Abstract
Three-dimensional printing has an increasing number of clinical applications in pediatric cardiology. Time required for dataset segmentation and conversion to stereolithography (STL) format remains a significant limitation. We investigated the impact of semi-automated cardiovascular-specific segmentation software on time and reproducibility of segmentation. Magnetic resonance angiograms (MRAs) of 19 patients undergoing intervention for right ventricular outflow lesions were segmented to demonstrate the right heart. STLs were created by two independent clinicians using semi-automated cardiovascular segmentation (SAS) and traditional manual segmentation (MS). Time was recorded and geometric STL disagreement was determined (0 % = no disagreement, 100 % = complete disagreement). MRA datasets were categorized as clean when only right heart structures were present in the MRA, or contaminated when left heart structures were also present and required removal. Eighteen (seven clean and 11 contaminated) cases were successfully segmented with both methods. Time to STL for clean datasets was faster with MS than SAS [median 209 s (IQR 192–252) vs. 296 s (272–317), p = 0.018] while contaminated datasets were faster with SAS [455 s (384–561) vs. 866 s (310–1429), p = 0.033]. Interobserver STL geometric disagreement was significantly lower using SAS than MS overall (0.70 ± 1.15 % vs. 1.31 ± 1.52 %, p = 0.030), and for the contaminated subset (0.81 ± 1.08 % vs. 1.75 ± 1.57 %, p = 0.036). Most geometric disagreement occurred at areas where left heart contamination was removed. Semi-automated segmentation was faster and more reproducible for contaminated datasets, while MS was faster but equally reproducible for clean datasets. Semi-automated segmentation methods are preferable for contaminated datasets and continued refinement of these tools should be supported.
Keywords: Stereolithography, 3D printing, Magnetic resonance angiography, Cardiac segmentation
Introduction
Three-dimensional (3D) printing is an evolving technology that is used increasingly in pediatric cardiology, especially in relation to interventional planning [1–3]. Use of patient-specific imaging for the creation of 3D printed models (3DPMs) requires the conversion of the imaging dataset into a stereolithography (STL) file, including the process of image segmentation. Segmentation is often time-consuming, and the consistency of the results of segmentation has not been thoroughly investigated in pediatric cardiology [4].
Patients with tetralogy of Fallot, double outlet right ventricle, truncus arteriosus, and other right ventricular outflow tract (RVOT) lesions often undergo reintervention on the RVOT as they grow older. For these patients, cardiac MRI (CMR) is indicated to help determine the need for intervention through accurate determination of right ventricular size and function [5–10], for improved visualization of the RVOT before going to the cardiac catheterization lab for interventions [11], and to determine the relationship of the coronaries to the RVOT [12–14]. During these CMR studies, patients usually undergo contrast-enhanced magnetic resonance angiography (MRA) to create accurate images of the vasculature; these MRA images can then be used to create 3DPMs that can be used in pre-procedural planning [15]. The use of multiple dynamics of the MRA often allows isolation of the right and left sides of the heart, but this process is not always successful; therefore, some right heart MRA can become contaminated with left heart or other extraneous structures.
We compared a standard manual segmentation (MS) method to a semi-automated segmentation (SAS) method, designed specifically for cardiac segmentation, in terms of time to and variation of RVOT STL datasets. We also investigated the differences for clean vs. contaminated datasets.
Materials and methods
Patient selection
We identified patients who had undergone both cardiac MRI and cardiac catheterization due to right heart lesions at a single institution (UT Southwestern Medical Center/Children’s Medical Center Dallas) over the last 5 years. Patients were included if they had undergone contrast-enhanced MRA as part of their CMR.
MRA dataset preparation
As a retrospective cohort, there was some variability in the MRA acquisition. The studies generally employed the high-temporal resolution “keyhole” MRA, with representative settings: TR 4–5.5 ms, TE 1.2–1.6 ms, flip angle 35–45°, CENTRA k-space filling, acquired voxel size 1.3–2 × 1.3–2 × 1.3–2 mm3, reconstructed voxel size 0.65–1 × 0.65–1 × 0.65–1 mm3, number of dynamics 6–8, keyhole percentage 25–38 %, two-dimensional SENSE factor 2 × 1–2, yielding approximately a 6 s dynamic time [16]. 0.1 mmol/kg of gadolinium (gadoteridol or gadobutrol) was injected at a rate of 2–3 mL/s.
Each MRA was visually analyzed to determine, qualitatively, which dynamic was the highest quality angiogram of the right heart structures, with the least contamination by left heart or other structures. The CMR dataset was exported anonymously from the PACS system and loaded into Mimics (18.0, Materialise, Leuven, Belgium). Using the thresholding tool in Mimics, a threshold was determined for each patient image dataset that qualitatively allowed maximal visualization of right heart structures with minimal left heart and extraneous selection. The dynamic and threshold chosen were noted and used for both MS and semi-automated segmentation. Datasets were categorized by a consensus of both reviewers as clean if the thresholded MRA dataset included only right heart structures, while datasets were categorized as contaminated if the thresholded MRA dataset included any left heart or other structures that required cropping.
Segmentation preparation
A single computer was used for analysis (HP ProBook 530, Hewlett Packard, Palo Alto, CA) running Windows 10 Pro 64-bit and Excel 2010 (Microsoft, Seattle, WA). No other programs were actively running on the computer to allow for equal available processing power. The order of the segmentation of the datasets was randomized, and half the datasets were first segmented manually, and half semi-automatically. Both MS and semi-automated segmentation were performed by two pediatric cardiologists (AT and TH) with 2 and 9 years of experience in CMR and image postprocessing, respectively.
Manual segmentation
MS and STL preparation were performed using Mimics and 3-matic (18.0.0.524 and 10.0.0.212, Materialise, Leuven, Belgium). The timer was started with the Mimics and 3-matic screens blank and ended when the STL file was saved. The correct patient was loaded and the correct dynamic selected. Thresholding was applied with the levels determined as above. A region growing seed was placed in the right ventricle to create the first mask. The mask was inspected in 3D view to see if any extraneous data, e.g. left heart structures, needed to be removed. If so, the 3D lasso tool, multiple slice edit tool, and region growing tools were repeated until each cardiologist determined that the right heart structures had been isolated and no left heart structures remained. The mask was converted to a solid 3D mesh object using the Calculate 3D tool with high quality setting, and then converted to an STL file using the STL + tool. The STL file was opened in 3-matic and the shell was selected, inverted, and deleted to create a single object. The fix wizard to correct shell geometry for printing was applied to this object and corrections automatically applied. Finally, the STL file was saved.
Semi-automated segmentation
Semi-automatic segmentation was performed using in Print (1.0.0.156 beta, Materialise, Leuven, Belgium). The timer was started with the in Print screen blank and ended when the STL file was saved. The correct patient was loaded and the correct dynamic selected. Thresholding was applied with the levels determined as above. Four seed points, used to identify cardiac chambers and start the segmentation process, were used per chamber when possible. The right atrial seed points included the right atrial-superior vena cava junction; the os of the right atrial appendage; the atrial side of the tricuspid valve; and a fourth seed in the body of the right atrium. The right ventricular seed points were placed just distal to the tricuspid valve; in the RVOT just proximal to the pulmonary valve or RV-PA conduit; at the right ventricular apex; and in the body of the right ventricle. The pulmonary seed points were placed just distal to the pulmonary valve or in the proximal RV-PA conduit; at the bifurcation of the pulmonary arteries; and one each in the distal right and left pulmonary arteries. If left heart structures were present in the dataset after thresholding, seed points were placed in these structures as well (e.g. in the pulmonary veins and left atrium, left ventricle, and aorta); automatically segmenting the left heart structures made it easier to later crop them out. The automatic region identification was performed. Left heart structure masks were deleted, and then the right heart structures were combined into a single right heart blood pool mask. If further segmentation was needed, 3D lasso tools were used to remove unwanted areas. A solid part was created from the result. The part was fixed to correct shell geometry for printing, and finally the STL file was saved.
Stereolithography file comparison
The created STL files were compared using 3-matic. A coefficient of difference, (1-Dice similarity coefficient) [17], was calculated between observers for SAM and MS.
Statistical analysis
Since time to STL had a nonparametric distribution, it was described with median and interquartile range and analyzed using Wilcoxon rank-sum test. Interobserver STL geometric disagreement was described using mean and standard deviation, and analyzed using paired t tests. A p value of <0.05 was considered statistically significant. Statistical analyses were performed using IBM SPSS Statistics (Version 21.0. Armonk, NY: IBM Corp.).
Results
Patient characteristics
Nineteen patients were analyzed based on the inclusion criteria. Eighteen datasets were successfully segmented using both methods. The dataset that failed SAS had both main and left pulmonary artery stents. The patients ranged in age at time of CMR from 4.4 to 21.7 years (see Table 1).
Table 1.
Patient demographics and diagnoses
| Age (years) | Sex | Diagnosis | BSA (m2) | HR (b/min) | Clean vs. contaminated |
|---|---|---|---|---|---|
| 4.4 | F | TOF/PS/MAPCAs, s/p RVOT stenting s/p melody s/p melody extraction and PVR | 0.68 | 62 | Clean |
| 4.6 | F | TOF, s/p RVOT patch | 0.68 | 84 | Contaminated |
| 7.8 | M | Truncus arteriosus, s/p repair and RV-PA conduit × 2 | 0.91 | 96 | Clean |
| 8.7 | M | Truncus arteriosus s/p RV-PA conduit × 2 | 1.13 | 105 | Contaminated |
| 9.0 | F | DORV, s/p rastelli and RV-PA conduit | 0.93 | 67 | Contaminated |
| 9.5 | M | DORV, s/p arterial switch with LeCompte maneuver | 1.18 | 107 | Clean |
| 10.5 | F | TOF/PA/MAPCAs, s/p RV-PA conduit | 1.06 | 59 | Clean |
| 10.9 | M | Aortic stenosis, s/p ross s/p RV-PA conduit | 1.56 | 75 | Contaminated |
| 12.1 | F | DORV, subaortic VSD, sub-PS, s/p transannular patch | 1.25 | 82 | Contaminated |
| 13.1 | M | TOF/PA, s/p RV-PA conduit | 2.07 | 81 | Contaminated |
| 14.0 | M | TOF/PA, s/p pulmonary homograft s/p homograft patch plasty s/p melody | 1.48 | 77 | Clean |
| 15.4 | M | TOF s/p valve-sparing repair | 1.54 | 89 | Contaminated |
| 16.4 | M | TOF, s/p transannular patch | 1.34 | 99 | Clean |
| 18.8 | M | TOF, s/p transannular patch | 1.91 | 78 | Clean |
| 18.9 | M | L-TGA, AVSD, PS, s/p LV-PA conduit | 2.21 | 97 | Contaminated |
| 20.5 | M | TOF, s/p PVR s/p melody | 1.72 | 49 | Contaminated |
| 21.4 | M | Aortic stenosis, s/p ross procedure s/p RV-PA conduit replacement | 1.99 | 58 | Contaminated |
| 21.7 | F | PS, s/p pulmonary homograft | 1.7 | 82 | Contaminated |
AVSD atrioventricular septal defect, DORV double outlet right ventricle, L-TGA l-transposition of the great arteries, LV-PA left ventricle to pulmonary artery, MAPCAs multiple aortopulmonary collaterals, PA pulmonary atresia, PS pulmonary stenosis, PVR pulmonary valve replacement, RVOT right ventricular outflow tract, RV-PA right ventricle to pulmonary artery, s/p status post, TOF tetralogy of fallot
Time to STL
Time to STL for clean datasets was faster with MS than SAS [median 209 s (IQR 192–252) vs. 296 s (272–317), p = 0.018], meaning that MS was 29 %, or 1.5 min, faster; contaminated datasets were faster with SAS [455 s (384– 561) vs. 866 s (310–1429), p = 0.033, see Fig. 1], a difference of 47 % or 6.9 min.
Fig. 1.
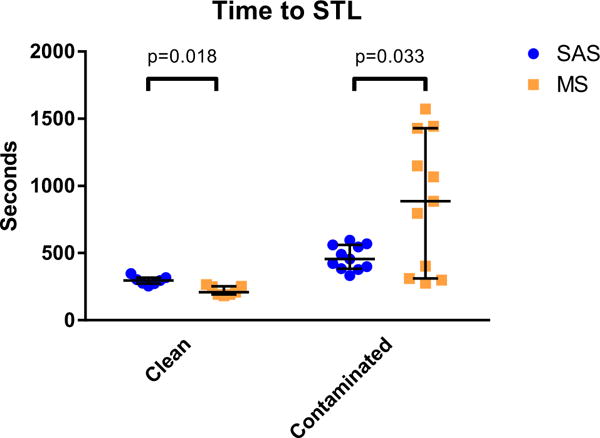
Time to STL. Shown are the times to STL, with median and interquartile ranges. Time to STL for clean datasets was faster with MS than SAS while contaminated datasets were faster with SAS. SAS semi-automated segmentation, MSmanual segmentation
STL geometric disagreement
Interobserver STL geometric disagreement was significantly lower using SAS than MS overall (0.70 ± 1.15 % vs. 1.31 ± 1.52 %, p = 0.030), and for the contaminated subset (0.81 ± 1.08 % vs. 1.75 ± 1.57 %, p = 0.036, Fig. 2). However, there was no significant difference between techniques for clean datasets (0.53 ± 1.40 % vs. 0.60 ± 1.36 %, p = 0.24, Fig. 2). Visual displays of differences demonstrate that most of the geometric disagreement occurred at areas where left heart contamination was removed (Fig. 3).
Fig. 2.
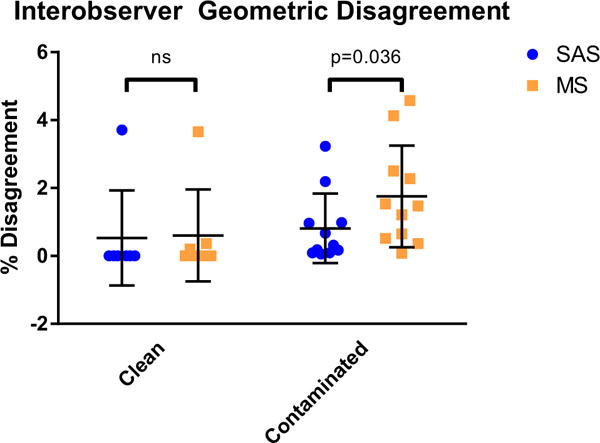
Interobserver geometric disagreement. Shown are the percentage disagreement (1-Dice similarity coefficient), with mean and standard deviations. Interobserver STL geometric disagreement was significantly lower using SAS than MS for the contaminated subset; however, there was no significant difference between techniques for clean datasets
Fig. 3.

A clean (a–d) and contaminated (e–h) dataset. a, e The anatomy with the SAS segmentation in blue and the MS segmentation in pink overlay. b–d, f–h A heat map of the thickness of any geometric disagreement (if present) for SAS and MS (b, f), interobserver SAS (c, g), and interobserver MS (d, h), scale 0–0.6 mm for b–d, 0–6 mm for f–h. Viewing angle was chosen to maximize visibility of geometric disagreement and is the same in a–d and e–h
Discussion
The time to right heart STL was largely dependent on whether the MRA dataset was clean or contaminated. For clean datasets, the time to right heart STL was under 5 min for almost all patients. Using MS with clean datasets required a single seed point and no further segmentation, leading to a very fast segmentation process. Using SAS with clean datasets still required numerous seed points, prolonging the segmentation process. In comparison, using MS with contaminated datasets required significantly more time given that much manual cropping was required. In SAS with contaminated datasets, the left heart structures were automatically segmented individually, thus allowing faster cropping and a significantly faster time to STL as compared to MS. Clinically, the differences in segmentation for clean datasets between MS and SAS are likely negligible, but for contaminated datasets, the time advantage of SAS is clear.
For clean datasets, we found no significant difference in the interobserver geometric disagreement between SAS and MS, likely because the amount of post-processing and cropping required was minimal. For contaminated datasets, which required more post-processing, we found that SAS has lower interobserver geometric disagreement. We believe that this is because SAS segmented left heart structures more consistently than MS, allowing easier cropping. SAS has less disagreement than MS, which is preferred.
Another method of speeding the segmentation process would be to obtain MRA datasets with less contamination. To that end, multiple techniques are being developed and optimized to improve the temporal resolution of MRA, such as the keyhole technique [16, 18], compressed sensing, and golden radial techniques [19], along with image subtraction [20]. Improvement in these techniques to improve image contrast will allow for cleaner datasets from which to begin segmentation. In particular, improved temporal resolution will result in shorter dynamic times with less chance of contamination between angiographic phases. These newer techniques will also enable the operator to decide between acquiring and reconstructing high temporal or high spatial resolution to get the most diagnostically helpful images. Although more research is required in the optimum angiography technique, this study has shown that cleaner datasets require less post-processing, and therefore will likely lead to faster and better segmentation. This then would lead to a quicker progression from CMR MRA datasets to 3DPMs.
The MS (Mimics) STL files were invariably larger in volume than the SAS (inPrint) datasets, despite using the same thresholds. In retrospect, we recognized that the major source of size discrepancy between Mimics and inPrint was due to differences in the accuracy setting during mesh creation between the two methods; this yielded MS STL files that were thicker than the SAS STLs (range of median thickness 0.19–0.47 mm, range of maximum thickness 0.64–12.39 mm). This likely overwhelmed any differences in the size discrepancies due to the segmentation process itself. This size discrepancy suggests that alongside a consistent thresholding and segmentation process, a consistent mesh generation setting must be used that allows the most reproducible results. Thus, much work yet needs to be done to assure that STL files, and therefore 3DPMs, accurately represent the underlying anatomy.
Taken as a whole, we believe that our results suggest that SAS is superior to MS for cardiac segmentation, especially for contaminated datasets. We believe that these results will be useful for the development of future segmentation tools. We purposely tested a segmentation task that we believe will be commonly used in the clinical setting in the near future. We also believe our findings will be applicable to whole heart segmentation, especially if specific structures are to be segmented individually.
Limitations
There are limitations to this study. Though settings were used to try and keep the segmentation as similar as possible between SAS and MS, different accuracy settings were used to generate the STL mesh. This, however, illustrates an important consideration for the segmentation process, namely that there are numerous possible methods that may not all yield the same results. Segmentation tools from a single software manufacturer were used, though this was an attempt to standardize segmentation processes as much as possible. A beta version of the SAS software was used, so the performance of the retail version may differ from that in our study. We specifically sought to create right heart datasets given the clinical scenario; results may be different depending on the goal of segmentation. Finally, we used contrast-enhanced MRA datasets to generate the segmentations; other source datasets may yield different results.
Conclusion
Semi-automated segmentation techniques can decrease the time needed to go from contaminated patient MRA datasets to 3D STLs, and make the process less variable and more reproducible; for clean datasets, both methods seem acceptable. Given that 3DPMs are promising tools for improving preprocedural planning and education, the development of more robust segmentation tools would be beneficial. Further study of different software packages and different SAS techniques are required to continue to push the field further.
Acknowledgments
AT, TH, and GFG were supported by the Early Career Research Award, Children’s Clinical Research Advisory Committee, Children’s Medical Center, Dallas.
Footnotes
Author contribution
AT and TH performed the segmentation. SZ, TH, and AT performed data analysis. All authors contributed to project design, drafting the article, and critical revision of the article. All authors approve of the content of the article.
Complaince with ethical standards
Conflicts of interest
All authors report no conflict of interest. AT, AKD, JMD, GFG, and TH all work at Children’s Medical Center, Dallas. We have full control of the primary data and agree to allow review of the data if requested.
References
- 1.Dankowski R, Baszko A, Sutherland M, Firek L, Kalmucki P, Wroblewska K, Szyszka A, Groothuis A, Siminiak T. 3D heart model printing for preparation of percutaneous structural interventions: description of the technology and case report. Kardiol Pol. 2014;6:546–551. doi: 10.5603/KP.2014.0119. [DOI] [PubMed] [Google Scholar]
- 2.Olivieri L, Krieger A, Chen MY, Kim P, Kanter JP. 3D heart model guides complex stent angioplasty of pulmonary venous baffle obstruction in a mustard repair of D-TGA. Int J Cardiol. 2014;2:e297–e298. doi: 10.1016/j.ijcard.2013.12.192. [DOI] [PubMed] [Google Scholar]
- 3.O’Neill B, Wang DD, Pantelic M, Song T, Guerrero M, Greenbaum A, O’Neill WW. Transcatheter caval valve implantation using multimodality imaging: roles of TEE, CT, and 3D printing. J Am Coll Cardiol Cardiovasc Imaging. 2015;2:221–225. doi: 10.1016/j.jcmg.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Mathur M, Patil P, Bove A. The role of 3D printing in structural heart disease: all that glitters is not gold. J Am Coll Cardiol Cardiovasc Imaging. 2015;8:987–988. doi: 10.1016/j.jcmg.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Babar JL, Jones RG, Hudsmith L, Steeds R, Guest P. Application of MR imaging in assessment and follow-up of congenital heart disease in adults. Radiographics. 2010;4:1145. doi: 10.1148/rg.e40. [DOI] [PubMed] [Google Scholar]
- 6.Fratz S, Chung T, Greil GF, Samyn MM, Taylor AM, Valsangiacomo Buechel ER, Yoo SJ, Powell AJ. Guidelines and protocols for cardiovascular magnetic resonance in children and adults with congenital heart disease: SCMR expert consensus group on congenital heart disease. J Cardiovasc Magn Reson. 2013;15(1):51. doi: 10.1186/1532-429X-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heathfield E, Hussain T, Qureshi S, Valverde I, Witter T, Douiri A, Bell A, Beerbaum P, Razavi R, Greil GF. Cardiovascular magnetic resonance imaging in congenital heart disease as an alternative to diagnostic invasive cardiac catheterization: a single center experience. Congenit Heart Dis. 2013;4:322–327. doi: 10.1111/chd.12032. [DOI] [PubMed] [Google Scholar]
- 8.Kilner PJ, Geva T, Kaemmerer H, Trindade PT, Schwitter J, Webb GD. Recommendations for cardiovascular magnetic resonance in adults with congenital heart disease from the respective working groups of the European Society of Cardiology. Eur Heart J. 2010;7:794–805. doi: 10.1093/eurheartj/ehp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ntsinjana HN, Hughes ML, Taylor AM. The role of cardiovascular magnetic resonance in pediatric congenital heart disease. J Cardiovasc Magn Reson. 2011;13(1):51. doi: 10.1186/1532-429X-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valsangiacomo Buechel ER, Grosse-Wortmann L, Fratz S, Eichhorn J, Sarikouch S, Greil GF, Beerbaum P, Bucciarelli-Ducci C, Bonello B, Sieverding L, Schwitter J, Helbing WA, Eacvi, Galderisi M, Miller O, Sicari R, Rosa J, Thaulow E, Edvardsen T, Brockmeier K, Qureshi S, Stein J. Indications for cardiovascular magnetic resonance in children with congenital and acquired heart disease: an expert consensus paper of the Imaging Working Group of the AEPC and the Cardiovascular Magnetic Resonance Section of the EACVI. Eur Heart J Cardiovasc Imaging. 2015;3:281–297. doi: 10.1093/ehjci/jeu129. [DOI] [PubMed] [Google Scholar]
- 11.Valverde I, Parish V, Hussain T, Rosenthal E, Beerbaum P, Krasemann T. Planning of catheter interventions for pulmonary artery stenosis: improved measurement agreement with magnetic resonance angiography using identical angulations. Catheter Cardiovasc Interv. 2011;3:400–408. doi: 10.1002/ccd.22695. [DOI] [PubMed] [Google Scholar]
- 12.Chung R, Taylor AM. Imaging for preintervention planning: transcatheter pulmonary valve therapy. Circ Cardiovasc Imaging. 2014;1:182–189. doi: 10.1161/CIRCIMAGING.113.000826. [DOI] [PubMed] [Google Scholar]
- 13.Goreczny S, Eicken A, Ewert P, Morgan GJ, Fratz S. A new strategy to identify potentially dangerous coronary arterial patterns before percutaneous pulmonary valve implantation. Postep Kardiol Interwencyjnej. 2014;4:294–297. doi: 10.5114/pwki.2014.46773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khambadkone S. Percutaneous pulmonary valve implantation. Ann Pediatr Cardiol. 2012;1:53–60. doi: 10.4103/0974-2069.93713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schievano S, Migliavacca F, Coats L, Khambadkone S, Carminati M, Wilson N, Deanfield JE, Bonhoeffer P, Taylor AM. Per-cutaneous pulmonary valve implantation based on rapid prototyping of right ventricular outflow tract and pulmonary trunk from MR data. Radiology. 2007;2:490–497. doi: 10.1148/radiol.2422051994. [DOI] [PubMed] [Google Scholar]
- 16.Hussain T, Greil GF, Bilska K, Miller OI. Toward defining pulmonary sequestration in infancy using two-dimensional echocardiography and novel high temporal resolution “keyhole” three-dimensional magnetic resonance angiography. Congenit Heart Dis. 2011;5:488–491. doi: 10.1111/j.174-0803.2011.00551.x. [DOI] [PubMed] [Google Scholar]
- 17.Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, Haker SJ, Wells WM, 3rd, Jolesz FA, Kikinis R. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;2:178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willinek WA, Hadizadeh DR, von Falkenhausen M, Urbach H, Hoogeveen R, Schild HH, Gieseke J. 4D time-resolved MR angiography with keyhole (4D-TRAK): more than 60 times accelerated MRA using a combination of CENTRA, keyhole, and SENSE at 3.0T. J Magn Reson Imaging. 2008;6:1455–1460. doi: 10.1002/jmri.21354. [DOI] [PubMed] [Google Scholar]
- 19.Prieto C, Uribe S, Razavi R, Atkinson D, Schaeffter T. 3D undersampled golden-radial phase encoding for DCE-MRA using inherently regularized iterative SENSE. Magn Reson Med. 2010;2:514–526. doi: 10.1002/mrm.22446. [DOI] [PubMed] [Google Scholar]
- 20.Rapacchi S, Han F, Natsuaki Y, Kroeker R, Plotnik A, Lehrman E, Sayre J, Laub G, Finn JP, Hu P. High spatial and temporal resolution dynamic contrast-enhanced magnetic resonance angiography using compressed sensing with magnitude image subtraction. Magn Reson Med. 2014;5:1771–1783. doi: 10.1002/mrm.24842. [DOI] [PMC free article] [PubMed] [Google Scholar]


