Abstract
Background
The Mycobacterium tuberculosis (M.tb) protein kinase B (PknB) which is now proved to be essential for the growth and survival of M.tb, is a transmembrane protein with a potential to be a good drug target. However it is not known if this target remains conserved in otherwise resistant isolates from clinical origin. The present study describes the conservation analysis of sequences covering the inhibitor binding domain of PknB to assess if it remains conserved in susceptible and resistant clinical strains of mycobacteria picked from three different geographical areas of India.
Methods
A total of 116 isolates from North, South and West India were used in the study with a variable profile of their susceptibilities towards streptomycin, isoniazid, rifampicin, ethambutol and ofloxacin. Isolates were also spoligotyped in order to find if the conservation pattern of pknB gene remain consistent or differ with different spoligotypes. The impact of variation as found in the study was analyzed using Molecular dynamics simulations.
Results
The sequencing results with 115/116 isolates revealed the conserved nature of pknB sequences irrespective of their susceptibility status and spoligotypes. The only variation found was in one strains wherein pnkB sequence had G to A mutation at 664 position translating into a change of amino acid, Valine to Isoleucine. After analyzing the impact of this sequence variation using Molecular dynamics simulations, it was observed that the variation is causing no significant change in protein structure or the inhibitor binding.
Conclusions
Hence, the study endorses that PknB is an ideal target for drug development and there is no pre-existing or induced resistance with respect to the sequences involved in inhibitor binding. Also if the mutation that we are reporting for the first time is found again in subsequent work, it should be checked with phenotypic profile before drawing the conclusion that it would affect the activity in any way. Bioinformatics analysis in our study says that it has no significant effect on the binding and hence the activity of the protein.
Keywords: PknB, M. tuberculosis, Conservation, Multidrug resistant, Mutation, Sequencing
Background
The emergence of multi-drug and extensively drug resistant strains (MDR & XDR) of Mycobacterium tuberculosis (M.tb) has highlighted the need for new drugs to treat tuberculosis (TB). As the targets of current drugs are modified by the resistant organisms, newer targets which remain conserved in clinical isolates remain the choice for drug development. Serine/threonine protein kinases (STPK) remain one such target that plays important roles in mycobacterial cellular processes, including cell division, cell wall synthesis, cell metabolism, etc. [1]. Protein kinases A and B, encoded by pknA and pknB, respectively, are part of the operon carrying cistrons coding for protein phosphatase pstP, rodA (involved in cell shape control), and pbpA (involved in peptidoglycan synthesis). This locus is found near the origin of replication throughout the Mycobacterium genus [2]. Both these kinases have been found to be essential based on transposon-insertion experiments [3]. A role for PknB as a replication switch in response to hypoxia has been proposed with its essential role in reactivation of cells from the hypoxic state [4]. Hence, the Ser/Thr protein kinase PknB is proved to be essential for sustaining mycobacterial growth and this support the idea of the development of protein kinase inhibitors as new potential anti-tuberculosis drugs. However, to the best of our knowledge, not much work has been done for exploring the conservation of these genes in clinical isolates. In the present study, we probed the conservation pattern of the target region by sequencing this gene in clinical strains of M.tb and verified the same by deciphering the role of mutation in the drug-protein interactions in the PknB protein of M.tb using bioinformatics analysis. These isolates were either susceptible or resistant to the drugs being used currently and had different spoligotypes.
Methods
Ethics permission
The study was approved by the institutional ethics committee and all the experiments were undertaken by the trained staff at BSL3 facility.
Isolates used
A total of 116 Mycobacterium tuberculosis isolates were obtained from three different geographical regions of India, viz., Pune (Western India), Varanasi (Northern India) and Chennai (Southern India). Also, two sets of isolates were obtained from Pune wherein set 1 was an older collection of the isolates (2006–2007) and set 2 was a recent collection (2012–2013). Isolates from Chennai and Varanasi were collected during 2013–2014 (Table 1).
Table 1.
Demographic and laboratory profile of the isolates used
| Sr. no. | Study no. | HIV | Age | Sex | Collection date | Drug susceptibility profile (MGIT) | Spoligotype | Mutations in pknB gene |
|---|---|---|---|---|---|---|---|---|
| 1 | N1 | N | 35 | F | 6/3/2006 | Susceptible | T1-53 | Nil |
| 2 | N2 | N | 50 | F | 17/03/2006 | Susceptible | Orphan | Nil |
| 3 | N3 | N | 30 | M | 24/03/2006 | Susceptible | EAI5-355 | Nil |
| 4 | N4 | N | 50 | M | 27/03/2006 | Susceptible | EAI3_IND-11 | Nil |
| 5 | N5 | N | 22 | F | 29/03/2006 | Susceptible | CAS1_DELHI-26 | Nil |
| 6 | N6 | N | 38 | M | 28/04/2006 | Susceptible | CAS1_DELHI-26 | Nil |
| 7 | N7 | N | 55 | F | 8/5/2006 | Susceptible | CAS-486 | Nil |
| 8 | N8 | N | 29 | F | 15/06/2006 | Susceptible | CAS-357 | Nil |
| 9 | N9 | N | 23 | M | 16/06/2006 | Susceptible | MANU2-1634 | Nil |
| 10 | N10 | N | 23 | M | 19/06/2006 | Susceptible | EAI5-126 | Nil |
| 11 | N11 | N | 40 | F | 27/06/2006 | Susceptible | CAS1_DELHI-427 | Nil |
| 12 | N12 | N | 28 | M | 29/06/2006 | Susceptible | T3-37 | Nil |
| 13 | N13 | N | 40 | M | 29/06/2006 | Susceptible | CAS1_DELHI-25 | Nil |
| 14 | N14 | N | 24 | M | 10/10/2006 | Susceptible | CAS1_DELHI-26 | Nil |
| 15 | N15 | N | 22 | M | 20/10/2006 | Susceptible | EAI1_SOM-48 | Nil |
| 16 | N16 | N | 25 | F | 11/5/2006 | S,H | T2-52 | 664 (GTC-ATC) |
| 17 | N17 | N | 32 | M | 5/7/2006 | S,H,R | Orphan | Nil |
| 18 | N18 | N | 30 | M | 12/9/2006 | S,H,R | CAS2-288 | Nil |
| 19 | N19 | N | 21 | M | 22/09/2006 | S,H | T3-37 | Nil |
| 20 | N20 | N | 27 | M | 27/09/2006 | S | Orphan | Nil |
| 21 | N21 | N | 42 | M | 31/10/2006 | S | EAI5-126 | Nil |
| 22 | N22 | N | 26 | M | 15/01/2007 | S,H | Orphan | Nil |
| 23 | N23 | N | 21 | M | 18/01/2007 | H | H3-99 | Nil |
| 24 | N24 | N | 38 | M | 22/01/2007 | S,H,E | CAS1_DELHI-26 | Nil |
| 25 | N25 | N | 45 | F | 31/01/2007 | H | CAS1_DELHI-26 | Nil |
| 26 | N26 | P | 32 | M | 7/2/2007 | S,H,R | T2-1077 | Nil |
| 27 | N27 | P | 40 | M | 7/3/2007 | S,H | CAS2-288 | Nil |
| 28 | N28 | N | 50 | M | 8/3/2007 | S | H3-99 | Nil |
| 29 | N29 | P | 35 | M | 8/3/2007 | H | Orphan | Nil |
| 30 | S1 | NT | 35 | M | 27/03/2013 | Susceptible | CAS1_DELHI-26 | Nil |
| 31 | S2 | NT | 27 | M | 3/4/2013 | Susceptible | T1-53 | Nil |
| 32 | S3 | NT | 50 | M | 3/4/2013 | Susceptible | Orphan | Nil |
| 33 | S4 | NT | 24 | M | 3/4/2013 | H,R,E,OFX | Orphan | Nil |
| 34 | S5 | NT | 38 | M | 4/4/2013 | S,H,R,E | BEIJING-1 | Nil |
| 35 | S6 | NT | 45 | M | 5/4/2013 | R,H | T1-53 | Nil |
| 36 | S7 | NT | 31 | M | 8/4/2013 | Susceptible | T1-53 | Nil |
| 37 | S8 | NT | 50 | M | 3/4/2013 | R,H | CAS1_DELHI-1789 | Nil |
| 38 | S9 | NT | 35 | M | 8/4/2013 | Susceptible | Orphan | Nil |
| 39 | S10 | NA | 55 | M | 8/4/2013 | OFX | MANU1-100 | Nil |
| 40 | S11 | NA | NA | NA | NA | S,H,E,OFX | BEIJING-1 | Nil |
| 41 | S12 | P | 40 | F | 2012–2013 | Susceptible | EAI3_IND-11 | Nil |
| 42 | S13 | P | NA | NA | 2012–2013 | Susceptible | CAS1_DELHI-26 | Nil |
| 43 | S14 | P | 35 | M | 2012–2013 | Susceptible | CAS1_DELHI-26 | Nil |
| 44 | S15 | P | 48 | F | 2012–2013 | H | CAS1_DELHI-26 | Nil |
| 45 | S16 | P | 26 | M | 2012–2013 | H,R | CAS1_DELHI-26 | Nil |
| 46 | S17 | P | 30 | F | 2012–2013 | H | CAS1_DELHI-26 | Nil |
| 47 | S18 | P | 31 | M | 2012–2013 | H,R,OFX | Orphan | Nil |
| 48 | S19 | P | 36 | M | 2012–2013 | H | Orphan | Nil |
| 49 | S20 | P | 23 | M | 2012–2013 | H | Orphan | Nil |
| 50 | C1 | NT | NA | NA | 2013 | R,H, OFX | T5-1268 | Nil |
| 51 | C2 | NT | NA | NA | 2013 | R,H | EAI3_IND-11 | Nil |
| 52 | C3 | NT | NA | NA | 2013 | R,H | BEIJING-1 | Nil |
| 53 | C4 | NT | NA | NA | 2013 | R,H,OFX | Orphan | Nil |
| 54 | C5 | NT | NA | NA | 2013 | R,H | Orphan | Nil |
| 55 | C6 | NT | NA | NA | 2013 | R,H,OFX | CAS1_DELHI-381 | Nil |
| 56 | C7 | NT | NA | NA | 2013 | R,H | T1–T2-78 | Nil |
| 57 | C8 | NT | NA | NA | 2013 | R,H,OFX | T2-52 | Nil |
| 58 | C9 | NT | NA | NA | 2013 | R,H,OFX | Orphan | Nil |
| 59 | C10 | NT | NA | NA | 2013 | R,H,OFX | Orphan | Nil |
| 60 | C11 | NT | NA | NA | 2013 | R,H | T1-53 | Nil |
| 61 | C12 | NT | NA | NA | 2013 | R,H | Orphan | Nil |
| 62 | C13 | NT | NA | NA | 2013 | S,H,R,E,OFX | Orphan | Nil |
| 63 | C14 | NT | NA | NA | 2013 | Susceptible | CAS1_DELHI-26 | Nil |
| 64 | C15 | NT | NA | NA | 2013 | S,H,E | T2-52 | Nil |
| 65 | C16 | NT | NA | NA | 2013 | S,H,R,E | Bejing-1 | Nil |
| 66 | C17 | NT | NA | NA | 2013 | S,E | CAS1_DELHI-26 | Nil |
| 67 | C18 | NT | NA | NA | 2013 | S,E | Orphan | Nil |
| 68 | C19 | NT | NA | NA | 2013 | S | Orphan | Nil |
| 69 | C20 | NT | NA | NA | 2013 | S | Orphan | Nil |
| 70 | C21 | NT | NA | NA | 2013 | Susceptible | EA15-934 | Nil |
| 71 | C22 | NT | NA | NA | 2013 | S,H,R,E | EA15-934 | Nil |
| 72 | C23 | NT | NA | NA | 2013 | Susceptible | EAI3_IND-11 | Nil |
| 73 | C24 | NT | NA | NA | 2013 | Susceptible | Orphan | Nil |
| 74 | C25 | NT | NA | NA | 2013 | Susceptible | Bejing-1 | Nil |
| 75 | C26 | NT | NA | NA | 2013 | S,H,R,E | Nil | |
| 76 | C27 | NT | NA | NA | 2013 | Susceptible | T1-804 | Nil |
| 77 | C28 | NT | NA | NA | 2013 | H | EAI3_IND | Nil |
| 78 | C29 | NT | NA | NA | 2013 | Susceptible | EAI3_IND | Nil |
| 79 | C30 | NT | NA | NA | 2013 | S,H,R,E,OFX | Orphan | Nil |
| 80 | C31 | NT | NA | NA | 2013 | Susceptible | Orphan | Nil |
| 81 | C32 | NT | NA | NA | 2013 | Susceptible | CAS1_DELHI | Nil |
| 82 | C33 | NT | NA | NA | 2013 | S,H, OFX | Bejing-1 | Nil |
| 83 | C34 | NT | NA | NA | 2014 | Susceptible | EAI3_IND-11 | Nil |
| 84 | C35 | NT | NA | NA | 2014 | S,H,OFX | Orphan | Nil |
| 85 | C36 | NT | NA | NA | 2014 | OF | U-1391 | Nil |
| 86 | C37 | NT | NA | NA | 2014 | Susceptible | EAI3_IND-11 | Nil |
| 87 | C38 | NT | NA | NA | 2014 | Susceptible | EAI3_IND-11 | Nil |
| 88 | C39 | NT | NA | NA | 2014 | R | EAI3_IND-11 | Nil |
| 89 | C40 | NT | NA | NA | 2014 | Susceptible | Orphan | Nil |
| 90 | C41 | NT | NA | NA | 2014 | Susceptible | EAI3_IND-11 | Nil |
| 91 | B1 | N | 18 | F | 2013–2014 | S,H,R,E,OFX | BEIJING-1 | Nil |
| 92 | B2 | N | 20 | M | 2013–2014 | S,H,R,E,OFX | H1-47 | Nil |
| 93 | B3 | P | 25 | F | 2013–2014 | S | LAM2–LAM4-194 | Nil |
| 94 | B4 | N | 25 | M | 2013–2014 | S,H,R,E, OFX | LAM-9 | Nil |
| 95 | B5 | N | 40 | M | 2013–2014 | S,H | CAS2-288 | Nil |
| 96 | B6 | N | 25 | M | 2013–2014 | Susceptible | Orphan | Nil |
| 97 | B7 | N | 47 | M | 2013–2014 | S,H,R,E | Orphan | Nil |
| 98 | B8 | N | 30 | M | 2013–2014 | S,H,R,E | BEIJING-1 | Nil |
| 99 | B9 | N | 20 | M | 2013–2014 | S,H,R,E,OFX | T1-53 | Nil |
| 100 | B10 | N | 21 | M | 2013–2014 | H,R | Orphan | Nil |
| 101 | B11 | N | 45 | M | 2013–2014 | H,R,OFX | BEIJING-1 | Nil |
| 102 | B12 | N | 23 | M | 2013–2014 | S,H | T1-53 | Nil |
| 103 | B13 | N | 23 | M | 2013–2014 | Susceptible | EAI1_SOM-235 | Nil |
| 104 | B14 | N | 22 | F | 2013–2014 | S,H,R,OFX | CAS1_DELHI | Nil |
| 105 | B15 | N | 22 | F | 2013–2014 | S,H,R,E,OFX | CAS1_DELHI | Nil |
| 106 | B16 | N | 70 | M | 2013–2014 | H,R,OFX | Orphan | Nil |
| 107 | B17 | N | 9 | F | 2013–2014 | H,R,E,OFX | BEIJING-1 | Nil |
| 108 | B18 | N | 19 | M | 2013–2014 | H,R,E | Orphan | Nil |
| 109 | B19 | N | 25 | F | 2013–2014 | H | T1-53 | Nil |
| 110 | B20 | P | 45 | F | 2013–2014 | Susceptible | BEIJING-1 | Nil |
| 111 | B21 | N | 30 | F | 2013–2014 | S,H,R,E,OFX | BEIJING-1 | Nil |
| 112 | B22 | N | 18 | F | 2013–2014 | S,H,R,E,OFX | BEIJING-1 | Nil |
| 113 | B23 | N | 46 | F | 2013–2014 | S | Orphan | Nil |
| 114 | B24 | N | 50 | M | 2013–2014 | H,R,OFX | BEIJING-1 | Nil |
| 115 | B25 | N | NA | NA | 2013–2014 | OFX | T1-1567 | Nil |
| 116 | B26 | N | NA | NA | 2013–2014 | H,S | U-172 | Nil |
N1–N29 isolates obtained from NARI, Pune; S1–S20 isolates obtained from STDC, Pune, Maharashtra; C1–C40 isolates obtained from NIRT, Chennai, Tamil Nadu; B1–B26 isolates obtained from BHU, Varanasi, Uttar Pradesh
NIRT National Institute for Research in Tuberculosis, BHU Banaras Hindu University, STDC State TB Training and Demonstration Centre, S resistant to streptomycin, H resistant to isoniazid, R resistant to rifampicin, E resistant to ethambutol, OFX resistant to ofloxacin, N negative, NT not tested, NA information not available
Sub-culturing
Upon arrival to our laboratory, the isolates were sub-cultured on Löwenstein-Jensen (LJ) medium and incubated till sufficient growth was indicated by typical non-pigmented, rough, dry colonies.
Drug susceptibility testing
Isolates were subjected to drug susceptibility testing on BACTEC™ MGIT™ 960 Mycobacterial Detection System (BD, NJ, USA) using SIRE kit for the susceptibility testing against streptomycin (S), isoniazid (I), rifampin (R) and ethambutol (E). For susceptibility testing against ofloxacin, blank tubes of medium were used wherein the final concentration of ofloxacin was kept to 2 μg/ml as per the CLSI standards using filtered stock solution of the drug. The inoculum was prepared from fresh LJ medium by following the methods used by Devasia and co-workers [5]. A growth control containing no antibiotic and a sterile control without inoculation were also used with every set of experiment.
DNA isolation and amplification of pknB gene
Genomic DNA isolation was done from freshly sub-cultured isolates using a cetyltrimethyl ammonium bromide (CTAB) based modified method as reported by earlier workers [6, 7]. The DNA so obtained was quantitated using NanoDrop 1000 spectrophotometer (Thermo, USA) and purity and integrity were checked on 0.8% agarose gel after electrophoresis. A length of 876 bp covering the cytosolic portion of the protein was amplified using forward primer as AATGACCACCCCTTCCCA and reverse primer as TCGGCA TCGGTGAGCACTTT designed with Mycobacterium tuberculosis H37Rv (NC_000962;16594-17469) as a template. The amplification reaction was done using KAPA HiFi DNA Polymerase (KAPA Biosystems, USA) using manufacturer’s instructions and annealing at 65 °C.
Spoligotyping
Spoligotyping was performed using the standard method at National Institute for Research in Tuberculosis, Chennai [8]. In brief, the spacers between the direct repeats in the target region were amplified by using a primer set, primer Dra 5′-CC AAG AGG GGA CGG AAA C-3′ and biotinylated primer Drb 5′-GGT TTT GGG TCT GAC GAC-3′. The polymerase chain reaction (PCR) products were then hybridized to a Biodyne C membrane (Isogen Bioscience). This membrane contains immobilized synthetic oligomeric spacer sequences derived from the direct-repeat region of M. tuberculosis H37Rv and Mycobacterium bovis BCG. Hybridized DNA was detected using an enhanced chemiluminescence kit (Amersham International), with exposure to X-ray film producing a pattern or profile reminiscent of a bar code. These were matched with the SITVIT web database to get spoligotypes matched with available profiles (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/).
Sequencing and sequence analysis
For sequencing of the amplified gene of pknB, same primers which were used for amplification were employed. The sequencing of plasmid DNA was carried out using Big Dye Terminator v. 3.1 cycle sequencing kit and an ABI 3730xl DNA analyzer, according to the manufacturer’s protocol. Nucleotide sequence analysis and curing were done using Applied Biosystem SeqScape v.2.5. The cured sequences were compared for sequence similarities using Rv0014c from NC_000962 as a template in SeqScape software.
Bioinformatics analysis
Based on sequencing of the pknB gene, the sequence that showed mutation was taken for bioinformatics analysis to access the impact of the change. Molecular dynamics simulations (MD) tool have potential to provide detailed information on the fluctuations and conformational changes of proteins and hence was used here. MD-simulations were performed to the docked complex to evaluate the stability, conformational changes and getting insights into the natural dynamic in solutions. MD-simulations were performed using GROMACS version-4.6.4 [9–11], with the AMBER99SB force field [12]. The ligand was parameterized by applying the standard RESP procedure using Antechamber [13], where charges for free ligand were derived from HF/6-31G* calculation in Gaussian03 [14]. Histidine residues were assumed to be charged, with the ND and NE atoms protonated; Arginine and Lysine residues were assumed to be protonated. All systems were solvated using explicit TIP3P water model [15], in a cubic box with a margin of 10 Å and neutralized by adding Sodium counter-ions. The Particle-Mesh Ewald method was applied to calculate long-range electrostatic interactions with a cutoff distance range of 10 Å, [16] and a Lennard-Jones 6–12 potential was used to evaluate van-der-Waals interactions within the cut-off range of 10 Å. The LINCS algorithm of fourth order expansion was used to constrain bond lengths [17]. After solvation and neutralization, each system was energy minimized for 10,000 steps using steepest-descent optimization method to remove poor van-der-Waals contacts in the initial geometry. After minimization, two stages of equilibration were conducted. First, the system was equilibrated for 100 ps with position restraints of 10,000 kJ/mol on all heavy atoms. A constant temperature of 300 K was maintained using the V-rescale algorithm [18], with a coupling time of 0.1 ps and separate baths for the solute and the solvent. Second, the pressure was kept constant at 1 bar using the Parrinello–Rahman pressure coupling scheme with a time constant of 2 ps [19]. Initial velocities were generated randomly using a Maxwell–Boltzmann distribution corresponding to 300 K. Neighbor lists were updated every 10 fs using a group cut-off scheme. Finally, the production run was performed for 100 ns without restraints at 300 K in the isothermal-isobaric ensemble.
Interaction analysis was performed using g_dist tool in the GROMACS package to measure the inter-atomic distances. The g_rms tool of the GROMACS package was used to calculate the root mean square deviations (RMSD) during the trajectories taking the minimized crystal structure as a reference. Root mean square fluctuations (RMSF) of the backbone of each residue were calculated by g_rmsf while g_mmpbsa tool is used to calculate the binding energy.
Results
DST and spoligotypes
The isolates had a variety of characteristics in terms of drug resistance to the drugs (SIRE/SHRE) with ofloxacin and spoligotypes as elaborated in Table 1. As obtained in Table 1, isolates used in the study ranged from pan-susceptible to complete resistance towards the tested drugs. The same stands true for their spoligotypes, reflecting different dominances in different geographical locations with more of Beijing strains in recent isolates.
Sequencing results
No mutation, except in one, could be observed in the region sequenced. A single mutation that was found was G to A resulting in the change of amino acid from Valine (GTC) to Isoleucine (ATC) in isolate N16. The isolate showed a change in one nucleotide as compared with wild-type sequence at the 664th position from the start of the codon i.e. A (TG….); this corresponds to the position 16,807 of the template. The sequence was submitted to NCBI and have Genbank accession number as KX029321. This change is reported as Non-synonymous SNP in Polytb (http://pathogenseq.lshtm.ac.uk/polytb) and GMTV (http://mtb.dobzhanskycenter.org/) SNP databases. However, the isolation sites were other than India, spoligotypes were different and no comment on its impact on protein structure or function could be found.
MD simulations results
The conformation achieved from 100 ns of simulation is more stable and credible than docked conformations. This can be justified by the fact that docking methods have some inherent pitfalls and approximations. However, MD-simulations represent more realistic conditions with the physiological environment. Therefore, the binding orientation of Mitoxantrone (ligand) predicted through MD-simulations shows better correlation to their biologically active state. The structural and dynamical property of the complex has been analyzed through the trajectories obtained from the MD-simulations. The trajectory data of complex were plotted for energy, RMSD, and RMSF and the interaction analysis of the ligand have been done within the binding pocket.
The potential energy of the system is quite stable throughout the simulations as shown in Fig. 1. The trajectories reveal that secondary structure of the protein is maintained throughout the simulation. RMSD plot in Fig. 2 shows that after an initial equilibration period of about 25 ns the system reached a plateau and the RMSD value never crossed the mark of 3.5 Å from the initial structure in the entire simulation of 100 ns. The flexibility of the structure is measured in terms of RMSF. Figure 3 demonstrates the flexibility of backbone with respect to an average structure which is below 2 Å. Only for loops it is a little bit higher and reaches a maximum of 3 Å. The interactions which are responsible for keeping the ligand inside the pocket are analyzed by a number of H-bond interactions. Figure 4 reveals that average 3 H-bonds between 2FUM protein and the ligand, Mitoxantrone, is quite persistent and sometimes 4 H-bonds were also seen which reached a maximum of 6–8. Further, we analyzed the particular residue responsible for these interactions as well.
Fig. 1.
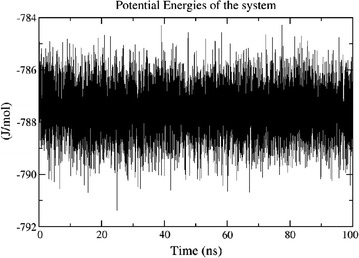
Energy plot of MD simulations for protein–ligand (Mitoxantrone) complex as a function of time
Fig. 2.
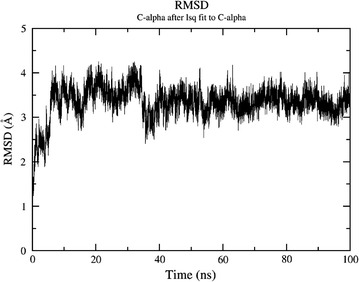
Root mean square deviation (RMSD) of Protein of C-alpha atoms with respect to the time over the course of 100 ns of MD simulation
Fig. 3.
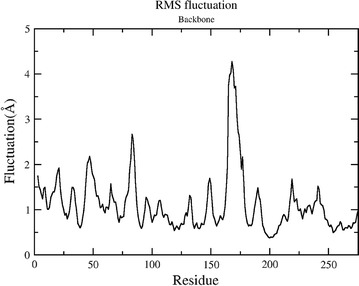
Root mean square fluctuation (RMSF) of residue backbone with respect to the average structure during the course of 100 ns of MD simulations
Fig. 4.
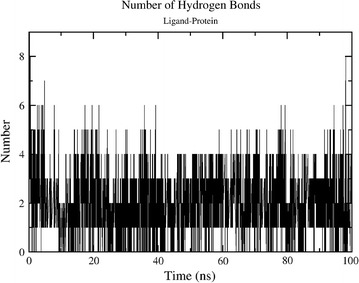
Average number of Hydrogen bond between ligand and protein during the entire 100 ns of simulations
Interaction analysis
The persistence of interaction is quantified by distance analysis between particular residues in the binding pocket with the specific atoms of Mitoxantrone. Figure 5 reveals the most persistent interaction found in simulations of 100 ns. Figure 5a shows the interaction of MET-155 with the ligand in which after initial equilibration period of 40 ns the interaction is quite stable and distance is below 3 Å while with VAL-25 it is quite persistent (about 3.75 Å) from the beginning of the simulation. The most surprising interaction found with ALA-142 with the ligand. Figure 5c shows that initially residue was distant (~7 Å apart) but after equilibration time the conformation of protein switched in a manner that ALA-142 started interacting with the ligand. Similarly, Fig. 5d also shows that initially the residue MET-92 was distant from the ligand but after equilibration time the conformation of protein switched in a manner that MET-92 started interacting with the ligand. These interactions were the major binding source for the ligand in the binding cavity.
Fig. 5.
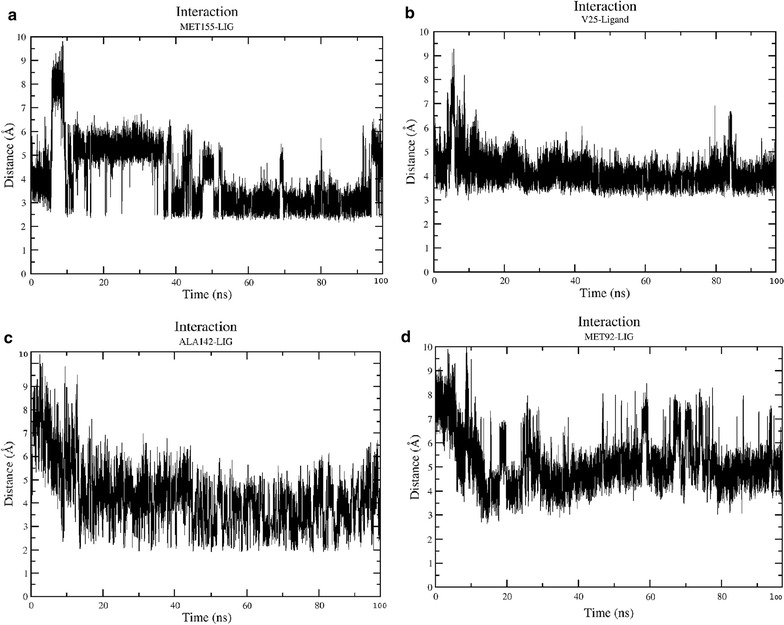
Interaction of residues with the ligand. a Interaction of Methionine-155 with the ligand. b Interaction of Valine-25 with the ligand. c Interaction of Alanine-142 with the ligand d Interaction of Methionine-142 with the ligand
MMPBSA results
The binding energy of the ligand is calculated by MMPBSA method which is allowing the calculation of non-bonded interactions i.e., Van-der-Waal and the electrostatic terms between two sets of atoms along the length of trajectory. Each snapshot from every nanosecond was taken for our calculations. The method also allowed the calculation of polar solvation energy and SASA energy as well. In our calculation, the overall binding energy of ligand with the protein is −71.564 kJ/mol, where negative term confirms substantial affinity of the receptor for the ligand. It also confirms our molecular dynamics simulation results. In residue-wise contribution in the binding, MET-155 plays a key role and shows maximum contribution. Our interaction analysis shows persistent interaction with the ligand (Table 2). This critical residue may provide guidance for the rational design to discover a more potent drug (Fig. 6).
Table 2.
MMPBSA calculation results
| Energetic contributions | KJ/mol |
|---|---|
| Van-der-Waal energy | −225.117 ± 11.889 |
| Electrostatic energy | −93.523 ± 15.703 |
| Polar solvation energy | 268.278 ± 23.834 |
| SASA energy | −21.201 ± 0.721 |
| Binding energy | −71.564 ± 19.084 |
Fig. 6.
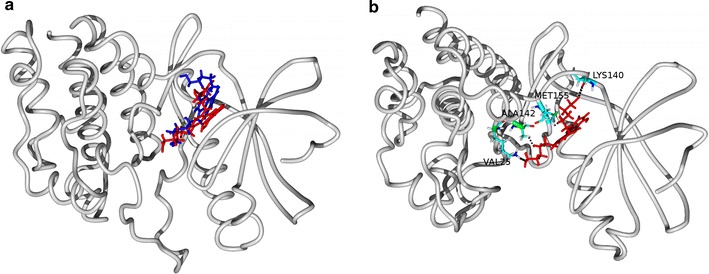
The orientation of Mitoxantrone before simulation in blue while after a simulation in red. Interaction of residue shown with side chains. Protein is represented in tube (grey colour) in both the figures (a, b)
Discussion
The M.tb genome encodes 11 serine/threonine protein kinases (STPK) which offer attractive targets for inhibitor development [20]. PknB is part of the 11 eukaryotic-like serine–threonine protein kinases listed as an essential protein in the M.tb TraSH and NGS datasets [21]. The necessity of pknB was verified by gene knockdown experiments, which showed a remarkable decrease in bacterial viability in liquid culture upon depletion of the kinase [22]. According to Ortega and co-workers, pknB provides a highly potential drug target against Mtb as it controls both the active replicating and latent nonreplicating forms of Mtb [4]. They tested the role of serine/threonine protein kinases—signaling molecules and found that kinase depletion led to decrease in bacterial viability and they specifically identified the PknB kinase as a critical regulator of the oxygen-dependent replication switch. These data show that signaling through PknB modulates the growth and replication state of Mtb in response to oxygen. Therefore, it can also be a valuable drug target for latent TB infection.
PknB has autophosphorylation properties [23], with a cross-phosphorylation potential of other STPKs such as PknA, and PknG [24]. Moreover, PknB seems to control major metabolic pathways directly via phosphorylation of multiple protein substrates, such as the protein regulator GarA that halts the TCA cycle [25], the KasA/B proteins involved in the biosynthesis of the essential mycolic acid [26], and the Wag31 proteins involved in cell division and morphology [27]. Likewise, virulence factors SigH and RshA [28], and cell wall biosynthetic enzymes GlmU or PBPA are also influenced [29, 30]. Thus, PknB is established as an important participant and a regulator of cell division and growth in M. tuberculosis proving its essentiality. However, there are no studies, to the best of our knowledge to say about the conservation status of this gene in clinical isolates with existing pressure of drugs being used for other targets.
This study tried to survey the same by sequencing the most important region of the cytoplasmic domain that participates in ligand/inhibitor binding. The crystal structure studies demonstrated that mitoxantrone is an ATP-competitive inhibitor of PknB and Leu17, Gly18, Val25, Ala38, Met 92, Glu93, Tyr94 and Val95 in the N-terminal lobe, Met145 and Met155 in the C-terminal lobe are the important residues taking part in the complex formation [31]. Moreover, we used high fidelity enzyme for amplification of pknB gene from genomic DNA to ensure error-free amplification before sequencing.
Conclusions
In our study, pknB was confirmed to be conserved, even in clinical isolates with different drug resistance and spoligotype profile. This was observed in sequencing experiments with 116 clinical isolates having susceptibility profiles ranging from multidrug resistant isolates with resistance to individual drugs including fluoroquinolones to the ones with complete susceptibility towards tested drugs. The choice of clinical strains of M.tb to probe the conservation pattern of the target region is the best strategy because of different drug exposures to them along with host’s immune pressure. Such isolates are under pressure to acquire drug resistance by the modification of the drug targets. Bioinformatics studies using hypothetical mutations in binding sites were also done to access the mutations which may disturb the drug binding and induce resistance. The absence of mutations in overall sequence including putative drug binding domains proved that pknB remain a good target for drug development. Even the highly resistant isolates did not have any mutations in this region. This also reveals that sequences in pknB are not influenced by the change in other genomic regions. However, a study is required to see if constant usage of inhibitors of this particular protein exerts a selection pressure and how susceptible is the target for mutations under specific drug pressure.
Authors’ contributions
AG: Execution of the work in the laboratory, manuscript review. SKP: Bioinformatics work, manuscript review. DP: Execution of the work in the laboratory. NAF: Execution of the work in the laboratory. SR: Execution of the work in the laboratory. DS: Bioinformatics work. SS: Spoligotyping of the isolates. PS: Collection and identification of isolates from BHU. MP: Collection and identification of isolates and their details from NARI. SB: Collection and identification of isolates and their details from STDC. GS: Collection and identification of isolates and their details from NIRT. KRU: Manuscript review; isolates and their details from NIRT. SA: Manuscript review; isolates and their details from BHU. VN: Principle Investigator of the project, conception of the work, manuscript writing and review. All authors read and approved the final manuscript.
Acknowledgements
Authors are thankful to Director, NARI, Director, NIRT, Joint Director of Health Services (Leprosy and TB), Pune and CMO, STDC, Pune for their kind support in obtaining isolates and related information for this study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets generated and/or analysed during the current study are available with the PI of the study and segregated data about the isolates is available with the authors from different institutes as listed in the author’s list. A detailed report with data has been submitted to funding agency (ICMR) also.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by institutional ethics committee.
Source of funding
This work was supported by Indian Council of Medical Research (extramural Grant No. AMR/45/2011-ECD-I).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Anamika Gupta and Sudhir K. Pal contributed equally to this work
Najneen A. Fakir and Sunita Rathod contributed equally to this work
Contributor Information
Anamika Gupta, Email: anamikamicrobiology@gmail.com.
Sudhir K. Pal, Email: sudhirkumarpal001@gmail.com
Divya Pandey, Email: divivittory@gmail.com.
Najneen A. Fakir, Email: najuka_aku@rediffmail.com
Sunita Rathod, Email: sunitarathod11@gmail.com.
Dhiraj Sinha, Email: meetdhirajsinha@gmail.com.
S. SivaKumar, Email: shanmugamsiva@nirt.res.in
Pallavi Sinha, Email: pallavisinhamicro@gmail.com.
Mycal Periera, Email: pereiramhc@yahoo.com.
Shilpa Balgam, Email: shilpavedant@gmail.com.
Gomathi Sekar, Email: gomathi.sekar@gmail.com.
K. R. UmaDevi, Email: umadevi.r@nirt.res.in
Shampa Anupurba, Email: shampa_anupurba@yahoo.co.in.
Vijay Nema, Phone: +91-20-27331200, Email: dr.vijaynema@gmail.com, Email: vnema@nariindia.org.
References
- 1.Chawla Y, Upadhyay S, Khan S, Nagarajan SN, Forti F, Nandicoori VK. Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J Biol Chem. 2014;289(20):13858–13875. doi: 10.1074/jbc.M114.563536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narayan A, Sachdeva P, Sharma K, Saini AK, Tyagi AK, Singh Y. Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol Genom. 2007;29(1):66–75. doi: 10.1152/physiolgenomics.00221.2006. [DOI] [PubMed] [Google Scholar]
- 3.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 4.Ortega C, Liao R, Anderson LN, Rustad T, Ollodart AR, Wright AT, Sherman DR, Grundner C. Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch. PLoS Biol. 2014;12(1):e1001746. doi: 10.1371/journal.pbio.1001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devasia RA, Blackman A, May C, Eden S, Smith T, Hooper N, Maruri F, Stratton C, Shintani A, Sterling TR. Fluoroquinolone resistance in Mycobacterium tuberculosis: an assessment of MGIT 960, MODS and nitrate reductase assay and fluoroquinolone cross-resistance. J Antimicrob Chemother. 2009;63(6):1173–1178. doi: 10.1093/jac/dkp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Short protocols in molecular biology. New York: Wiley; 2002. pp. 2–10. [Google Scholar]
- 7.Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43(6):2996–2997. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PW, Martin C, Palittapongarnpim P, Plikaytis BB, Riley LW, Yakrus MA, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37(8):2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 10.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Comm. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 11.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 14.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, et al. GAUSSIAN 03 (revision C.02) Wallingford: Gaussian, Inc.; 2004. [Google Scholar]
- 15.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 16.Darden T, York D, Pedersen L, Ewald P. An N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 17.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comp Chem. 1997;18(12):1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 18.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 19.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 20.Prisic S, Husson RN. Mycobacterium tuberculosis serine/threonine protein kinases. Microbiol Spectr. 2014;2(5). [DOI] [PMC free article] [PubMed]
- 21.Le Chevalier F, Cascioferro A, Majlessi L, Herrmann JL, Brosch R. Mycobacterium tuberculosis evolutionary pathogenesis and its putative impact on drug development. Future Microbiol. 2014;9(8):969–985. doi: 10.2217/fmb.14.70. [DOI] [PubMed] [Google Scholar]
- 22.Forti F, Crosta A, Ghisotti D. Pristinamycin-inducible gene regulation in mycobacteria. J Biotechnol. 2009;140(3–4):270–277. doi: 10.1016/j.jbiotec.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Mieczkowski C, Iavarone AT, Alber T. Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. EMBO J. 2008;27(23):3186–3197. doi: 10.1038/emboj.2008.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19(14):1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, Pritsch O, Obal G, Baumgartner J, Vialaret J, Johnsson K, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70(6):1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 26.Veyron-Churlet R, Molle V, Taylor RC, Brown AK, Besra GS, Zanella-Cleon I, Futterer K, Kremer L. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J Biol Chem. 2009;284(10):6414–6424. doi: 10.1074/jbc.M806537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154(Pt 3):725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 28.Park ST, Kang CM, Husson RN. Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105(35):13105–13110. doi: 10.1073/pnas.0801143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J Mol Biol. 2009;386(2):451–464. doi: 10.1016/j.jmb.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152(Pt 2):493–504. doi: 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- 31.Wehenkel A, Fernandez P, Bellinzoni M, Catherinot V, Barilone N, Labesse G, Jackson M, Alzari PM. The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett. 2006;580(13):3018–3022. doi: 10.1016/j.febslet.2006.04.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available with the PI of the study and segregated data about the isolates is available with the authors from different institutes as listed in the author’s list. A detailed report with data has been submitted to funding agency (ICMR) also.


