with the data of Valente et al. (12), who demonstrated mitochondrial accumulation of N-terminally tagged PINK1 in transfected cells. However, many mitochondrial leader peptides are cleaved from the mature protein after import into the mitochondria by mitochondrial processing peptidases (26), suggesting that intact N-terminal tagged protein is likely to be a preprotein. Therefore, before examining the effects of mutations, we explored the possible cleavage of the N-terminal targeting sequence.
Cells transfected with PINK1 tagged at the N terminus with myc showed a single band with an apparent molecular mass of 65 kDa. This finding is close to the theoretical molecular mass of the 63-kDa myc tag and consistent with the results of Valente et al. (12). An additional band with an apparent molecular mass of 55 kDa was seen by using a C-terminal tagged construct (Fig. 2a). Similar results were seen with the larger GFP tag (Fig. 2b), where the preprotein was ≈90 kDa, and the smaller fragment was ≈80 kDa. The decrease in size is consistent with proteolysis of an ≈10-kDa fragment from the N terminus of the protein. Additionally, we saw a band of ≈30 kDa in cells transfected with N-terminal GFP-PINK1 protein. This band probably represents GFP plus the mitochondrial targeting sequence before the cleavage site. With transient transfections driven from the strong CMV promoter, it is very likely that amount of PINK1 is much higher than it would normally be in the cell. Therefore, it is also likely the remaining peptides are not fully degraded in the mitochondria after cleavage; this conclusion is consistent with the observation that there is an amount of preprotein remaining with both constructs. Cleavage of constructs containing a C-terminal V5/His6 tag was also seen (data not shown). These results suggest that PINK1 is processed at the N terminus after import into the mitochondria and that different fusion partners do not interfere with processing.
Fig. 2.
Processing of PINK1 protein in mammalian cells. COS-7 cells were transfected with myc-tagged (a) or GFP-tagged (b) constructs. N-terminal (lane 1) and C-terminal (lane 2) tagged versions were used, and either untransfected cells (lane 3 in a) or GFP-transfected cells (lane 3 in b) were included as controls. Arrows show preprotein (before cleavage of the N terminus), and filled arrowhead indicates mature peptide. Open arrowhead in the GFP constructs shows a possible N-terminal fragment (see Results); *, cross-reactivity seen with myc monoclonal in untransfected cells. (c) Subcellular fractionation. COS-7 cells were transfected with C-terminal (lanes 2 and 5) or N-terminal (lanes 3 and 6) GFP-tagged PINK1 or untransfected (lanes 1 and 4) and cell lysates separated into a 10,000 × g pellet (P10, lanes 4–6) and 100,000 × g supernatant (S100, lanes 1–3) fractions. Blot was reprobed with antibodies to Hsp70 and VDAC1 in Middle and Bottom, respectively (open arrowheads). Data are representative of more than two experiments for each construct. Markers on the right of all blots are in kilodaltons.
We also performed subcellular fractionation (Fig. 2c) by using the GFP-tagged proteins. The preprotein, represented by a single band in the N-terminally tagged construct, was enriched in the 10,000 × g pellet (P10) that included mitochondrial proteins, although an amount was also present in the 100,000 × g supernatant (S100). As mitochondrial proteins are often translated in the cytosol, this result likely reflects protein produced before import. Surprisingly, we also found that a proportion of the mature peptide (the lower band with C-terminal constructs) was also in the S100 fraction (Fig. 2c).
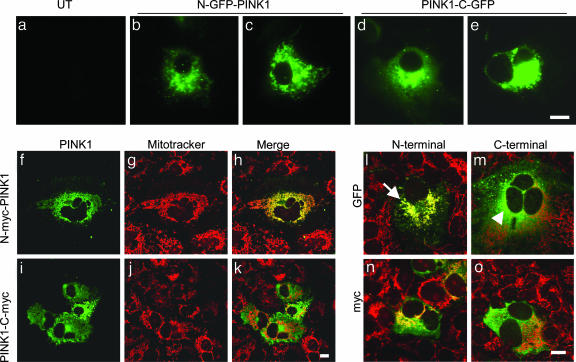
To confirm these data, we performed subcellular localization by using several different constructs. GFP-tagged PINK1 was observed directly in transfected cells without fixation. N-terminal GFP-tagged PINK1 was present in structures reminiscent of mitochondria, whereas C-terminal tagged protein was present in the cytosol (Fig. 3 a–e). In some cells, some of the PINK1 signal accumulated in multiple small cytosolic punctae or in single large perinuclear inclusion, behavior reminiscent of relatively insoluble aggregating protein (Fig. 3e). We also stained cells with MitoTracker to positively identify mitochondria. Using confocal microscopy, we saw that although the signal for N-terminally tagged PINK1 overlapped with MitoTracker (Fig. 3 f–h), signals for mitochondria and C-terminally tagged proteins were generally separated (Fig. 3 i–k). Similar results were seen for both myc and GFP-tagged proteins (Fig. 3 l–o) and for several different fixation methods, including methanol fixation (data not shown).
Fig. 3.
Localization of proteins in transfected cells. (a–e) COS-7 cells were transfected with GFP-tagged PINK1 constructs, at either the N terminus (b and c) or the C terminus (d and e) and imaged without fixation or staining. Untransfected cells taken at the same exposure settings are shown as a control in a.(f–k) Cells were transfected, N-terminal (f–h) or C-terminal (i–k) myc-tagged PINK1 and counterstained with MitoTracker. After fixation, myc staining was revealed by using anti-myc monoclonal antibody (green in f and i) and compared with MitoTracker (red in g and h). Merged images are shown in h and k.(l–o) GFP-vs. myc-tagged proteins, showing merged images only. The arrow in l indicates the area of overlap between N-GFP-PINK1 and MitoTracker compared with a cell transfected with PINK1-C-GFP where the signals remain separated (arrowhead in m). (Scale bars: 10 μm.) Each image is representative of at least three experiments performed in duplicate for each construct.
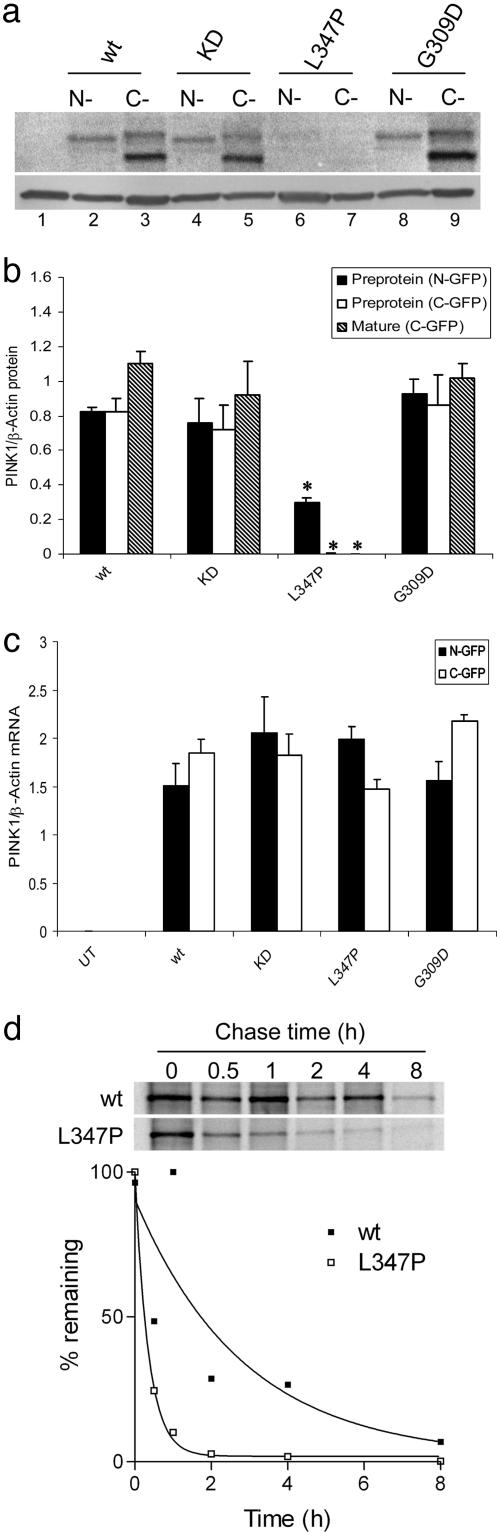
L347P PINK1 Is Unstable in Mammalian Cells. We next transfected cells with variant PINK1 constructs tagged with GFP at either end. Although mutation of the important kinase residues had no effect on steady-state levels of transfected protein, the amount of transfected PINK1 was drastically decreased by using the L347P mutant (Fig. 4a). Quantification showed that there was a very dramatic decrease in the amount of L347P protein (Fig. 4b) but not mRNA (Fig. 4c). The level of L347P protein was significantly (P < 0.01) less than that of wild type or other constructs when measuring either preprotein or mature protein, whereas there were no statistically significant differences between mRNA levels (P = 0.23 by ANOVA; n = 3), arguing that transfection efficiencies were similar for all constructs. No effect was noted for G309D constructs, similar to the report by Valente and colleagues (12). Constructs containing the L347P mutation but tagged with V5-His at the C terminus were also unstable (data not shown). The loss of protein with constant mRNA levels is highly reminiscent of the behavior of L166P DJ-1 (23) and is consistent with instability of the protein due to degradation by proteases in the cell. To test that the low steady-state levels were due to increased turnover of L347P, we performed pulse–chase analyses (Fig. 4d). We estimated that the half-life of wild-type N-terminal GFP-tagged PINK1 was ≈2 h, whereas the L347P mutation had a half-life of 0.25 h.
Fig. 4.
Effects of mutations on PINK1 protein stability in mammalian cells. (a) COS-7 cells were transfected with N-terminal (lanes 2,4,6 and 8) or C-terminal (lanes 3, 5, 7, and 9) GFP-tagged PINK1 variants or left untransfected as a control (lane 1). Steady-state protein levels were decreased for L347P (lanes 6 and 7) but not G309D (lanes 8 and 9) recessive mutant PINK1 compared with wild type (lanes 2 and 3) control or the artificial kinase-dead construct (lanes 4 and 5). The arrow indicates the position of the preprotein, the filled arrowhead shows the mature protein, and the open arrowhead shows the position of β-actin used as a loading control. We quantified both protein (b) and mRNA (c) expression for all constructs from n = 3 experiments. For protein data, we measured the intensity of the preprotein band from the N-terminal construct (filled bars) with both the preprotein (open bars) and mature protein (hatched bars) from the C-terminal construct. *, protein levels were significantly different from wild-type construct (P < 0.01 by ANOVA). (d) Pulse–chase analysis of protein stability. COS-7 cells were transiently transfected with N-terminal GFP-tagged wild-type (Upper) or L347P (Lower) PINK1, labeled with 35S-Met/Cys and chased with unlabeled media for 0–8 h as indicated above each lane. Quantification of this experiment is shown, with curves fitted to a single-phase exponential decay function. Similar estimates of stability were obtained in duplicate experiments.
We also examined the localization of mutant proteins. Kinasedead PINK1 localized as a wild-type protein, with N-terminal tagged constructs overlapping with MitoTracker and C-terminal tagged constructs being present in the cytoplasm, occasionally as inclusions (Fig. 5 a and b, which is published as supporting information on the PNAS web site). Consistent with the lower steady-state levels of protein observed by using blotting, L347P PINK1 gave much lower levels of immunostaining, especially for the C-terminal constructs (Fig. 5 c and d). G309D PINK1, however, appeared to be normally localized (Fig. 5 e and f) and, like wild-type or kinase-dead proteins, was present in cytosolic inclusions in some cells (data not shown).
Discussion
In this study, we sought to establish whether mutations in PINK1 affected any of several parameters relevant to the known properties of the protein. As PINK1 is thought to be a mitochondrially directed protein kinase, we compared the kinase activity and localization of wild-type PINK1 with G309D and L347P mutants. Additionally, as controls we generated a triple kinase-dead artificial mutant and established the effects of various mutations on protein stability in mammalian cells.
These data confirm (20) that PINK1 is a kinase by using the relatively crude assay of self-directed phosphorylation. Although probably not entirely physiological, the assay is useful in that it allowed us to quantify the effects of mutations on activity in the folded enzyme. Consistent with descriptions in ref. 20, full-length PINK1 was not soluble and could not be stably expressed, and although we have had difficulties with degradation of smaller fragments of PINK1, we were able to purify active kinase. We were able to inhibit activity of the protein by mutating key residues (K219, D362, and D384) predicted to be important in orienting the ATP and carrying out the phosphate-transfer reaction to acceptor residues on the substrate. We chose to mutate these residues to alanine residues to minimize secondary structure disruption, which is likely to have been the case because the protein was efficiently expressed in bacteria and mammalian cells (see below). Both recessive, disease-associated mutations tended to decrease activity, although G309D had a less dramatic effect on activity than L347P. Consistent with our predictions from modeling, L347P was quite unstable even when expressed as a fusion protein in E. coli.
As well as having a predicted kinase domain, PINK1 has a mitochondrial targeting peptide (12). In our comparisons of N- and C-terminally tagged proteins in mammalian cells, we demonstrated that ≈8–10 kDa is cleaved from the N terminus, although in overexpression studies, not all of the protein is processed. Predicting the precise cleavage sequence is difficult because mitochondrial processing peptidases have rather loose consensus sequences for cleavage sites, sometimes (but not always) directed to amino acids placed two to three residues after an arginine. Examination of the primary sequence of human PINK1 also reveals a RFFRQSVAGL motif, which is consistent with the consensus for the mitochondrial intermediate peptidase [RX(F/L/I)XX(T/S/G)XXXX], a secondary peptidase that acts on a subset of inner matrix proteins. Therefore, there may be multiple sites of degradation of human PINK1 within the first 100 amino acids of the protein. Further experiments are warranted to identify the exact sequences that are cleaved from the N-terminal portion of PINK1.
These results are consistent with mitochondrial targeting of PINK1, as suggested elsewhere in ref. 12. For the N-terminal constructs, we confirmed mitochondrial localization. However, at least in transfected cells, a proportion of mature PINK1 appears to be exported to the cytosol. Because this polypeptide is the processed form of the protein, it is likely that the protein has already been acted on by mitochondrial peptidases. We have also seen accumulation of C-terminally tagged protein in inclusion bodies, reminiscent of proteins with relatively poor solubility when expressed at high levels by using heterologous promoters. These considerations suggest that PINK1 may be prone to aggregation or loss of solubility, highlighting the importance of further studies by using endogenous protein.
Although unusual, there are precedents for proteins that behave in this manner. For example, yeast fumarase is folded in the mitochondria but can be exported to the cytoplasm by retrograde transport (27). In mammalian cells, some glutatione S-transferase isoenzymes can be targeted to the cytosol or to mitochondria in a phosphorylation-dependent fashion (28). It is not yet clear whether the differential localization of preproteins and mature PINK1 is a physiological response or whether it might be modulated under different conditions. For example, the cytosolic location might be an attempt by the cell to degrade the excess protein. We confirmed the localization by using two different C-terminal tags, so it is unlikely to be the presence of (for example) GFP, which drives cytosolic accumulation. Again, this result requires confirmation by using endogenous levels of protein. The anti-peptide antibody used for detection of GST-fusion proteins did not recognize endogenous protein (A.B. and M.RC., unpublished observations). However, if there is a cytosolic pool of mature, kinase active PINK1, then this observation complicates the assumption that PINK1 mutations support a mitochondrial hypothesis for the disease process in recessive parkinsonism (21, 22).
An unambiguous result from these studies is that the L347P mutation produces low steady-state levels of protein in cells compared with wild-type PINK1. Although the mechanism is not yet identified, this mutation may have a similar effect to the L166P mutation in DJ-1, where misfolding of structurally important α-helix leads to poor stability. In mammalian cells, this misfolding leads to recognition by either the ubiquitin-proteasome system (23) or other proteolytic machineries (29). Irrespective of the mechanism(s) involved, the very low steady state of L347P PINK1 implies that this mutation is causal. If so, with a carrier frequency of 8%, this finding implies that this mutation will account for many cases of recessive parkinsonism in the Philippines. The loss of function mechanism of the G309D mutation is not identical because this protein is relatively stable. Previously, Valente and colleagues demonstrated that this mutant lacks protective function in transfected cells. Whether the small decrease in kinase activity observed in this study would account for this loss of protective activity is not clear. However, a reasonable hypothesis for recessive PINK1 mutations would be that they either destabilize the protein and/or decrease kinase activity below that required to support neuronal survival in vulnerable groups. Future studies to establish that the kinase activity of PINK1 is necessary for neuronal survival and to identify the natural substrates of the protein are required.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Singleton (National Institute on Aging) for helpful discussions regarding the L347P mutation.
Author contributions: S.K., D.W.M., and M.R.C. designed research; A.B., M.V.D.B., and R.A. performed research; S.K., D.W.M., and G.A.P. contributed new reagents/analytic tools; A.B., M.V.D.B., D.W.M., and M.R.C. analyzed data; and A.B. and M.R.C. wrote the paper.
Abbreviation: PINK1, PTEN-induced putative kinase 1.
References
- 1.Kitada, T., Asakawa, S., Hattori, N., Matsumine, H., Yamamura, Y., Minoshima, S., Yokochi, M., Mizuno, Y. & Shimizu, N. (1998) Nature 392, 605–608. [DOI] [PubMed] [Google Scholar]
- 2.Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256–259. [DOI] [PubMed] [Google Scholar]
- 3.Hilker, R., Klein, C., Ghaemi, M., Kis, B., Strotmann, T., Ozelius, L. J., Lenz, O., Vieregge, P., Herholz, K., Heiss, W. D. & Pramstaller, P. P. (2001) Ann. Neurol. 49, 367–376. [PubMed] [Google Scholar]
- 4.Broussolle, E., Lucking, C. B., Ginovart, N., Pollak, P., Remy, P. & Durr, A. (2000) Neurology 55, 877–879. [DOI] [PubMed] [Google Scholar]
- 5.Scherfler, C., Khan, N. L., Pavese, N., Eunson, L., Graham, E., Lees, A. J., Quinn, N. P., Wood, N. W., Brooks, D. J. & Piccini, P. P. (2004) Brain 127, 1332–1342. [DOI] [PubMed] [Google Scholar]
- 6.Khan, N. L., Brooks, D. J., Pavese, N., Sweeney, M. G., Wood, N. W., Lees, A. J. & Piccini, P. (2002) Brain 125, 2248–2256. [DOI] [PubMed] [Google Scholar]
- 7.Dekker, M., Bonifati, V., van Swieten, J., Leenders, N., Galjaard, R. J., Snijders, P., Horstink, M., Heutink, P., Oostra, B. & van Duijn, C. (2003) Movement Disorders 18, 751–757. [DOI] [PubMed] [Google Scholar]
- 8.Hering, R., Strauss, K. M., Tao, X., Bauer, A., Woitalla, D., Mietz, E. M., Petrovic, S., Bauer, P., Schaible, W., Muller, T., et al. (2004) Hum. Mutat. 24, 321–329. [DOI] [PubMed] [Google Scholar]
- 9.Cookson, M. R. (2003) Curr. Biol. 13, R522–R524. [DOI] [PubMed] [Google Scholar]
- 10.Cookson, M. R. (2003) Neuron 37, 7–10. [DOI] [PubMed] [Google Scholar]
- 11.Canet-Aviles, R. M., Wilson, M. A., Miller, D. W., Ahmad, R., McLendon, C., Bandyopadhyay, S., Baptista, M. J., Ringe, D., Petsko, G. A. & Cookson, M. R. (2004) Proc. Natl. Acad. Sci. USA 101, 9103–9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valente, E. M., Abou-Sleiman, P. M., Caputo, V., Muqit, M. M., Harvey, K., Gispert, S., Ali, Z., Del Turco, D., Bentivoglio, A. R., Healy, D. G., et al. (2004) Science 304, 1158–1160. [DOI] [PubMed] [Google Scholar]
- 13.Hatano, Y., Li, Y., Sato, K., Asakawa, S., Yamamura, Y., Tomiyama, H., Yoshino, H., Asahina, M., Kobayashi, S., Hassin-Baer, S., et al. (2004) Ann. Neurol. 56, 424–427. [DOI] [PubMed] [Google Scholar]
- 14.Healy, D. G., Abou-Sleiman, P. M., Gibson, J. M., Ross, O. A., Jain, S., Gandhi, S., Gosal, D., Muqit, M. M., Wood, N. W. & Lynch, T. (2004) Neurology 63, 1486–1488. [DOI] [PubMed] [Google Scholar]
- 15.Rogaeva, E., Johnson, J., Lang, A. E., Gulick, C., Gwinn-Hardy, K., Kawarai, T., Sato, C., Morgan, A., Werner, J., Nussbaum, R., et al. (2004) Arch. Neurol. (Chicago) 61, 1898–1904. [DOI] [PubMed] [Google Scholar]
- 16.Rohe, C. F., Montagna, P., Breedveld, G., Cortelli, P., Oostra, B. A. & Bonifati, V. (2004) Ann. Neurol. (Chicago) 56, 427–431. [DOI] [PubMed] [Google Scholar]
- 17.Bentivoglio, A. R., Cortelli, P., Valente, E. M., Ialongo, T., Ferraris, A., Elia, A., Montagna, P. & Albanese, A. (2001) Movement Disorders 16, 999–1006. [DOI] [PubMed] [Google Scholar]
- 18.Khan, N. L., Valente, E. M., Bentivoglio, A. R., Wood, N. W., Albanese, A., Brooks, D. J. & Piccini, P. (2002) Ann. Neurol. (Chicago) 52, 849–853. [DOI] [PubMed] [Google Scholar]
- 19.Unoki, M. & Nakamura, Y. (2001) Oncogene 20, 4457–4465. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima, A., Kataoka, K., Hong, M., Sakaguchi, M. & Huh, N. H. (2003) Cancer Lett. 201, 195–201. [DOI] [PubMed] [Google Scholar]
- 21.Shen, J. & Cookson, M. R. (2004) Neuron 43, 301–304. [DOI] [PubMed] [Google Scholar]
- 22.Giasson, B. I. (2004) Sci. Aging Knowledge Environ. 2004, pe42. [DOI] [PubMed] [Google Scholar]
- 23.Miller, D. W., Ahmad, R., Hague, S., Baptista, M. J., Canet-Aviles, R., McLendon, C., Carter, D. M., Zhu, P. P., Stadler, J., Chandran, J., et al. (2003) J. Biol. Chem. 278, 36588–36595. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez, R., Chinea, G., Lopez, N., Pons, T. & Vriend, G. (1998) Comput. Appl. Biosci. 14, 523–528. [DOI] [PubMed] [Google Scholar]
- 25.Baptista, M. J., O'Farrell, C., Daya, S., Ahmad, R., Miller, D. W., Hardy, J., Farrer, M. J. & Cookson, M. R. (2003) J. Neurochem. 85, 957–968. [DOI] [PubMed] [Google Scholar]
- 26.Neupert, W. (1997) Annu. Rev. Biochem. 66, 863–917. [DOI] [PubMed] [Google Scholar]
- 27.Knox, C., Sass, E., Neupert, W. & Pines, O. (1998) J. Biol. Chem. 273, 25587–25593. [DOI] [PubMed] [Google Scholar]
- 28.Robin, M. A., Prabu, S. K., Raza, H., Anandatheerthavarada, H. K. & Avadhani, N. G. (2003) J. Biol. Chem. 278, 18960–18970. [DOI] [PubMed] [Google Scholar]
- 29.Gorner, K., Holtorf, E., Odoy, S., Nuscher, B., Yamamoto, A., Regula, J. T., Beyer, K., Haass, C. & Kahle, P. J. (2004) J. Biol. Chem. 279, 6943–6951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.