Abstract
Background
Manipulating maternal nutrition during specific periods of gestation can result in re-programming of fetal and post-natal development. In this experiment we investigated how a feed restriction of 85% compared with 140% of total metabolizable energy requirements, fed to cows during mid-to-late gestation, influences phenotypic development of fetuses and mRNA expression of growth (Insulin-Like Growth Factor family and Insulin Receptor (INSR)), myogenic (Myogenic Differentiation 1 (MYOD1), Myogenin (MYOG), Myocyte Enhancer Factor 2A (MEF2A), Serum Response Factor (SRF)) and adipogenic (Peroxisome Proliferator Activated Receptor Gamma (PPARG)) genes in fetal longissimus dorsi (LD) and semitendinosus (ST) muscle. DNA methylation of imprinted genes, Insulin Like Growth Factor 2 (IGF2) and Insulin Like Growth Factor 2 Receptor (IGF2R), and micro RNA (miRNA) expression, were also examined as potential consequences of poor maternal nutrition, but also potential regulators of altered gene expression patterns.
Results
While the nutrient restriction impacted dam body weight, no differences were observed in phenotypic fetal measurements (weight, crown-rump length, or thorax circumference). Interestingly, LD and ST muscles responded differently to the differential pre-natal nutrient levels. While LD muscle of restricted fetal calves had greater mRNA abundances for Insulin Like Growth Factor 1 and its receptor (IGF1 and IGF1R), IGF2R, INSR, MYOD1, MYOG, and PPARG, no significant differences were observed for gene expression in ST muscle. Similarly, feed restriction had a greater impact on the methylation level of IGF2 Differentially Methylated Region 2 (DMR2) in LD muscle as compared to ST muscle between treatment groups. A negative correlation existed between IGF2 mRNA expression and IGF2 DMR2 methylation level in both LD and ST muscles. Differential expression of miRNAs 1 and 133a were also detected in LD muscle.
Conclusions
Our data suggests that a nutrient restriction of 85% as compared to 140% of total metabolizable energy requirements during the 2nd half of gestation can alter the expression of growth, myogenic and adipogenic genes in fetal muscle without apparent differences in fetal phenotype. It also appears that the impact of feed restriction varies between muscles suggesting a priority for nutrient partitioning depending on muscle function and/or fiber composition. Differences in the methylation level in IGF2, a well-known imprinted gene, as well as differences in miRNA expression, may be functional mechanisms that precede the differences in gene expression observed, and could lead to trans-generational epigenetic programming.
Keywords: Beef cattle, fetal programming, nutrient restriction, liver, muscle, IGF2, methylation, microRNA
Background
Maternal nutritional stimuli or insult during gestation can lead to genetic and phenotypic changes on the developing fetus that may also affect post-natal development of the calf [1]. Fetal developmental programming can have implications for the beef industry, as the impacts of these stimuli may influence fertility, health and immunity, growth potential, carcass yield, and overall muscle mass and composition, all of which impacts profitability [2].
Previous work in cattle and other ruminants have often focused on nutritional insults in early-to-mid gestation [3–5]. However, during mid-to-late gestation the fetus undergoes rapid growth, representing a more significant metabolic toll on the dam [6]. Vital organs such as brain and heart, and to some extent liver, remain relatively immune to the effects of prenatal maternal nutrition during the 1st trimester of fetal development in the bovine [7, 8], and appear to have priority over skeletal muscle for nutrient partitioning. Therefore, since growth predominates during the latter half of gestation, and is of a lower priority for nutrient partitioning in the fetus, sub-optimal maternal nutrition at this stage could be detrimental for fetal growth and muscle development [2, 9]. Previous research has shown that heifers born from cows receiving supplementation in the last trimester of gestation were heavier at weaning, pre-breeding and at pregnancy confirmation compared to unsupplemented cows [10], and that steers born from dams managed on a higher plane of nutrition (improved pasture) as compared to a low plane of nutrition (native range), had improved meat tenderness characteristics, animal growth, and carcass composition [11]. Therefore proper late gestation nutrition can support fetal programming events towards improved growth and meat quality traits.
Fetal muscle development has been shown to be sensitive to nutrient abundance. The number, type and size of muscle fibres are the main determinant of muscle mass and total fibre number is positively correlated to growth potential [12, 13]. Interestingly, while muscle fibre hypertrophy occurs mostly after birth, muscle fibre number and type is determined in utero ([14], and as reviewed by [7]). In cattle fetuses, by day 240 of gestation, total muscle fibre number has been fixed consequently predetermining the lifetime potential of an animal [13]. Moreover, other meat characteristics such as marbling can be impacted prenatally [9]. Changes in offspring total muscle fibre number and/or in muscle fibre type have been observed as a consequence of maternal under- and over-nutrition in ewes, sows and rats [5, 15–17]. Therefore nutritional restriction has the potential to hamper the performance and muscle development of its offspring
Muscle development in utero is a complex process involving proliferation and differentiation of the myogenic precursor cells [12]. IGF2 is a potent mitogen and inducer of cell differentiation known to impact fetal growth and muscle development [13]. It is known to be transcribed from at least 4 different promoters, generating several transcript variants, each possessing their own spatio-temporal expression signature during development and growth [18]. It is also a well-known imprinted gene in several species and interestingly, its methylation status has been found to be affected by prenatal nutrition in humans [19]. The latter is particularly important because changes in DNA methylation will not only affect the tissue composition of the animal but it has a greater chance to be inherited by subsequent generations affecting their offspring phenotype.
A group of myogenic regulatory factors (MRF) including MYOD1 and MYOG are important transcription factors involved in myogenesis [12]. While MYOD1 has been shown to be important for formation and survival of myoblasts, MYOG and MYOD1 have also been shown to be involved in terminal differentiation of myotubes. Any impacts upon muscle development in utero may in concert affect the expression of these MRF.
Understanding the molecular mechanisms underlying the differences in fetal skeletal muscle development in response to maternal nutrition is of considerable practical significance. Therefore, the objectives of this experiment were to determine the consequences of moderate maternal nutrient restriction during the 2nd half of gestation: 1) on fetal phenotypic characteristics, 2) on the expression of growth, myogenic and adipogenic genes and miRNA in the fetal muscles and 3) on DNA methylation levels in the imprinted genes IGF2 and IGF2R in two muscle types.
Methods
Animals, experimental design and dietary treatments
This experiment was conducted in accordance with the guidelines from the Canadian Council on Animal Care and with the approval of the University of Guelph Animal Care Committee. The experiment was a continuation of a study investigating maternal metabolism in mid-to-late gestation described previously by Wood et al. [20]. Briefly, 24 mature (3–6 yrs. old) multiparous pregnant Angus-Simmental cross-bred cows were used in a randomized complete block design where cows were blocked (n = 6) by expected date of parturition. The cows were divided into 2 dietary groups (n = 12 cows/diet), and fed one of 2 levels of nutrition; 1) ad-libitum intake (~140% of metabolizable energy requirements [21, 22]; HIGH), and 2) restricted to 85% of metabolizable energy requirements (LOW), as calculated from dry matter intake (DMI) of the cows described by Wood et al. [20]. The two levels of energy supplied to the cattle were chosen to represent a moderate energy restriction verses the energy intake a cattle producer would realize if cows were fed ad libitum on the same diet as used in this study. The diet consisted of a haylage-based total mixed ration containing 20% wheat straw and contained a commercially available trace mineral and vitamin supplement [20]. All cattle started the feeding trial simultaneously at 147 ± 15 days of gestation and were slaughtered over 6 weeks (blocks), in groups of 4 (2 cows from both groups/week) at 247 ± 10 days of gestation. DMI was measured for individual animals using Calan gates (American Calan, Inc., Northwood, NH). Intakes were adjusted every 14 d based on body weight.
Sample Collection and Carcass Measurements
The pregnant cows were slaughtered using captive-bolt stunning and exsanguination at the University of Guelph Meat Laboratory, and fetal phenotypic evaluation was performed. Fetal weight, crown-rump length and thorax circumference were measured immediately after obtaining the fetuses. Fetal organs including heart, liver, placenta and kidney as well as the cow uterus were dissected free of connective tissue and weighed. The placenta and uterus were then further dissected to isolate the caruncles and cotyledons. The fetal LD and ST muscles were also dissected free of connective and fat tissue. Two samples of approximately 5 g of heart, liver, caruncle, cotyledon, LD and ST muscles were collected, rinsed in ice-cold saline, snap frozen in liquid nitrogen and stored at −80 °C. Two cows, one from both the HIGH and LOW treatments, were carrying twin fetuses. Due to potential differences in nutrient demand from twins as opposed to singleton pregnancies, data from these cows and fetuses were removed leaving the number of individuals equal to eleven for both treatment groups for all subsequent analyses.
RNA extraction and real-time PCR
Heart, liver, caruncle, cotyledon, and LD and ST muscle samples were ground under liquid nitrogen using a mortar and pestle. Total RNA was extracted from 30 to 50 mg of heart, caruncle and cotyledon using TRIzol reagent (Life technologies) following the manufacturer’s instructions. The total RNA was precipitated with 0.5 volume of isopropanol and resuspended in nuclease-free H2O (Ambion, Foster City, CA, USA). Total RNA from liver, LD and ST muscle was extracted from 30 to 50 mg of tissue using the miRNeasy micro Kit (Qiagen, Germantown, MD, USA) according to manufacturer’s instructions. All total RNA samples were quantified using a spectrophotometer ND-1000 (NanoDrop, Wilmington, DE, USA), evaluated for RNA integrity (RIN) using an Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA, USA) and stored at -80 °C until cDNA synthesis. The RIN value of RNAs isolated from all tissues were >7, with the exception of fetal liver and maternal caruncle, which were >6.5.
Gene expression analysis
Total RNA (2 μg) from each individual calf and tissue was reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Life Technologies Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. RNaseOUT (Life Technologies Inc.) was also added to the reaction at a concentration of 2 U/μl. After reverse transcription (RT), the cDNA were diluted to 0.5 ng/μl with nuclease-free H2O (Ambion). Real-Time PCR analysis was performed in duplicate using 1 ng of cDNA in 96-well fast plates using the SYBR fast master mix ABI prism (D-Mark Biosciences) and the Step-One Plus Real-time PCR system (Life Technologies Inc.). A blank sample and a minus RT were added to control for nonspecific amplification. Relative standard curves, made from a serial dilution of pooled cDNA from the tissue of interest and ranging from 20 to 0.02 ng, were used to determine the relative quantity of each sample. The primers were designed in Primer3 [23–25] using species-specific sequences found in GenBank, were designed to cover exon-exon junctions when possible, and ran with a annealing/extension temperature in the real-time PCR reaction of 60 °C (Table 1). The amplification efficiency for each gene was determined using serial dilution of tissue specific cDNA and was found to be 100 ± 10% for all genes (data not shown). The resulting quantitative PCR (qPCR) amplicon were also sequenced to confirm their identity (data not shown). For each tissue, 2–4 endogenous controls were tested and the best individual or combination of endogenous control was chosen using NormFinder [26]. Therefore, Peptidylprolyl Isomerase A (PPIA), Hydroxymethylbilane Synthase (HMBS), Eukaryotic Translation Elongation Factor 1 Alpha 2, (EEF1A2), Tyrosine 3-Monooxygenase/Tryptophan (YWHAZ) and the geometric mean of YWHAZ/Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) were used as the endogenous control to correct for RNA extraction and reverse transcription efficiency in liver, heart, muscle (LD and ST), cotyledon, and caruncle, respectively. The endogenous controls were also tested for any treatment effect and were found to be stable among samples within each tissue type confirming their usefulness as suitable endogenous controls. For all genes only the expression of transcripts and miRNAs that could be reliably detected in each tissue type are reported.
Table 1.
Primer sequences and amplification conditions for gene expression measured by real-time PCR
| Gene | GenBank Accession # | Primer | Sequence 5′ - 3’ | Product size (bp) |
|---|---|---|---|---|
| ACTB | NM_173979.3 | Fwd | CTCTTCCAGCCTTCCTTCCT | 245 |
| Rev | CCAATCCACACGGAGTACTTG | |||
| EEF1A2 | NM_001037464.1 | Fwd | AGTTCACGTCCCAGGTCATC | 149 |
| Rev | CTCCAACTTCTTGCCAGAGC | |||
| FLT1 | NM_001191132.2 | Fwd | AGGGAAGAAGGTGGTCATCC | 185 |
| Rev | TGACTGTTGTCTCGCAGGTC | |||
| GAPDH | NM_001034034.1 | Fwd | TGACCCCTTCATTGACCTTC | 143 |
| Rev | GATCTCGCTCCTGGAAGATG | |||
| HMBS | NM_001046207.1 | Fwd | CTACTTCGCTGCATTGCTGA | 105 |
| Rev | CAGGTACAGTTGCCCATCCT | |||
| IGF1 | NM_001077828 | Fwd | GATGCTCTCCAGTTCGTGTG | 141 |
| Rev | CTCCAGCCTCCTCAGATCAC | |||
| IGF1R | NM_001244612 | Fwd | CAAAGGCAATCTGCTCATCA | 139 |
| Rev | CAGGAAGGACAAGGAGACCA | |||
| IGF2 | NM_174087.3 | Fwd | CCAGCGATTAGAAGTGAGCC | 95 |
| Rev | AGACCTAGTGGGGCGGTC | |||
| IGF2R a | NM_174352 | Fwd | GCAATGCTAAGCTTTCGTATTACG | 188 |
| Rev | GGTGTACCACCGGAAGTTGTATG | |||
| INSR | XM_002688832 | Fwd | CCTATGCCCTGGTGTCACTT | 114 |
| Rev | GCTGCCTTAGGTTCTGGTTG | |||
| KDR | NM_001110000.1 | Fwd | ATCGAAGTTTCCTGCACAGC | 133 |
| Rev | TCACCCTGCGGATAGTTAGG | |||
| MEF2A | NM_001083638 | Fwd | CAATGCCAACTGCCTACAAC | 130 |
| Rev | TGTCCTAAATGGTGCTGCTG | |||
| MYOD1 | NM_001040478 | Fwd | GAACACTACAGCGGCGACTC | 121 |
| Rev | AGTAAGTGCGGTCGTAGCAG | |||
| MYOG | NM_001111325 | Fwd | CAGTGAATGCAGCTCCCATA | 164 |
| Rev | CGACATCCTCCACTGTGATG | |||
| PGF | NM_173950 | Fwd | GGAACATTTCATCGGAGGTG | 136 |
| Rev | CAGGATGGGCTGAATAGATG | |||
| PPARG | NM_181024,2 | Fwd | ACCACCGTTGACTTCTCCAG | 137 |
| Rev | ACAGGCTCCACTTTGATTGC | |||
| PPIA | NM_178320.2 | Fwd | GTCAACCCCACCGTGTTCT | 132 |
| Rev | TCCTTTCTCTCCAGTGCTCAG | |||
| SLC2A1 | NM_174602.2 | Fwd | ACACAGCCTTCACTGTCGTG | 156 |
| Rev | TGCTCAGGTAGGACATCCAG | |||
| SLC2A3 | NM_174603 | Fwd | CCTCTGATCTTCGCCATCTC | 170 |
| Rev | AAGGACCACAGGGATGTGAG | |||
| SLC7A1 | NM_001135792.1 | Fwd | AGCAGCAGCGATTCTCAGAC | 158 |
| Rev | AGGCAGAAGGTGATGACCAG | |||
| SRF | NM_001206016 | Fwd | CGGCTTTGAAGAGACAGACC | 101 |
| Rev | GCAGGTTGGTGACGGTAAAC | |||
| VEGFA | NM_174216.1 | Fwd | AGACCCTGGTGGACATCTTC | 185 |
| Rev | TATGTGCTGGCTTTGGTGAG | |||
| YWHAZ | NM_174814.2 | Fwd | AGACGGAAGGTGCTGAGAAA | 123 |
| Rev | CGTTGGGGATCAAGAACTTT |
aSpicer et al. [48]
MicroRNA analysis
For miRNA analysis, 200 ng of total RNA from LD muscle was reverse transcribed with miRNA specific primers using the TaqMan microRNA Reverse Transcription Kit (Life Technologies Inc.) according to manufacturer’s instructions. The cDNA was then diluted to 0.25 ng/μl with nuclease-free H2O (Ambion). Real-time PCR for miRNA analysis in LD samples was performed in duplicate using 0.5 ng of cDNA as described previously but using the TaqMan Universal Master Mix (Life Technologies Inc.), and the miRNA specific primers. A relative standard curve ranging from 5 to 0.005 ng was also used to determine the relative quantity of each sample. Three miRNA endogenous controls (microRNA 16b (miR-16b), microRNA-191 (miR-191), and RNA, U6 Small Nuclear 6 (RNU6B)) were tested and the best individual or combination of endogenous control was chosen using NormFinder (24). The geometric mean of miR-16b and miR-191 was used to normalize the data. Amplification efficiency and endogenous control stability was also determined as described previously.
DNA methylation analysis
Genomic DNA extraction
Genomic DNA (gDNA) was extracted from LD and ST muscle tissue by overnight digestion at 50 °C in digestion buffer [100 mM NaCl, 10 mM Tris, 25 mM EDTA, 0.5% (w/v) SDS, 80 μg/ml RNaseA and 0.1 mg/ml Proteinase K]. The gDNA was then extracted with Phenol:Chloroform:Isoamyl Alcohol (25:24:1 (v/v)) using the Maxtract Tube (Qiagen), precipitated with 0.5 volume of 7.5 M ammonium acetate and 2 volume 100% ethanol, resuspended in nuclease-free H2O (Ambion) and stored at -20 °C. The gDNA was then quantified using the Quant-iT PicoGreen dsDNA assay kit (Life technologies) according to manufacturer’s instructions and its integrity was evaluated on a 0.8% (w/v) agarose gel.
Methylation analysis
DNA methylation analysis of partial regions from the IGF2 DMR2 (UCSC_Btau_4.6.1_Chr29:51,079,954–51,080,478), IGF2/H19 intergenic control region (ICR) (UCSC_Btau_4.6.1_Chr29:51,165,094–51,165,593), and IGF2R DMR2 (UCSC_Btau_4.6.1_Chr9:100,467,361–100,467,716) was performed by McGill University and Genome Quebec Innovation Centre using EpiTYPER® DNA methylation analysis technology (Agena Bioscience) according to their standard procedure. Briefly, 1 μg of gDNA was treated with sodium bisulfite using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions, except the bisulfite conversion was performed with 15 μl of sample, 5 μl of M-Dilution Buffer and 130 μl of CT-Conversion Reagent followed by 16 cycles of 95 °C for 30 s and 50 °C for 1 h. Bisulfite treated DNA was eluted from the Silicon-A Binding plate in 70 μl of M-Elution Buffer. The bisulfite treated DNA was then PCR amplified, treated with Shrimp Alkaline Phosphatase, in vitro transcribed and submitted to base-specific RNA cleavage according to the manufacturer’s instructions (Agena Bioscience). The primers used in this experiment were designed using EpiDesigner (Agena Bioscience; Table 2). The transcription cleavage products were then conditioned with CLEAN resin and transferred to a SpectroCHIP® using the MassARRAY nanodipenser (Agena Bioscience). The MALDI-TOF mass spectra were acquired using the MassARRAY analyser and the data were processed with the EpiTYPER software (Agena Bioscience). Non-quantifiable and ambiguous CpG were excluded from the analysis.
Table 2.
Primers used to amplify each of the three DMRs investigated
| Gene | Location | Primer | Sequence 5′ - 3’ | Product size (bp) | Total CpG sites (Analysable site) |
|---|---|---|---|---|---|
| IGF2R DMR2 | UCSC_Btau_4.6.1 Chr9:100,467,361–100,467,716 | Fwd | aggaagagagGAGTAAAGATTTGAAATTGAAAGTAG | 356 | 17 (11) |
| Rev | cagtaatacgactcactatagggagaaggctAACCTTCTCAACACCTTACTCAAAA | ||||
| IGF2 DMR2 | UCSC_Btau_4.6.1 Chr29:51,079,954–51,080,478 | Fwd | aggaagagagGTATTTTGGGTATTTGGGGTAGTTT | 525 | 22 (10) |
| Rev | cagtaatacgactcactatagggagaaggctATTCTAATCCCCTCAACCAAATAAA | ||||
| IGF2/H19 ICR | UCSC_Btau_4.6.1 Chr29:51,165,094–51,165,593 | Fwd | aggaagagagGGATAGGAGATTAGGTTTAGAGGGG | 500 | 17 (14) |
| Rev | cagtaatacgactcactatagggagaaggctAAAAAAAACTATAAAATCCTCCTACCC |
Statistical analysis
Prior to the statistical analysis, the data from 2 cows (1 from each dietary treatment) and their corresponding fetuses were removed from the analysis because of the confounding effect of being pregnant with twins, leaving eleven fetuses/treatment group. The cow and fetal phenotypic data, the real-time PCR data and the methylation data were analysed using the MIXED procedure of SAS (SAS Institute Inc., Cary, NC). The model for the experiment included block (date of parturition) and diet as the fixed independent variables, and pen nested within block and treatment as the random variable. The interaction of fetal sex by treatment was also tested in the model but was found to be non-significant and was removed. Differences between means were analysed using a Least Significant Difference (LSD) test at a 95% confidence level. When necessary, the data were transformed to meet the assumption of normality and homoscedasticity. Correlation analyses were also performed in LD and ST muscle to investigate relationships between fetal weight and the mRNA abundance for each gene, between IGF2 and the myogenic genes and between DNA methylation and IGF2 and IGF2R expression.
Results
Phenotypic data
The cows used in this experiment showed no differences in body weight between treatments at the beginning of the feeding trial (Table 3; as also reported by Wood et al., [20]). During and at the end of the feeding trial HIGH cows had higher dry matter intake (DMI) and average daily gain (ADG) (P < 0.01), which resulted in greater total body weight gain and heavier body weight at the end of trial (P < 0.05, Table 3). On the other hand, at slaughter the fetuses obtained from HIGH and LOW cows showed no differences in age at slaughter, body weight, crown-rump length, thorax circumference, and organ weight (Table 3). Similarly, no differences in uterine and placental weight were observed between the HIGH and LOW diet treatments.
Table 3.
The impact of mild nutrient restriction or no restriction during mid-to-late gestation on cow weight gain, and fetal weight and organ weight, and uterine and placental weight
| Item | Treatment | P-value | ||
|---|---|---|---|---|
| High dieta | Low dieta | |||
| Cowb | Start weight (kg) | 645.5 ± 21.8 | 633.4 ± 20.9 | 0.726 |
| Final weight (kg) | 749.3 ± 18.7 | 686.3 ± 19.3 | 0.0438 | |
| Weight gain (kg) | 103.8 ± 7.6 | 52.9 ± 8.2 | 0.0015 | |
| Average daily gain (kg/d) | 1.06 ± 0.08 | 0.53 ± 0.07 | 0.0012 | |
| Average dry matter intake (kg/d) | 11.00 ± 0.28 | 6.46 ± 0.18 | < 0.0001 | |
| Uterine Weight (g) | 4827.4 ± 206.7 | 4463.2 ± 284.1 | 0.338 | |
| Fetus | Age at slaughter (d) | 247.8 ± 4.1 | 245.7 ± 1.2 | 0.743 |
| Weight | 31.9 ± 1.7 | 28.5 ± 2.4 | 0.332 | |
| Crown-rump length (cm) | 78.2 ± 2.4 | 75.9 ± 3.8 | 0.696 | |
| Thorax circumference (cm) | 65.6 ± 1.9 | 62.2 ± 2.0 | 0.339 | |
| Heart weight (g) | 210.9 ± 11.9 | 196.7 ± 17.7 | 0.607 | |
| Liver weight (g) | 806.7 ± 47.0 | 683.6 ± 66.2 | 0.199 | |
| Kidney weight (g) | 89.4 ± 6.1 | 80.2 ± 5.9 | 0.253 | |
| Placental weight (g) | 3562.1 ± 297.7 | 3837.1 ± 362.6 | 0.533 | |
aData are expressed as lsmeans ± SEM
bAlso reported in Wood et al. [20]
Gene and miRNA expression
Analysis of the expression of insulin-like growth factor family members in liver tissue revealed increased IGF1 and reduced IGF1R mRNA abundance in fetuses from HIGH cows (P < 0.05, Fig. 1a). In the heart, a reduction in IGF2R mRNA abundance in the HIGH fetuses was observed (P < 0.05, Fig. 1b), while no differences in expression of myogenic genes MEF2A and SRF were observed (Fig. 1b).
Fig. 1.

mRNA abundance for Insulin-like growth factors and myogenesis related genes in liver (a) and heart (b, c) tissue from fetal calf (n = 22) exposed to a high or low diet in utero during the 2nd half of gestation. Different letters represent significant differences (P < 0.05 a,b)
In fetal LD muscle, gene expression analysis of the insulin-like growth factors, and the myogenic and adipogenic families, revealed many differences associated with maternal HIGH and LOW diets (Fig. 2a-c). IGF1, IGF1R, IGF2R and INSR mRNA had greater abundance in the muscle from LOW fetuses (P < 0.05, Fig. 2a). Similarly, MYOD1, MYOG and PPARG mRNA also had greater abundance in the LD muscle of LOW fetuses (P < 0.05, Fig. 2b, c). Interestingly, LD expression of IGF1 (r = −0.55, P ≤ 0.01), IGF1R (r = −0.48, P ≤ 0.05), IGF2 (r = −0.65, P ≤ 0.01), IGF2R (r = −0.65, P ≤ 0.01), MYOG (r = −0.48, P ≤ 0.05) and SRF (r = −0.61, P ≤ 0.01) were negatively correlated with fetal weight (Fig. 3a-f). In addition, in LD muscle IGF2 mRNA abundance was positively correlated with that of MYOD1 (r = 0.56, P ≤ 0.01), MYOG (r = 0.76, P ≤ 0.0001), and SRF (r = 0.56, P ≤ 0.01) (Fig. 3g-i). The results differed in the ST muscle where only trends for increased IGF2 and IGF2R mRNA abundance were observed in LOW fetuses (P < 0.1, Fig. 2d-f). Nonetheless, in ST negative correlations were also observed between IGF1 (r = −0.48, P ≤ 0.05), IGF1R (r = −0.52, P ≤ 0.05), IGF2 (r = −0.57, P ≤ 0.01), IGF2R (r = −0.59, P ≤ 0.01) and MYOD1 (r = −0.72, P ≤ 0.001) with fetal weight (Fig. 4a-e). In ST muscle, IGF2 mRNA abundance was positively correlated with MYOD (r = 0.56, P ≤ 0.01), MYOG (r = 0.70, P ≤ 0.001) and MEF2A (r = 0.44, P ≤ 0.05) (Fig. 4f-h).
Fig. 2.
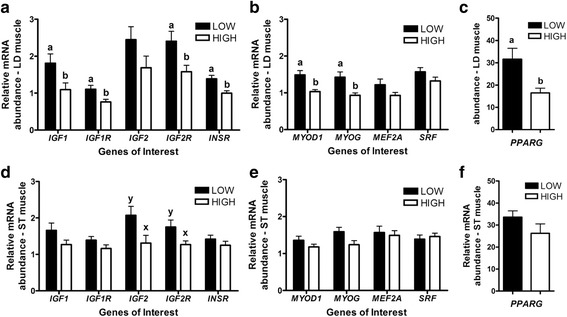
mRNA abundance for Insulin-like growth factors, and myogenesis and adipogenesis related genes, in longissimus dorsi (a, b, c) and semitendinosus (d, e, f) muscle from fetal calf (n = 22) exposed to a high or low diet in utero during the 2nd half of gestation. Different letters represent significant differences (P < 0.05 a,b; P < 0.1×,y)
Fig. 3.

Correlation analysis between fetal weight and IGF1 (a), IGF1R (b), IGF2 (c), IGF2R (d), MYOG (e) and SRF (f) mRNA abundance and between IGF2 and MYOD1 (g), MYOG (h) and SRF (i) mRNA abundance in fetal LD muscle. Only significant (P < 0.05) correlations are reported
Fig. 4.
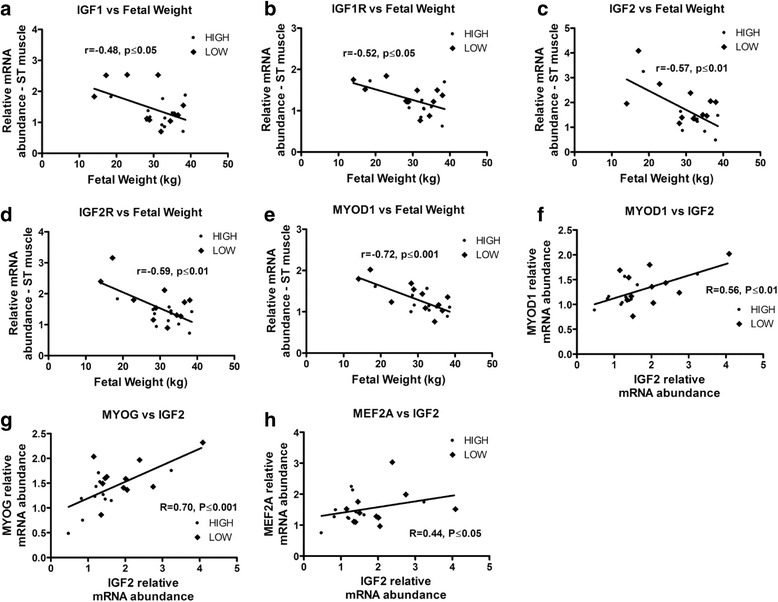
Correlation analysis between fetal weight and IGF1 (a), IGF1R (b), IGF2 (c), IGF2R (d) and MYOD1 (e) mRNA abundance and between IGF2 and MYOD1 (f), MYOG (g) and MEF2A (h) mRNA abundance in fetal ST muscle. Only significant (P < 0.05) correlations are reported
Analysis of microRNA 1 (miR-1) and microRNA 133a (miR-133a) expression also revealed that both microRNAs abundance was lower in the LD muscle of LOW fetuses (P < 0.05, Fig. 5). Lastly, although trends for differential mRNA abundance of the angiogenic factors Vascular Endothelial Growth Factor A (VEGFA) and Fms Related Tyrosine Kinase 1 (FLT1), and the glucose transporter Solute Carrier Family 2 Member 3 (SLC2A3) were observed in caruncles of HIGH and LOW cows (increased in HIGH), no other significant differences were observed in the placenta (Fig. 6).
Fig. 5.

microRNA abundance for miR-1 and miR-133a in the LD muscle of fetal calves (n = 22) exposed to a high or low diet in utero during the 2nd half of gestation. Different letters represent significant differences (P < 0.05a,b)
Fig. 6.

mRNA abundance for Insulin-like growth factors, angiogenesis and solute carrier related genes in the placenta (Caruncle (a, b, c); Cotyledon (d, e, f)) of fetal calves (n = 22) exposed to a high or low diet in utero during the 2nd half of gestation. Different letters represent differences at P < 0.1x,y
DNA methylation
Methylation analysis of the IGF2/H19 ICR in LD and ST muscle did not reveal any differences in methylation level between HIGH and LOW fetuses. Similarly, only the CpG group 23–24 of the IGF2R DMR was differently methylated in the LD muscle of HIGH and LOW fetuses (0.344 vs 0.363 respectively; P = 0.039). No correlations were observed between the CpG group 23–24 and IGF2R mRNA abundance. On the other hand, several CpG and CpG groups of the IGF2 DMR2 were differentially methylated in the LD muscle of HIGH and LOW diet fetuses (Table 4). In all instances, the methylation levels of these CpG or CpG groups were higher in the muscle of HIGH fetuses and the differences in methylation level observed between the two groups varied between 2.5 and 10.1% (Table 4). Moreover, the overall methylation level for DMR2 in LD muscle was greater in the HIGH fetuses (P < 0.05), and while a similar trend was observed in ST muscle the difference was not significant (Table 4). Although HIGH fetuses had greater methylation levels in all quantifiable CpG in IGF2 DMR2 in both LD and ST muscle, only in the LD muscle were the differences significant at P < 0.05. This observation is consistent with the observed gene expression differences between the two muscle types. The methylation level of several CpG and CpG groups in the DMR2 of IGF2 was negatively correlated with IGF2 mRNA abundance in both muscle tissues (Table 4). Using the web-based prediction program TFBIND (http://tfbind.hgc.jp) [27], we identified several putative transcription factor binding sites in the IGF2 DMR2 region including potential binding sites for MYOD, SRF, Sp1 Transcription Factor (SP1) and MEF2A (data not shown).
Table 4.
Impact of maternal nutrition during mid-to-late gestation on the methylation status of IGF2 DMR2 as assessed by Sequenom Epityper analysis
| Amplicon | CpG | Position in amplicon | High dieta | Low dieta | P-value | Correlation with IGF2 |
|---|---|---|---|---|---|---|
| Longissimus dorsi methylation (%) | ||||||
| IGF2 DMR2 Chr29:51,079,954–51,080,478 | 1, 2, 3 | 29, 38, 47 | 29.1 ± 1.4 | 27.2 ± 0.9 | 0.38 | R = −0.40, p = 0.066 |
| 10,11 | 173, 183 | 52.3 ± 1.6 | 48.6 ± 1.3 | 0.11 | R = −0.67, P = 0.0007 | |
| 12, 13, 14 | 189, 192, 198 | 40.4 ± 1.5 | 35.7 ± 0.9 | 0.019 | R = −0.70, P = 0.0003 | |
| 15, 16 | 202, 206 | 45.8 ± 1.4 | 41.9 ± 1.0 | 0.07 | R = −0.70, P = 0.0003 | |
| 20 | 257 | 68.1 ± 1.5 | 63.3 ± 1.4 | 0.039 | R = −0.73, P = 0.0001 | |
| 21, 22 | 287, 293 | 48.9 ± 1.4 | 47.5 ± 0.6 | 0.44 | R = −0.57, P = 0.0059 | |
| 28 | 384 | 67.9 ± 2.4 | 63.0 ± 3.3 | 0.13 | R = −0.25, P = 0.26 | |
| 29 | 390 | 89.1 ± 1.8 | 82.5 ± 1.3 | 0.014 | R = −0.64, P = 0.0013 | |
| 34 | 482 | 76.7 ± 4.4 | 71.6 ± 2.4 | 0.31 | R = −0.25, P = 0.29 | |
| 35 | 493 | 68.4 ± 1.0 | 65.9 ± 0.7 | 0.073 | R = −0.71, P = 0.0002 | |
| All | na | 51.3 ± 1.3 | 47.4 ± 0.9 | 0.025 | R = −0.65, P = 0.001 | |
| Semitendinosus methylation (%) | ||||||
| IGF2 DMR2 Chr29:51,079,954–51,080,478 | 1, 2, 3 | 29, 38, 47 | 31.3 ± 0.8 | 29.9 ± 0.7 | 0.28 | R = −0.66, P = 0.0008 |
| 10,11 | 173, 183 | 55.0 ± 1.2 | 51.6 ± 1.0 | 0.042 | R = −0.68, P = 0.0005 | |
| 12, 13, 14 | 189, 192, 198 | 41.8 ± 1.3 | 38.1 ± 0.8 | 0.06 | R = −0.78, P ≤ 0.0001 | |
| 15, 16 | 202, 206 | 46.3 ± 0.8 | 43.7 ± 0.8 | 0.056 | R = −0.74, P ≤ 0.0001 | |
| 20 | 257 | 69.4 ± 1.3 | 66.4 ± 1.1 | 0.16 | R = −0.70, P = 0.0003 | |
| 21, 22 | 287, 293 | 50.3 ± 1.2 | 47.3 ± 1.2 | 0.11 | R = −0.49, P = 0.019 | |
| 28 | 384 | 71.7 ± 3.7 | 61.6 ± 1.9 | 0.075 | R = −0.63, P = 0.0017 | |
| 29 | 390 | 87.8 ± 1.5 | 85.8 ± 1.8 | 0.46 | R = −0.44, P = 0.038 | |
| 34 | 482 | 80.1 ± 3.6 | 75.2 ± 4.1 | 0.53 | R = −0.56, P = 0.011 | |
| 35 | 493 | 68.5 ± 1.0 | 66.6 ± 0.7 | 0.12 | R = −0.69, P = 0.0004 | |
| All | na | 52.8 ± 1.1 | 49.6 ± 0.9 | 0.086 | R = −0.79, P ≤ 0.0001 | |
aData represent the methylation percentage for CpG or CpG group and are expressed as lsmeans ± SEM
Discussion
The influence of maternal nutrition during gestation on fetal programming and more specifically on muscle development has been previously studied in beef cattle [2, 6]. However, our understanding of the molecular mechanisms leading to phenotypic changes in the offspring remains limited. The present study investigated the effect of maternal nutrition during the second half of pregnancy on fetal muscle development and demonstrates that differential nutrient levels can potentially permanently influence the fetus via DNA methylation and gene expression differences even if no obvious differences in phenotypes are observed.
Restricting feed intake to approximately 85% of metabolizable energy requirements had the expected impact on the cow. Restricted cows were lighter at the end of the treatment period because of lower average daily weight gain. Interestingly, the difference in maternal weight gain did not appear to impact fetal growth as fetuses were similar in weight, crown-rump length, and thorax circumference from the HIGH and LOW maternal diet treatments. Our results are in contrast with previous studies in beef cattle that showed that nutritional restriction during the 2nd half of gestation led to lighter birth weights [28, 29]. However, these discrepancies can likely be explained by the milder nutrient restriction imposed in this experiment compared to the more severe nutrient restrictions treatments applied to other ruminant experiments in the literature. On a side note, weaning weights of calves born from cows exposed the same pre-natal dietary treatments as the fetal collection calves were investigated and no differences due to treatment were found (data not shown). Although the authors would like to inform that due to death loss, the presence of twins, and the influences of subsequent research projects which lead to three sex groups among the weaned calves (bulls, steers, and heifers), no concrete conclusions should be made with regard to the weaning data.
Although there may be instances where no apparent fetal phenotypic responses to pre-natal maternal diets are observed, fetuses may show adaptation on the molecular level. In order to assess if the fetuses responded at the molecular level to differential pre-natal nutritional levels, expression of insulin-like growth factors family members was examined in the fetal liver. Our results indicated that restricted fetuses had lower abundance of IGF1 mRNA, consistent with the hypothesized effect that lower nutrient resources would elicit a metabolic response in the liver of restricted fetuses. Indeed, a study in baboons showed that maternal dietary restriction lead to lower expression of IGF1 mRNA and protein in fetal liver [30], as well, IGF1 mRNA was lower in livers of nutrient-restricted fetal sheep when measured at the end of the restriction period [31]. Our results indicate that the fetuses used in this experiment were sensitive to the changes in nutrient supply.
Relative to essential organs such as the heart, skeletal muscle has lower priority for nutrient partitioning in the developing fetus [6]. However, recent work using a KEGG analysis of mRNA-Sequencing results from an experiment that investigated maternal nutrient sources in late gestation on LD mRNA expression in sheep found that energetic metabolism pathways, including insulin signalling, displayed the largest proportion of differentially expressed genes [32]. As such, we evaluated the expression of insulin-like growth factor and myogenic genes in various tissues of HIGH and LOW fetuses. As expected, very few differences were observed in the heart with the exception of a significant increase in IGF2 in restricted fetuses. We made similar observations in semitendinosus muscle where no significant differences in IGF2 and IGF2R mRNA abundance were detected. On the other hand, increased mRNA abundance of IGF1, IGF1R, IGF2R, INSR, MYOD1 and MYOG were observed in the longissimus dorsi muscle of restricted fetuses. This is in contrast with a previous study in cattle that showed that a low protein diet during the 1st and/or 2nd trimester altered IGF1, IGF1R, IGF2 and IGF2R in the ST muscle of restricted animals but not in the LD [33], although several differences exist between the 2 studies including timing of the restriction (early to mid-gestation vs mid- to late gestation), type (total metabolizable energy verses protein) and degree of restriction, and breed of cattle (Angus-Simmental cross-bred vs composite breeds including Bos indicus). Differences in gene expression observed between the LD and ST muscle of the fetuses used in the current study suggest that different muscles are affected to varying degrees by nutrient restriction. Just as essential organs have priority over skeletal muscle for nutrient partitioning, it is possible that muscles with different functions, for example postural vs locomotion, also have different priority for nutrient partitioning. This hypothesis is attractive as muscle with different functions may also differ in muscle fiber number, size and composition [34], which would be reflected in differential gene expression.
It is interesting to note that all differentially expressed genes in LD muscle have greater abundance in the muscle of LOW fetuses. This led us to hypothesize that muscle development in LOW fetuses was delayed as compared to development in HIGH fetuses, but also that the muscle of the HIGH diet fetuses had transitioned from myogenesis to adipogenesis by the time of sample collection. To address the latter hypothesis, we designed primers for PPARG and CCAAT/Enhancer Binding Protein Alpha (CEPBA) which are known inducers of adipogenesis [35]. Using two different sets of primers we could not successfully amplify CEPBA in LD and ST muscle. However, PPARG mRNA abundance was higher in the LD muscle of LOW fetuses suggesting that the fetuses fed the HIGH diet had not increased adipogenesis. Further investigation into the literature revealed potential roles for PPARG expression in skeletal muscle regulating its internal lipid metabolism [36]. Increased expression of PPARG in skeletal muscle fibres in offspring has been shown to be associated with pre-natal consumption of a maternal cafeteria diet in rats [16]. Although no direct evidence exists linking pre-natal maternal undernutrition to increased PPARG expression in skeletal muscle, other groups have hypothesized that mobilization of fatty acids in the maternal circulation from maternal adipose reserves, brought about by maternal undernutrition, might elicit similar responses in PPARG expression in fetal muscle as is seen with the maternal overnutrition model [37]. Therefore higher expression of PPARG in LOW diet fetuses might be an appropriate response to the nutritional treatment applied to their dams in the present experiment.
MicroRNAs have been shown to be important for various biological processes including muscle development [38, 39]. The miRNA miR-1 and miR-133a are known to modulate proliferation and differentiation in skeletal muscle [38]. Their expression is also modulated by myogenic transcription factors including MEF2A, SRF and MYOD1. Our results revealed that miR-1 and miR-133a expression is increased in the longissimus dorsi muscle of the HIGH diet fetuses. Interestingly, in human fetal skeletal muscle, miR-1 and miR-133a expression were found to increase during the late stages of development [39]. This strengthened our previous hypothesis suggesting that the LD muscle of the LOW diet fetuses are developmentally delayed compared to that of the HIGH fetuses.
Our results suggest that differential pre-natal nutrient level during the 2nd half of pregnancy can alter the methylation status of IGF2 as shown by the difference in methylation of several CpG units in the DMR2 of IGF2. This observation is curious since no differences in IGF2 mRNA abundance were observed in LD and ST muscles. However, the standard error for IGF2 mRNA expression is relatively large suggesting a wide range of response between fetuses belonging to the same treatment. Correlation analysis indicate a negative relationship between IGF2 mRNA abundance and CpG methylation in DMR2, supporting our hypothesis that increased methylation reduces IGF2 expression. This is in contrast with the suggested mechanism for IGF2 expression since the DMR2 is known to contain a methylation sensitive activator that increases IGF2 expression with increasing methylation [40]. Analysis of our DMR2 partial sequence with the web-based prediction program TFBIND revealed several predicted transcription factor binding sites, including MYOD1, MEF2A, SRF and SP1 that are known regulators of myogenesis. It is therefore tempting to speculate that increased methylation in that region altered transcription factor or enhancer binding, leading to reduced gene expression. IGF2 and MYOD1 are known to mutually modulate their expression in muscle [41]. Therefore the positive correlation observed between IGF2 and MYOD1 mRNA abundance certainly reinforces our hypothesis.
In the current study, we also examined the expression of growth, angiogenic, and solute carrier family members in the uterus and placenta of animals exposed to HIGH and LOW diets. Our observations reveal no significant differences in gene expression in the caruncle and cotyledon suggesting that the placentome of these cattle was not altered by the nutritional treatment at the time of tissue collection. This is also corroborated by the lack of differences in placental and uterine weight, fetal weight, crown-rump length, and thorax circumference observed between the HIGH and LOW fetuses. Our results are in contrast with previous studies that have shown that nutritional restriction during the 1st and 2nd half of pregnancy can be detrimental to placental development. In cattle, nutritional restriction from day 30 to 125 of gestation was shown to decrease placentome weight [41, 42], while in the ewe, maternal undernutrition during early- to mid-gestation led to increased placental weight [43]. On the other hand, nutrient restriction during the 2nd half of gestation in rats has been shown to decrease the placental:fetal weight ratio [44]. Maternal undernutrition can also alter placental vascularity leading to reduction in blood flow and consequently affecting nutrient transfer to the fetus. This was shown in cattle where nutrient restriction during early- to mid-gestation affected placental vascularity and expression of VEGFA mRNA [42, 45]. In cows, development of the placentome is established in early to mid-gestation and therefore the impact of a mild nutrient restriction during mid-to-late gestation is likely to have limited impacts on the uterus and placenta [1, 46]. Therefore, the changes in gene expression observed in the muscle in the current study are unlikely to originate from major alterations in placental development or gene expression related to nutrient transfer to the fetus.
Conclusions
The findings of the present study demonstrate that differential pre-natal nutritional level during the 2nd half of gestation can have impacts on fetal development even if no obvious phenotypic differences are observed in late term fetuses. This work illustrates the importance of maternal nutrition during gestation for cattle, even when nutrition restriction is mild. However, further research is needed to determine if these differences in expression in growth-factor-related genes, and genes involved in muscle development in the fetus, translate to observable phenotypic differences later in life. It would be particularly important if DNA methylation and gene expression differences result in changes in growth performance, feed efficiency, and carcass characteristics. Our data also suggests that different muscle groups respond differently to nutrient insults, likely as a consequence of their function and muscle fiber type composition. Further research is needed to determine if the changes in DNA methylation observed in the DMR2 of IGF2 are transient or are heritable. Using fetal programming as a management tool for beef producers may provide opportunities for improvements for growth and production traits in cattle.
Acknowledgements
The authors wish to acknowledge the staff of Elora Beef Research Station and the University of Guelph and Meat Laboratory for technical assistance with this experiment.
Funding
Funding for the project was provided by the Canadian Beef Science Cluster (Beef Cattle Research Council), Ontario Ministry of Agriculture, Food and Rural Affairs, and the Beef Farmers of Ontario. The funding bodies did not play a role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
The raw data supporting findings in this study will be made available from UAL Dataverse (https://dataverse.library.ualberta.ca/dataverse/ualib): https://doi.org/10.7939/DVN/7CV6C3.
Abbreviations
- ACTB
Beta-Actin
- ADG
average daily gain
- CEPBA
CCAAT/Enhancer Binding Protein Alpha
- DMI
dry matter intake
- DMR2
Differentially Methylated Region 2
- EEF1A2
Eukaryotic Translation Elongation Factor 1 Alpha 2
- FLT1
Fms Related Tyrosine Kinase 1
- GAPDH
Glyceraldehyde-3-Phosphate Dehydrogenase
- gDNA
Genomic DNA
- HIGH
cows fed to ad-libitum intake (~140% of metabolizable energy requirements (NRC, 1996)) during the second half of gestation
- HMBS
Hydroxymethylbilane Synthase
- ICR
intergenic control region
- IGF1
Insulin Like Growth Factor 1
- IGF1R
Insulin Like Growth Factor 1 Receptor
- IGF2
Insulin Like Growth Factor 2
- IGF2R
Insulin Like Growth Factor 2 Receptor
- INSR
Insulin Receptor
- KDR
Kinase Insert Domain Receptor
- LD
longissimus dorsi
- LOW
cows restricted to 85% of metabolizable energy requirements (NRC, 1996) during the second half of gestation
- LSD
Least Significant Difference
- MEF2A
Myocyte Enhancer Factor 2A
- miR-1
microRNA 1
- miR-133a
microRNA 133a
- miR-16b
microRNA 16b
- miR-191
microRNA-191
- miRNA
micro RNA
- MRF
myogenic regulatory factors
- MYOD1
Myogenic Differentiation 1
- MYOG
Myogenin
- PGF
Placental Growth Factor
- PPARG
Peroxisome Proliferator Activated Receptor Gamma
- PPIA
Peptidylprolyl Isomerase A
- qPCR
quantitative PCR
- RIN
RNA Integrity Number
- RNU6B
RNA U6 Small Nuclear 6
- RT
reverse transcription
- SLC2A1
Solute Carrier Family 2 Member 1
- SLC2A3
Solute Carrier Family 2 Member 3
- SLC7A1
Solute Carrier Family 7 Member 1
- SP1
Sp1 Transcription Factor
- SRF
Serum Response Factor
- ST
semitendinosus
- VEGFA
Vascular Endothelial Growth Factor A
- YWHAZ
Tyrosine 3-Monooxygenase/Tryptophan
Authors’ contributions
FP, KW, SP, BM, KS, CF contributed to the design of the experiment; KW, BM, KS, CF implemented the experiment and sample collection; FP and CF conducted the molecular analysis; FP, KW, CF analyzed and interpreted the results; FP, KW, and CF prepared the manuscript, and all authors read and approved of final manuscript.
Ethics approval
Animal use and ethics approval for this trial was officially granted by the University of Guelph Animal Care Committee (Animal Use Protocol # 1700-10R093), under the recommendations outlined by the set forth by the Canadian Council of Animal Care [47]. All cattle were sourced from the breeding herd of the University of Guelph Elora Beef Research Station.
Consent for publication
not applicable.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Francois Paradis, Email: fparadis@ualberta.ca.
Katie M. Wood, Email: kwood@uoguelph.ca
Kendall C. Swanson, Email: kendall.swanson@ndsu.edu
Stephen P. Miller, Email: smiller@angus.org
Brian W. McBride, Email: bmcbride@uoguelph.ca
Carolyn Fitzsimmons, Phone: 1-780-248-1700, Email: cfitzsim@ualberta.ca.
References
- 1.Reynolds LP, Borowicz PP, Caton JS, Vonnahme KA, Luther JS, Hammer CJ, Maddock Carlin KR, Grazul-Bilska AT, Redmer DA. Developmental programming: The concept, large animal models, and the key role of uteroplacental vascular development. J Anim Sci. 2010;88(13 suppl):E61–E72. doi: 10.2527/jas.2009-2359. [DOI] [PubMed] [Google Scholar]
- 2.Funston RN, Larson DM, Vonnahme KA. Effects of maternal nutrition on conceptus growth and offspring performance: Implications for beef cattle production. J Anim Sci. 2010;88(13 Suppl):E205–E215. doi: 10.2527/jas.2009-2351. [DOI] [PubMed] [Google Scholar]
- 3.Nordby DJ, Field RA, Riley ML, Kercher CJ. Effects of maternal undernutrition during early pregnancy on growth, muscle cellularity, fiber type and carcass composition in lambs. J Anim Sci. 1987;64:1419–1427. doi: 10.2527/jas1987.6451419x. [DOI] [PubMed] [Google Scholar]
- 4.Long NM, Prado-Cooper MJ, Krehbiel CR, DeSilva U, Wettemann RP. Effects of nutrient restriction of bovine dams during early gestation on postnatal growth, carcass and organ characteristics, and gene expression in adipose tissue and muscle. J Anim Sci. 2010;88(10):3251–3261. doi: 10.2527/jas.2009-2512. [DOI] [PubMed] [Google Scholar]
- 5.Zhu MJ, Ford SP, Nathanielsz PW, Du M. Effect of maternal nutrient restriction in sheep on the development of fetal skeletal muscle. Biol Reprod. 2004;71(6):1968–1973. doi: 10.1095/biolreprod.104.034561. [DOI] [PubMed] [Google Scholar]
- 6.Ferrell CL, Garrett WN, Hinman N. Growth, development and composition of the udder and gravid uterus of beef heifers during pregnancy. J Anim Sci. 1976;42:1477–1489. doi: 10.2527/jas1976.4261477x. [DOI] [PubMed] [Google Scholar]
- 7.Long NM, Vonnahme KA, Hess BW, Nathanielsz PW, Ford SP. Effects of early gestational undernutrition on fetal growth, organ development, and placentomal composition in the bovine. J Anim Sci. 2009;87:1950–1959. doi: 10.2527/jas.2008-1672. [DOI] [PubMed] [Google Scholar]
- 8.Meyer AM, Reed JJ, Vonnahme KA, Soto-Navarro SA, Reynolds LP, Ford SP, Hess BW, Caton JS. Effects of stage of gestation and nutrient restriction during early to mid-gestation on maternal and fetal visceral organ mass and indices of jejunal growth and vascularity in beef cows. J Anim Sci. 2010;88:2410–2424. doi: 10.2527/jas.2009-2220. [DOI] [PubMed] [Google Scholar]
- 9.Du M, Tong J, Zhao J, Underwood KR, Zhu M, Ford SP, Nathanielsz PW. Fetal programming of skeletal muscle development in ruminant animals. J Anim Sci. 2010;88(13 Suppl):E51–E60. doi: 10.2527/jas.2009-2311. [DOI] [PubMed] [Google Scholar]
- 10.Martin JL, Vonnahme KA, Adams DC, Lardy GP, Funston RN. Effects of dam nutrition on growth and reproductive performance of heifer calves. J Anim Sci. 2007;85(3):841–847. doi: 10.2527/jas.2006-337. [DOI] [PubMed] [Google Scholar]
- 11.Underwood KR, Tong JF, Price PL, Roberts AJ, Grings EE, Hess BW, Means WJ, Du M. Nutrition during mid to late gestation affects growth, adipose tissue deposition, and tenderness in cross-bred beef steers. Meat Sci. 2010;86:588–593. doi: 10.1016/j.meatsci.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Maltin CA, Delday MI, Sinclair KD, Steven J, Sneddon AA. Impact of manipulations of myogenesis in utero on the performance of adult skeletal muscle. Reproduction. 2001;122(3):359–374. doi: 10.1530/rep.0.1220359. [DOI] [PubMed] [Google Scholar]
- 13.Oksbjerg N, Gondret F, Vestergaard M. Basic principles of muscle development and growth in meat-producing mammals as affected by the insulin-like growth factor (IGF) system. Domest Anim Endocrin. 2004;27(3):219–240. doi: 10.1016/j.domaniend.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Stickland NC. A Quantitative study of muscle development in the bovine foetus (Bos indicus). Anat Histol Embryol1978; 7(3):193-205. [DOI] [PubMed]
- 15.Bee G. Effect of early gestation feeding, birth weight, and gender of progeny on muscle fiber characteristics of pigs at slaughter. J Anim Sci. 2004;82(3):826–836. doi: 10.2527/2004.823826x. [DOI] [PubMed] [Google Scholar]
- 16.Bayol SA, Simbi BH, Stickland NC. A maternal cafeteria diet during gestation and lactation promotes adiposity and impairs skeletal muscle development and metabolism in rat offspring at weaning. J Physiol. 2005;567(Pt 3):951–961. doi: 10.1113/jphysiol.2005.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu MJ, Ford SP, Means WJ, Hess BW, Nathanielsz PW, Du M. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J Physiol. 2006;575(Pt 1):241–250. doi: 10.1113/jphysiol.2006.112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curchoe C, Zhang S, Bin Y, Zhang X, Yang L, Feng D, O'Neill M, Tian XC. Promoter-specific expression of the imprinted IGF2 gene in cattle (Bos taurus) Biol Reprod. 2005;73(6):1275–1281. doi: 10.1095/biolreprod.105.044727. [DOI] [PubMed] [Google Scholar]
- 19.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood KM, Awda BJ, Fitzsimmons C, Miller SP, McBride BW, Swanson KC. Effect of moderate dietary restriction on visceral organ weight, hepatic oxygen consumption, and metabolic proteins associated with energy balance in mature pregnancy beef cows. J Anim Sci. 2013;91:4245–4255. doi: 10.2527/jas.2013-6421. [DOI] [PubMed] [Google Scholar]
- 21.NRC. Nutrient requirements of beef cattle. 7th ed. Natl. Acad. Press, Washington, DC. 1996.
- 22.Weiss WP, Conrad HR, St. Pierre NR. A theoretically based model for predicting total digestible nutrient values for forages and concentrates. Anim Feed Sci Technol. 1992;39:95–110. doi: 10.1016/0377-8401(92)90034-4. [DOI] [Google Scholar]
- 23.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press. p. 365–86. [DOI] [PubMed]
- 24.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 25.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3 –new capabilities and interfaces. Nucleic Acids Res. 2012;40(15 supp):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 27.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15(7–8):622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 28.Tygesen MP, Harrison AP, Therkildsen M. The effect of maternal nutrient restriction during late gestation on muscle, bone and meat parameters in five month old lambs. Livestock Sci. 2007;110(3):230–241. doi: 10.1016/j.livsci.2006.11.003. [DOI] [Google Scholar]
- 29.Larson DM, Martin JL, Adams DC, Funston RN. Winter grazing system and supplementation during late gestation influence performance of beef cows and steer progeny. J Anim Sci. 2009;87(3):1147–1155. doi: 10.2527/jas.2008-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Schlabritz-Loutsevitch NE, Hubbard GB, Han V, Nygard K, Cox LA, McDonald TJ, Nathanielsz PW. Effects of maternal global nutrient restriction on fetal baboon hepatic insulin-like growth factor system genes and gene products. Endocrinology. 2009;150(10):4634–4642. doi: 10.1210/en.2008-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brameld J, Mostyn A, Dandrea J, Stephenson T, Dawson J, Buttery P, Symonds M. Maternal nutrition alters the expression of insulin-like growth factors in fetal sheep liver and skeletal muscle. J Endocrinol. 2000;167(3):429–437. doi: 10.1677/joe.0.1670429. [DOI] [PubMed] [Google Scholar]
- 32.Peñagaricano F, Wang X, Rosa GJ, Radunz AE, Khatib H. Maternal nutrition induces gene expression changes in fetal muscle and adipose tissues in sheep. BMC Genomics. 2014;15(1):1–13. doi: 10.1186/1471-2164-15-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micke GC, Sullivan TM, McMillen IC, Gentili S, Perry VE. Protein intake during gestation affects postnatal bovine skeletal muscle growth and relative expression of IGF1, IGF1R, IGF2 and IGF2R. Mol Cell Endocrinol. 2011;332(1–2):234–241. doi: 10.1016/j.mce.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Klont RE, Brocks L, Eikelenboom G. Meat Consumption and Culture 44th International Congress of Meat Science and Technology: Muscle fibre type and meat quality. Meat Sci. 1998;49:S219–S229. doi: 10.1016/S0309-1740(98)90050-X. [DOI] [PubMed] [Google Scholar]
- 35.Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBPalpha induces adipogenesis through PPARgamma: A unified pathway. Genes Dev. 2002;16(1):22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lapsys NM, Kriketos AD, Lim-Fraser M, Poynten AM, Lowy A, Furler SM, Chisholm DJ, Cooney GJ. Expression of Genes Involved in Lipid Metabolism Correlate with Peroxisome Proliferator-Activated Receptor γ Expression in Human Skeletal Muscle. J Clin Endocrinol Metab. 2000;85:4293–4297. doi: 10.1210/jcem.85.11.6973. [DOI] [PubMed] [Google Scholar]
- 37.Rees WD, McNeil CJ, Maloney CA. The Roles of PPARs in the Fetal Origins of Metabolic Health and Disease. PPAR Res. 2008;2008:1–8. [DOI] [PMC free article] [PubMed]
- 38.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21(3):461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol. 2011;11:34. doi: 10.1186/1471-213X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murrell A, Heeson S, Bowden L, Constancia M, Dean W, Kelsey G, Reik W. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2001;2(12):1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montarras D, Aurade F, Johnson T, Ilan J, Gros F, Pinset C. Autonomous differentiation in the mouse myogenic cell line, C2, involves a mutual positive control between insulin-like growth factor II and MyoD, operating as early as at the myoblast stage. J Cell Sci. 1996;109(Pt 3):551–560. doi: 10.1242/jcs.109.3.551. [DOI] [PubMed] [Google Scholar]
- 42.Zhu MJ, Du M, Hess BW, Means WJ, Nathanielsz PW, Ford SP. Maternal nutrient restriction upregulates growth signaling pathways in the cotyledonary artery of cow placentomes. Placenta. 2007;28(4):361–368. doi: 10.1016/j.placenta.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Heasman L, Clarke L, Firth K, Stephenson T, Symonds ME. Influence of restricted maternal nutrition in early to mid-gestation on placental and fetal development at term in sheep. Pediatr Res. 1998;44(4):546–551. doi: 10.1203/00006450-199810000-00013. [DOI] [PubMed] [Google Scholar]
- 44.Belkacemi L, Chen CH, Ross MG, Desai M. Increased placental apoptosis in maternal food restricted gestations: Role of the Fas pathway. Placenta. 2009;30(9):739–751. doi: 10.1016/j.placenta.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Vonnahme KA, Zhu MJ, Borowicz PP, Geary TW, Hess BW, Reynolds LP, Caton JS, Means WJ, Ford SP. Effect of early gestational undernutrition on angiogenic factor expression and vascularity in the bovine placentome. J Anim Sci. 2007;85(10):2464–2472. doi: 10.2527/jas.2006-805. [DOI] [PubMed] [Google Scholar]
- 46.Vonnahme KA, Lemley CO, Shukla P, O’Rourke ST. 2011 and 2012 early careers achievement awards: Placental programming: How the maternal environment can impact placental function. J Anim Sci. 2013;91:2467–2480. doi: 10.2527/jas.2012-5929. [DOI] [PubMed] [Google Scholar]
- 47.Canadian Council on Animal Care. CCAC guidelines on: The care and use of farm animals in research, teaching, and testing. 2009. ISBN: 978-0-919087-50-7.
- 48.Spicer LJ, Aad PY. Insulin-like growth factor (IGF) 2 stimulates steroidogenesis and mitosis of bovine granulosa cells through the IGF1 receptor: Role of follicle-stimulating hormone and IGF2 receptor. Biol Reprod. 2007;77(1):18–27. doi: 10.1095/biolreprod.106.058230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting findings in this study will be made available from UAL Dataverse (https://dataverse.library.ualberta.ca/dataverse/ualib): https://doi.org/10.7939/DVN/7CV6C3.


