Abstract
The mammalian abasic endonuclease, APE1, has two distinct roles in the repair of oxidative DNA damage and in gene regulation. Here we show that both functions are essential for cell survival. Deletion of the APE1 gene causes embryonic lethality in mice, and no nullizygous embryo fibroblasts have been isolated. We have now established nullizygous embryo fibroblast lines from APE1–/– mouse embryos that are transgenic with the “floxed” human APE1 (hAPE1) gene. Removal of hAPE1 by Cre expression through nuclear microinjection elicited apoptosis in these cells within 24 h, which was blocked by coinjection of the wild-type hAPE1 gene. In contrast, mutant hAPE1 alleles, lacking either the DNA repair or acetylation-mediated gene regulatory function, could not prevent apoptosis, although the combination of these two mutants complemented APE deficiency induced by Cre. These results indicate that distinct and separable functions of APE1 are both essential for mammalian cells even in vitro and provide the evidence that mammalian cells, unlike yeast or Escherichia coli, absolutely require APE for survival, presumably to protect against spontaneous oxidative DNA damage.
Keywords: conditional gene inactivation, DNA repair, endogenous DNA damage, base excision repair
Abasic endonuclease (APE), a ubiquitous enzyme, plays a central role in repairing toxic and mutagenic abasic (AP) sites generated in genomes during the repair of oxidation and alkylation damage through the base excision repair (BER) pathway (1). Oxidative DNA lesions, including AP sites, are also spontaneously generated at an estimated rate of 1.5 × 105 residues·cell–1·day–1 (2). Unlike two distinct APEs present in Escherichia coli and Saccharomyces cerevisiae, only one active APE, APE1, an ortholog of E. coli xth and yeast APN2, has been identified in mammalian cells (3). Based on sequence homology, a second APE-like gene, APE2, was cloned from mammalian cells. However, we could not detect APE activity in the recombinant human APE2 (4), and hAPE2, unlike hAPE1, could not complement yeast APE mutants (5). Although APE-negative bacteria and yeast are viable, very early death (3.5–7.5 days after fertilization) was observed in APE1 nullizygous mouse embryos (6–8). Unlike other BER proteins, e.g., DNA polymerase β and X-ray cross complementation group 1, which are essential for embryonic survival but not for mouse embryonic fibroblasts (MEFs) cultured in vitro (9, 10), APE1-null MEF mutant lines have not been established. The mammalian APE1, independently identified as redox-enhancing factor 1 (Ref1), has a distinct regulatory function in reductively activating C-Jun, p53, and other transcription factors (3, 11) for which Cys-65 (Cys-64 in mouse APE1) was identified as the active site (12). The N-terminal region of the 36-kDa polypeptide, including Cys-65, is not conserved in the E. coli homolog exonuclease III. An additional regulatory function of APE1/Ref1 was identified in Ca2+-dependent down-regulation of the parathyroid hormone and renin genes containing negative Ca2+-response elements (nCaREs) (13, 14). These elements may also be present in many other genes (15). APE1 was identified as a component of the nCaRE-bound protein complexes. We have recently reported that acetylation of APE1 at Lys-6 or Lys-7 by the histone acetyltransferase p300 promotes its binding to nCaREs (16). Whether any one or all of these functions of APE1 are essential for embryos or somatic cells has not been explored. A recent report showing viability of C64S APE1 knock-in mouse mutant indicates that either the redox function of APE1 is not essential or it does not involve Cys 64 (17). It has also been suggested that the essentiality of APE1 is due to its regulatory roles unrelated to its DNA repair function. In any case, it has been difficult to examine individually the biological significance of the three different functions, namely DNA repair, reductive activation, and acetylation-mediated gene regulation. This report describes our success in establishing viable, APE1 nullizygous MEF cells that express floxed hAPE1 transgene. Using these MEF cell lines, we have shown that survival requires both DNA repair and acetylation-mediated gene regulation functions of APE1.
Materials and Methods
Generation of Transgenic Mice. A 6-kb DNA fragment containing the hAPE1 gene (HindIII-XbaI) was subcloned into the pBlue-script plasmid (Stratagene) with loxP elements at both ends. Transgenic (tg) mice carrying the hAPE1 gene were generated in the University of Texas Medical Branch's Transgenic Mouse Core Facility. The heterozygous APE1 mice generated by Ludwig et al. (7) were used to produce +/–tg mice that were subsequently mated to generate –/–tg embryos. The mice (using tail snips) and embryos were genotyped by Southern analysis and PCR with APE1 specific primers (see Fig. 2). Fibroblast cultures were established from 9.5 days after fertilization (E9.5) embryos after dissection and grown in DMEM supplemented with 10% FBS and streptomycin/penicillin. The primary cells were transformed with a SV40 T-antigen expression plasmid to establish MEF lines.
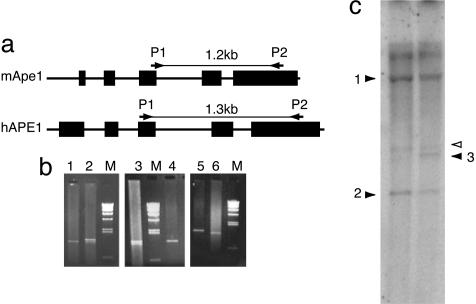
Fig. 2.
PCR and Southern blot analysis of MEF DNA. (a) PCR primers used for genotyping. Mouse and human APE1 genes are shown with exons (boxes). The primers anneal to both genes, but PCR generates different sized DNA fragments. (b) Representative results of PCR genotyping. Genomic DNA from mouse tails (lanes 1 and 6, normal; lanes 2 and 3, transgenic) or from the –/–tg MEF (lanes 4 and 5) were amplified by using the primers in a. (c) Southern blot analysis by using hAPE1 cDNA as a probe. Genomic DNA from –/–tg (lane 1) or +/–tg (lane 2) MEF were cut with BamHI before Southern hybridization. Filled arrowheads: 1, specific to the hAPE1 transgene; 2, specific to the targeted mAPE1 gene (neoresistance gene insert); 3, specific to normal mAPE1 gene. Open arrow indicates that the bands unaffected by the gene rearrangements. One or two copies of the transgene were integrated in this transgenic mouse and MEF strain, based on band intensity in the Southern analysis.
DNA Microinjection. The –/–tg MEF cells were plated with DMEM/F12 without phenol red at 24 h before microinjection. The plates were etched to identify injected cells. Injection needles were pulled from borosilicate capillaries by using a Flaming/Brown Micropipette Puller, model P-97 (Sutter Instruments, Novato, CA) with outer tip diameters of 2.5–3 μm, as determined by scanning electron microscopy (18). Immediately after dialysis of DNA in an injection buffer (114 mM KCl/0.5 mM KPO4, pH 7.4) for 10 min, the cells were injected with 5 fl of sample containing DNA (≈20 copies of each DNA per injection), together with Oregon Green dextran (0.4 mg/ml), as a marker of the injected cells by using the electronically interfaced Eppendorf Micromanipulator (model 5171) and Transjector (model 5246) (18). The injection was monitored under a phase-contrast Olympus (Melville, NY) IX70 inverted microscope equipped with a temperature-controlled stage maintained at 37°C. Only cells within an etched boundary were injected. To avoid any bias, microinjection was carried out by the University of Texas Medical Branch's Microinjection Core Facility personnel without prior knowledge about biological functions of APE1 or its mutants and without direct interest in this study.
Transient Transfection of DNA. The cre gene was cloned from P1 phage DNA through PCR into a phrGFP vector (Stratagene) to place the cre gene upstream of humanized recombinant GFP (Stratagene). The hAPE1 cDNA was then inserted into this vector (see Fig. 4). Thirty-six hours after transient transfection with Lipofectamine 2000 (Invitrogen), the cells were fixed with methanol, stained with primary antibodies specific for Cre (Novagen), APE1 (19), and activated caspase 3 (Cell Signaling Technology, Beverly, MA), followed by secondary antibodies conjugated to rhodamine (Chemicon), and finally analyzed by confocal microscopy (L510Meta, Zeiss) in University of Texas Medical Branch's Optical Image Laboratory. The MEFs were separately transfected with a vector carrying the Cre-EGFP fusion gene (20) (provided by B. Sauer, University of Kansas Medical Center), stained with propidium iodide without fixation, and then analyzed as above.
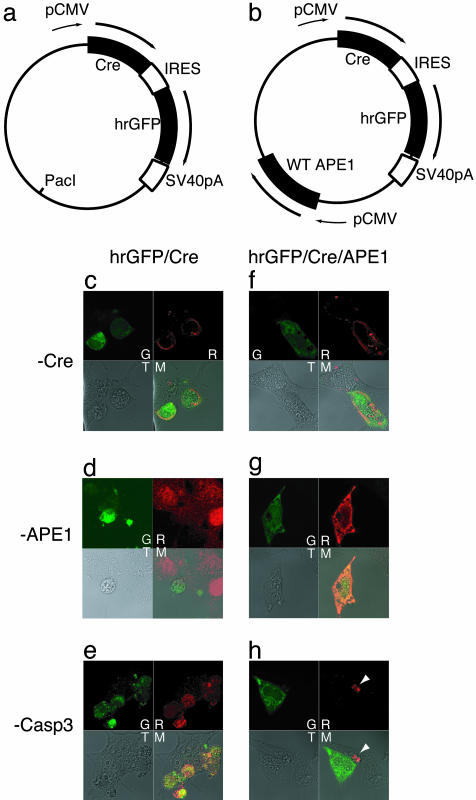
Fig. 4.
Caspase 3 activation after APE1 removal by Cre expression. (a and b) Bicistronic vectors for simultaneous expression of Cre and humanized recombinant GFP (hrGFP) proteins from a single pCMV promoter. (a) The cre gene was inserted into a phrGFP vector (Stratagene). An internal loxP site in the vector was replaced with a PacI site, which was then used to insert the WT hAPE1 cDNA with pCMV in b. Plasmids expressing Cre and hrGFP proteins (c–e), or expressing WT APE1 in addition to Cre and hrGFP (f–h) were used for transient transfection. After 36 h, the cells were fixed and stained with anti-Cre, anti-APE1, and anti-activated caspase 3 antibodies, followed by rhodamine-conjugated secondary rabbit antibody. G, GFP; R, rhodamine; T, transmission; M, merge. The white arrow in h denotes a dead cell that was untransfected (GFP negative) but caspase 3-positive.
Results
Isolation of MEF Cells Lacking the Mouse APE1 Gene. In an effort to establish conditional APE1 mouse mutants, we generated tg mice expressing hAPE1 from a 6-kb genomic clone including the promoter, (19) bracketed by loxP elements (Fig. 1a), which allows its Cre recombinase-catalyzed excision from the chromosomal integration site (20). After confirming stable expression of hAPE1 (Fig. 1a), we mated APE1 heterozygous tg mice with APE1 heterozygous knockout mice (+/–) (7) to generate heterozygous tg progeny (+/–; tg). Subsequent crosses between these mutant mice were carried out to generate nullizygous transgenic progeny that we expected to be viable because of hAPE1 expression. Surprisingly, none of the 109 progeny mice generated from two independent tg strains had the homozygous null genotype (Fig. 1b). We therefore concluded that the ectopic hAPE1 could not complement the deficiency of endogenous APE1, probably because of improper regulation of the transgene essential for embryo development. In utero examination also showed no embryos of –/–tg genotype at E13–14. However, at E12 we identified resorption bodies of the –/–tg genotype and succeeded in harvesting live, normal size embryos of this genotype at E9.5. These results showed that the transgenic APE1 could extend the life of APE-negative embryos by several days but not through the full term. We were able to culture MEF from E9.5 embryos and confirmed their combined transgenic/nullizygous genotype (Fig. 2). These MEFs grew normally in primary cultures from which we established immortalized cell lines by transformation with SV40 T-antigen.
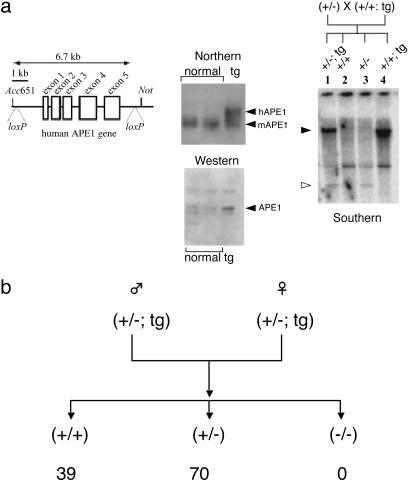
Fig. 1.
Generation of conditional Ape1 null mouse. (a) Construction of transgenic mice expressing hAPE1. (Left) A 6-kb human APE1 genomic DNA with intrinsic HindIII (upstream) and XbaI (downstream) sites was cloned into pBluescript SK(–) (Stratagene) and loxP elements were inserted at both ends. (Center) Expression of APE1 was confirmed with Northern and Western blotting, showing 2- to 3-fold higher expression in transgenic mouse livers. (Right) A typical Southern blot for genotyping transgenic mice after BamHI digestion and probing with hAPE1 cDNA. Filled arrowhead, transgene-specific band; open arrowhead, specific to the neointegrated mAPE1 gene. (b) Genotyping of progeny mice after intercrossing (+/–; tg). Numbers at the bottom indicate occurrence of the corresponding genotypes. With the total number of mice tested (109), the probability of finding no –/–tg mouse based on Mendelian law is <1 × 10–12.
Removal of the hAPE1 Gene from MEFs by Microinjection of the cre Gene. We tested the effect of deleting the hAPE1 transgene by microinjecting a Cre expression plasmid into the nuclei of these MEFs. Coinjection of Oregon Green marker with the plasmid allowed monitoring of the injected cells (Fig. 3 a and b). A majority of the cells underwent morphological changes typical of apoptosis, including blebbing and rounding, within 24 h of microinjection, a process initiated as early as 12 h (Fig. 3 a and c–e). After 48 h, the injected cells were found to have died without undergoing cell division. To test whether the death was the direct result of APE1 depletion, we coinjected an expression plasmid for the WT hAPE1 along with the Cre plasmid. The WT APE1 prevented abnormal cell morphology and death, and the microinjected cells divided normally (Fig. 3b). Furthermore, we observed normal cell divisions when the cre gene was injected into cytoplasm instead of nuclei (Fig. 3e).
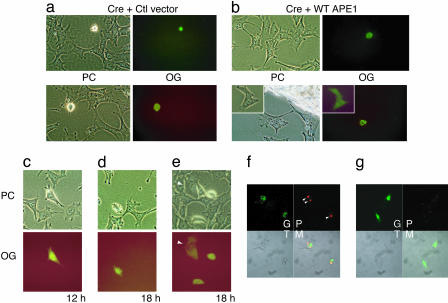
Fig. 3.
Effect of APE1 deletion on cell survival. (a–e) Cre expression vector was coinjected into nuclei with Oregon Green and a vector control (a and c–e) or a WT APE1 expression vector (b). Two representative results are shown in a and b, with an inset for divided cells (b). PC, phase contrast; OG, Oregon Green. The cells were photographed at 24 h after the injection (a and b) or at indicated times (c–e). The arrow in e indicates cells in which the cre plasmid was injected into the cytoplasm, which appeared normal and divided after 24 h. (f and g) Cell death induced by expression of Cre-EGFP fusion protein. The –/–tg MEF (f) or BALB/c 3T3 control cells (g) were transfected with the Cre-EGFP vector. Cells were stained with propidium iodide without fixation, and then fluorescence of EGFP (G) and propidium iodide (P) was visualized with confocal microscopy. Arrowheads indicate sites of chromatin condensation. T, transmission; M, merged.
Caspase 3 Activation in the MEF After Deletion of the hAPE1 Transgene. In an independent set of experiments, the MEF cells were transiently transfected with an expression plasmid for the Cre-EGFP fusion protein (20) and stained with propidium iodide without fixation to monitor dead or dying cells. Propidium iodide fluorescence was observed in EGFP-expressing cells as early as 18 h after transfection (Fig. 3f); these cells had a rounded appearance with chromatin condensation, characteristic of apoptosis, whereas a control BALB/c mouse line did not show any abnormality due to the Cre-EGFP expression (Fig. 3g). Similar results were obtained for spontaneously transformed cells in the absence of T antigen (data not shown), suggesting that apoptosis induced by APE1 inactivation is independent of p53 status (21). We also transiently transfected the MEFs with a bicistronic expression plasmid (Fig. 4 a and b) for expression of Cre and GFP (Fig. 4). The GFP-positive cells showed altered morphology 24–48 h after transfection, associated with Cre expression, loss of APE1, and appearance of activated caspase 3 (Fig. 4 c–e). In contrast, the cells appeared normal when WT APE1 was coexpressed with Cre and GFP (Fig. 4 f–h). We conclude from these results that APE1 is essential for viability of the MEF and that these cells underwent apoptosis within 24 h after APE1 inactivation. These results also predict a high turnover of APE1, consistent with our earlier studies (22).
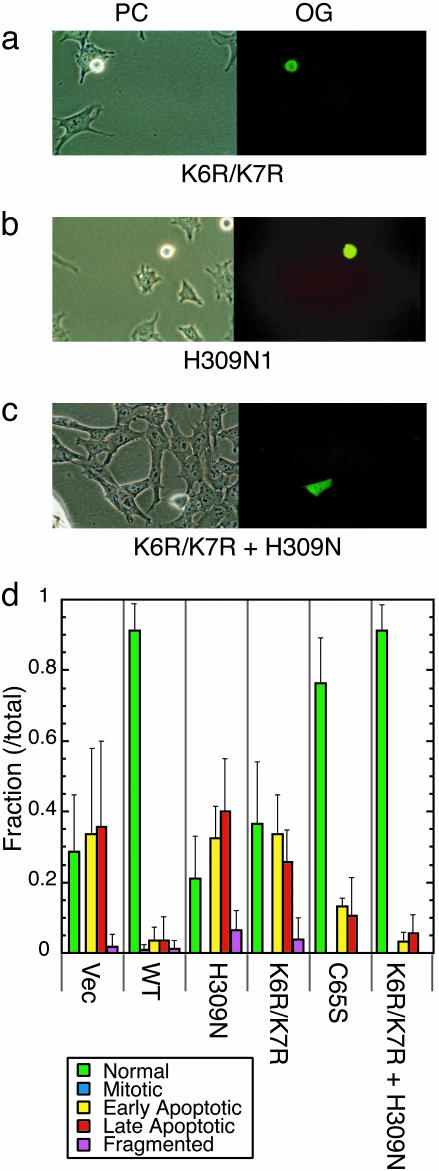
Microinjection of APE1 Alleles Lacking One of Its Functions. The microinjection procedure allows control of the number of DNA molecules introduced into cell nuclei. We carried out complementation experiments by microinjecting APE1 cDNA at a constant level with the Cre plasmid (≈20 copies per cell). We chose three hAPE1 mutants, each of which specifically lacks one of APE1's functions: (i) the C65S mutant previously reported to be deficient in redox gene regulation (12); (ii) the K6R/K7R mutant lacking two acetyl-acceptor lysines required for nCaRE-mediated gene regulation (16); and (iii) the H309N mutant lacking AP-endonuclease activity (23).
Ectopic Expression of the C65S mutant was as effective as the WT APE1 in preventing Cre-induced apoptosis (Fig. 5d). This observation, showing the nonessentiality of Cys-65, is consistent with the recent mAPE1 knock-in study (17). On the other hand, both K6R/K7R and H309N mutants failed to complement APE1 deficiency; the microinjected cells showed apoptotic morphological change and cell death at a similar time scale as with expression of Cre and the vector alone (Fig. 5 a, b, and d).
Fig. 5.
Mutational analysis of the protective effect of APE1. Survival analysis of MEF after microinjection with K6R/K7R (a), H309N (b), or K6R/K7R + H309N (c) plasmids and the Cre plasmid and Oregon Green. (d) Bar graph of cells with various morphologies related to apoptotic response, as described in the figure, which were visually assessed at 24 h after microinjection (when Oregon Green was visible even after mitosis). The experiments were carried out three to nine times (>60 total cell numbers for each APE1 cDNA). Error bars denote standard deviations.
The interpretation of the above results would be compromised if the H309N mutation had affected the acetylation mediatedgene regulation function, or the K6R/K7R double mutation changed APE1's DNA repair activity. Although structural analyses could not resolve the N-terminal 40 amino acids found on the APE surface (24), it appears unlikely that the N-terminal Lys residues directly interact with H309, which is buried inside of the protein. Also, the H309N mutation did not affect APE1's repressor function (data not shown) in a reporter gene assay described in ref. 16. To further ensure whether the repair and regulatory functions are separable in the APE1 polypeptide, a mixture of K6R/K7R and H309N APE1 mutant plasmids was injected into the cell nuclei along with the cre plasmid (Fig. 5 c and d). We observed that the APE1 mutants together completely blocked Cre-mediated cell death. We therefore conclude that the two functions of APE1 are independent and that both are essential for cell viability.
Discussion
Although three groups demonstrated independently that mouse embryos lacking APE1 die at a very early stage (6–8), it was not clear whether APE1 is also essential for the survival of cultured cells, because such cell lines were not isolated in the past. This report documents the successful isolation of MEFs lacking the mouse APE1 completely, albeit expressing the human APE1 gene. The fact that these cells underwent apoptosis soon after removal of the hAPE1 transgene indicates that APE1 is absolutely required for cellular survival. Importantly, this result also implies that the deletion of the APE1 gene, and not the disruption of expression of an adjacent gene, O-sialoglycoprotein endopeptidase, which shares its promoter with APE1 (25), was responsible for the embryonic lethality of APE1 nullizygous mice.
Using the microinjection assay we devised a “complementation test” for individually examining the essentiality of APE1's functions. It was perhaps not surprising that the C65S APE1 mutant allele behaved like the WT APE1 in preventing death because the C64A knock-in mutant mice showed normal viability (17). Although these results question authenticity of C65 as the active site residue for Ref1 function, the C65S APE1 mutant was previously shown to have an effect on APE1's Ref1 function by several groups (12, 26, 27). It may be worthwhile to test whether the C64A APE1 mutant mice show a delayed phenotype.
Acetylation of APE1 at the N-terminal K6/K7 residues is a posttranslational modification necessary for APE1 to repress genes containing nCaREs (16). This direct regulatory function of APE1 has not received as much attention as its role in redox regulation. However, its importance was clearly shown in regulation of both parathyroid hormone and renin genes (13, 14). Our results suggest that APE1 may be indispensable for controlling expression of many other genes too, because the nCaRE elements exist upstream of many other genes (15). If this assumption is indeed correct, APE1 deletion would cause changes in expression of numerous genes, which can be examined by using genomic and proteomic approaches with the MEF cells after deletion of the APE1 gene.
It is particularly intriguing that the endonuclease activity of mammalian APE1 is essential for survival, because E. coli and S. cerevisiae mutants lacking APEs are viable (28, 29). Although APE1 appears to be the only APE in mammalian cells, they express a significant level of polynucleotide kinase (PNK) with intrinsic DNA 3′ phosphatase activity that removes DNA 3′ phosphate termini (30). We have recently shown that endonuclease VIII-like (NEIL)1 and NEIL2, belonging to a distinct family of DNA glycosylases, which generate DNA 3′ phosphate termini after base excision and also at AP sites, are involved in an APE-independent, PNK-dependent pathway for repair of oxidized bases (4). Thus, a potential backup mechanism for AP site repair exists in mammalian cells. We suggest several possibilities to explain the unexpected essentiality of the repair function of APE1 in somatic cells. First, it is possible that the amount of spontaneous oxidative DNA damage (including AP sites) in mammalian cells is so high that it overwhelms the NEIL/PNK-dependent alternative repair system in the absence of APE1. Second, the repair of DNA strand-break termini with 3′-phosphoglycolate and oxidized AP sites could not use the alternative pathway and would require APE1 for their processing (31, 32). In the case of E. coli or S. cerevisiae, repair of oxidative DNA damage may be carried out through the nucleotide excision repair (NER) pathway in the absence of APEs, because the combined deficiency of APE and NER in these organisms is lethal (28, 29). Although it is not clear whether mammalian cells could similarly use the NER pathway, we raise an intriguing third possibility that the loss of mitochondria-specific APE, which is generated from APE1 after N-terminal truncation (R. Chattopadhyay, L. Wiederhold, B. Szczesny, T.I., T. K. Hazara, and S.M., unpublished data), and not of nuclear APE1, is responsible for triggering apoptosis. Mitochondrial DNA is more susceptible to reactive oxygen species than the nuclear genome, and mitochondria lack the NER pathway (33, 34). Furthermore, the NEILs may be absent in mitochondria (35).
The APE1 conditional mutant cells provide a system for better estimation of the rate of formation of spontaneous DNA damage, including AP sites, in both nuclear and mitochondrial genomes, for elucidating the mechanism of apoptosis induced by spontaneous DNA damage. The cells also provide an assay for structure-function analysis of mutant APEs to elucidate its regulatory mechanism.
Acknowledgments
We thank Mr. E. F. Willmore, Jr., for animal husbandry and excellent technical assistance; Dr. B. Sauer for the Cre-EGFP fusion vector, Dr. C. Yallampalli for guidance in embryo isolation; Dr. J. Ceci for generating the APE1 transgenic mice; Drs. D. Konkel, M. Weinfeld, and S. Adhya for critical reading of the manuscript; and Ms. W. Smith for secretarial help. This research was supported by U.S. Public Health Service Grants R01 ES08457, R01 CA53791, P01 06676, and R01 CA98664.
Author contributions: T.I. and S.M. designed research; T.I., D.B.B., C.V.N., K.K.B., and H.S. performed research; T.I., D.B.B., K.K.B., M.A.M., and D.J.C. contributed new reagents/analytic tools; T.I. and S.M. analyzed data; and T.I. and S.M. wrote the paper.
Abbreviations: AP, abasic; APE1, AP-endonuclease 1; En, n days after fertilization; MEF, mouse embryonic fibroblast; nCaRE, negative calcium-response element; Ref1, redoxenhancing factor 1; tg, transgenic.
References
- 1.Demple, B. & Harrison, L. (1994) Annu. Rev. Biochem. 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 2.Beckman, K. B. & Ames, B. N. (1997) J. Biol. Chem. 272, 19633–19636. [DOI] [PubMed] [Google Scholar]
- 3.Evans, A. R., Limp-Foster, M. & Kelley, M. R. (2000) Mutat. Res. 461, 83–108. [DOI] [PubMed] [Google Scholar]
- 4.Wiederhold, L., Leppard, J. B., Kedar, P., Karimi-Busheri, F., Rasouli-Nia, A., Weinfeld, M., Tomkinson, A. E., Izumi, T., Prasad, R., Wilson, S. H., et al. (2004) Mol. Cell 15, 209–220. [DOI] [PubMed] [Google Scholar]
- 5.Ribar, B., Izumi, T. & Mitra, S. (2004) Nucleic Acids Res. 32, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xanthoudakis, S., Smeyne, R. J., Wallace, J. D. & Curran, T. (1996) Proc. Natl. Acad. Sci. USA 93, 8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig, D. L., MacInnes, M. A., Takiguchi, Y., Purtymun, P. E., Henrie, M., Flannery, M., Meneses, J., Pedersen, R. A. & Chen, D. J. (1998) Mutat. Res. 409, 17–29. [DOI] [PubMed] [Google Scholar]
- 8.Meira, L., Devaraj, S., Kisby, G., Burns, D., Daniel, R., Hammer, R., Grundy, S., Jialal, I. & Friedberg, E. (2001) Cancer Res. 61, 5552–5557. [PubMed] [Google Scholar]
- 9.Thompson, L. H. & West, M. G. (2000) Mutat. Res. 459, 1–18. [DOI] [PubMed] [Google Scholar]
- 10.Sobol, R. W., Horton, J. K., Kuhn, R., Gu, H., Singhal, R. K., Prasad, R., Rajewsky, K. & Wilson, S. H. (1996) Nature 379, 183–186. [DOI] [PubMed] [Google Scholar]
- 11.Xanthoudakis, S., Miao, G., Wang, F., Pan, Y. C. & Curran, T. (1992) EMBO J. 11, 3323–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker, L. J., Robson, C. N., Black, E., Gillespie, D. & Hickson, I. D. (1993) Mol. Cell. Biol. 13, 5370–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki, T., Chung, U., Nishishita, T., Ebisu, S., Usuda, S., Mishiro, S., Xanthoudakis, S., Igarashi, T. & Ogata, E. (1994) J. Biol. Chem. 269, 27855–27862. [PubMed] [Google Scholar]
- 14.Fuchs, S., Philippe, J., Corvol, P. & Pinet, F. (2003) J. Hypertens. 21, 327–335. [DOI] [PubMed] [Google Scholar]
- 15.McHaffie, G. S. & Ralston, S. H. (1995) Bone (NY) 17, 11–14. [DOI] [PubMed] [Google Scholar]
- 16.Bhakat, K. K., Izumi, T., Yang, S. H., Hazra, T. K. & Mitra, S. (2003) EMBO J. 22, 6299–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ordway, J. M., Eberhart, D. & Curran, T. (2003) Mol. Cell. Biol. 23, 4257–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, D., Yallampalli, U., Owlia, A. & Singh, P. (2003) Endocrinology 144, 201–211. [DOI] [PubMed] [Google Scholar]
- 19.Izumi, T., Henner, W. D. & Mitra, S. (1996) Biochemistry 35, 14679–14683. [DOI] [PubMed] [Google Scholar]
- 20.Le, Y., Miller, J. L. & Sauer, B. (1999) Anal. Biochem. 270, 334–336. [DOI] [PubMed] [Google Scholar]
- 21.Vayssiere, J. L., Petit, P. X., Risler, Y. & Mignotte, B. (1994) Proc. Natl. Acad. Sci. USA 91, 11752–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramana, C. V., Boldogh, I., Izumi, T. & Mitra, S. (1998) Proc. Natl. Acad. Sci. USA 95, 5061–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barzilay, G., Mol, C. D., Robson, C. N., Walker, L. J., Cunningham, R. P., Tainer, J. A. & Hickson, I. D. (1995) Nat. Struct. Biol. 2, 561–568. [DOI] [PubMed] [Google Scholar]
- 24.Mol, C. D., Izumi, T., Mitra, S. & Tainer, J. (2000) Nature 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 25.Seki, Y., Ikeda, S., Kiyohara, H., Ayabe, H., Seki, T. & Matsui, H. (2002) Gene 285, 101–108. [DOI] [PubMed] [Google Scholar]
- 26.Fan, Z., Beresford, P. J., Zhang, D., Xu, Z., Novina, C. D., Yoshida, A., Pommier, Y. & Lieberman, J. (2003) Nat. Immunol. 4, 145–153. [DOI] [PubMed] [Google Scholar]
- 27.Vasko, M. R., Guo, C. & Kelley, M. R. (2005) DNA Repair (Amsterdam) 4, 367–379. [DOI] [PubMed] [Google Scholar]
- 28.Guillet, M. & Boiteux, S. (2003) Mol. Cell. Biol. 23, 8386–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saporito, S. M., Gedenk, M. & Cunningham, R. P. (1989) J. Bacteriol. 171, 2542–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehouse, C. J., Taylor, R. M., Thistlethwaite, A., Zhang, H., Karimi-Busheri, F., Lasko, D. D., Weinfeld, M. & Caldecott, K. W. (2001) Cell 104, 107–117. [DOI] [PubMed] [Google Scholar]
- 31.Izumi, T., Hazra, T. K., Boldogh, I., Tomkinson, A. E., Park, M. S., Ikeda, S. & Mitra, S. (2000) Carcinogenesis 21, 1329–1334. [PubMed] [Google Scholar]
- 32.Klungland, A. & Lindahl, T. (1997) EMBO J. 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohr, V. A., Stevnsner, T. & de Souza-Pinto, N. C. (2002) Gene 286, 127–134. [DOI] [PubMed] [Google Scholar]
- 34.Dobson, A. W., Kelley, M. R., Wilson, G. L. & LeDoux, S. P. (2002) Methods Mol. Biol. 197, 351–362. [DOI] [PubMed] [Google Scholar]
- 35.Hazra, T. K., Kow, Y. W., Hatahet, Z., Imhoff, B., Boldogh, I., Mokkapati, S. K., Mitra, S. & Izumi, T. (2002) J. Biol. Chem. 277, 30417–30420. [DOI] [PubMed] [Google Scholar]