Abstract
Infected cell protein 0 (ICP0) is a 775-residue multifunctional herpes simplex virus protein associated with numerous functions related to transactivation of gene expression and repression of host defenses to infection. We report that an uncharted domain of ICP0 located between residues 245 and 510 contains multiple SH3 domain binding motifs similar to those required for binding to CIN85, the Mr 85,000 protein that interacts with Cbl. CIN85 and Cbl are involved in endocytosis and negative regulation of numerous receptor tyrosine kinases. We report that ICP0 binds CIN85 in a reciprocal manner and that the complexes pulled down by ICP0 also contain Cbl. We tested the role of ICP0 in the down-regulation of receptor tyrosine kinases by using epidermal growth factor receptor (EGFR) as a prototypic receptor. In transfection assays, ICP0, in the absence of other viral genes, down-regulated EGF-dependent expression of a reporter gene (luciferase). ICP0 also down-regulated both total and cell surface levels of EGFR in EGF-independent manner. In wild-type virus-infected cells, the surface levels of EGFR were also decreased in the absence of EGF stimulation. Stimulation by EGF enhanced the decrease in surface EGFR. We conclude that ICP0 encodes SH3 domain binding sites that function to down-regulate signaling pathways associated with receptor tyrosine kinases. The results suggest that ICP0 precludes signaling to the infected cells through the receptor tyrosine kinases.
Keywords: endocytosis, receptor tyrosine kinase, ubiquitin ligase
The infected cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) is a multifunctional 775-aa protein expressed immediately after infection. As reviewed in detail elsewhere, in transduced cells, ICP0 enhances the expression of genes introduced by infection or transfection (reviewed in ref. 1). In infected cultured cells, ICP0 plays an important role at low multiplicities of infection but no obvious role at high multiplicities. ICP0 does play a key role in enabling viral replication and spread in experimental animal systems. ICP0 has been shown to interact with a large number of cellular proteins. Among these proteins are cyclin D3, cdc34, the transcriptional factor BMAL1, the translation elongation factor 1δ, and the ubiquitin specific protease USP7 (1). ICP0 is encoded in three exons of 19, 221, and 533 codons, respectively. The functions identified to date map in sequences encoded by exon 2 and the carboxyl terminal 160 amino acids of exon 3.
In this article, we report a function of ICP0 mapping in the uncharted domain of sequences encoded in exon 3. Specifically, we report that ICP0 contains several putative SH3 domain binding sites. The recurrent motif of these binding sites conforms to the PX(P/A)XXR recognition consensus of SH3 domains of CIN85 (2), the Mr 85,000 adapter protein that binds Cbl, a multifunctional oncoprotein that also functions as an E3 ubiquitin ligase (ref. 3 and references therein). We thus report that ICP0 binds CIN85 in a reciprocal manner and that the complex containing ICP0 and CIN85 also contains Cbl. The significance of this observation stems from the role of Cbl and CIN85 in the biology of cells. Receptor tyrosine kinases activated by binding to their cognate ligands are negatively regulated by endocytosis through either degradation or recycling (4). This process is mediated by the Cbl–CIN85 and endophilin (a regulatory component of clathrin-coated vesicles) complex (5, 6). In the studies reported here, we used the epidermal growth factor receptor (EGFR) as a prototypic receptor tyrosine kinase known to be regulated by the CIN85–Cbl complex to define the role of ICP0 in the EGFR signaling pathway. Exposure of cultured cells to EGF results in activation of the signaling pathway concurrent with the rapid disappearance of EGFR from the cell surface. In this report, we show that EGF-dependent gene activation is blocked in cells transfected with ICP0. ICP0 alone, in the absence of other viral genes, caused a reduction in the total and cell surface amounts of EGFR. Cell surface levels of EGFR were also reduced in cells infected with wild-type virus.
Materials and Methods
Cells and Viruses. HEK293 and HEp-2 cells were obtained from American Type Culture Collection (Manassas, VA). Cells were maintained in DMEM, supplemented with 10% FBS (HEK293) or 5% newborn calf serum (HEp-2 cells). HSV-1(F) is the prototype HSV-1 strain used in this laboratory. The HSV-1(F) derived mutants lacking UL13 (R7356) or US3 (R7041) were described in refs. 7 and 8.
Antibodies. The antibodies used in these studies were anti-CIN85 (Calbiochem, no. 231006), anti-Cbl (Santa Cruz Biotechnology, c-15), anti-GST (Santa Cruz Biotechnology), anti-EGFR (Upstate Biotechnology, Lake Placid, NY, no. 06-129), and monoclonal anti-ICP0 antibody (Goodwin Cancer Research Institute, Plantation, FL).
Plasmids. Reporter construct SRE-Luc expressing luciferase under the control of a promoter containing serum response element (SRE), pRK5-EGFR and pRK5-Cbl were obtained from Ivan Dikic (Goethe University Medical School, Frankfurt) (described in ref. 5). pcDNA4/TO-LacZ was purchased from Invitrogen. Mammalian expression vector MTS1 and ICP0 expression vector MTS1-ICP0 was described in refs. 9 and 10.
GST Fusion Proteins. The SH3 domains of CIN85 fused to GST, designated GST-SH3–1, GST-SH3–2, or GST-SH3–3, were described in ref. 2. GST-ICP0 constructs were made by fusing in-frame relevant ICP0 fragments (amino acids 1–19, amino acids 20–241, amino acids 245–395, amino acids 245–510, or amino acids 543–768) to GST and verified by DNA sequencing at the University of Chicago Cancer Research Center DNA Sequencing Facility. Competent Escherichia coli strain BL-21 cells were transformed with the GST fusion constructs, induced by isopropyl β-d-thiogalactoside and GST fusion proteins were immobilized to glutathione-Sepharose beads and purified according to the manufacturer's protocol (Amersham Pharmacia).
GST Pull-Down Assays. Cells were harvested in cold PBS, lysed by brief sonication in RIPA buffer (100 mM Tris, pH 8.0/140 mM NaCl/1% Triton X-100/0.1% SDS/1% deoxycholic acid/0.5 mM EDTA/protease and phosphatase inhibitors) and clarified by centrifugation in a Sorvall biofuge pico microcentrifuge at 13,000 rpm for 20 min at 4°C. Aliquots of total cell lysate were diluted in pull-down buffer (50 mM Tris, pH 7.5/100 mM NaCl/0.1% Nonidet P-40/1 mg/ml BSA) to 1 ml each and reacted overnight at 4°C with GST fusion proteins bound to glutathione-Sepharose beads. The GST beads were rinsed in pull-down buffer and resuspended in equal volume of 1× SDS loading buffer (50 mM Tris, pH 6.8/2.75% sucrose/5% 2-mercaptoethanol/2% SDS). The solubilized proteins were boiled and electrophoretically separated in a denaturing 10% polyacrylamide gel.
Immunoblots. Electrophoretically separated proteins were electrically transferred to a nitrocellulose membrane, blocked at room temperature with 5% nonfat dry milk in PBS and reacted with primary antibody diluted in PBS/1% BSA (anti-CIN85, 1:500; anti-Cbl, 1:500; anti-GST, 1:2,000; anti-ICP0, 1:2,000; anti-EGFR, 1:1,000), followed by an appropriate secondary antibody conjugated to either peroxidase (Sigma) or alkaline phosphatase (Bio-Rad). Reactive protein bands were visualized with either enhanced chemiluminescence (Amersham Pharmacia) or 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (Denville Scientific, Metuchen, NJ), according to the manufacturer's instructions.
Reporter Gene Assays. The cotransfection protocol was adapted from ref. 5. Briefly, triplicate sets of HEK293 12 well-plate cultures were transfected with lipofectamine reagent (GIBCO/BRL), incubated for 12 h in standard medium and for an additional 24 h in serum-free DMEM, then mock stimulated or stimulated with EGF (100 ng/ml, Sigma). The cells were then harvested and lysed, and luciferase and β-galactosidase activities were assayed with the Dual-Light Combined Reporter Gene Assay System (Applied Biosystems) and Promega Turner TD-20/20 Luminometer. Luciferase activity was normalized against β-galactosidase for each transfection, and EGF-induced increase (fold increase = (+)EGF/(–)EGF – 1) in luciferase activity was quantified for every pair in the triplicate and was expressed as the average induction (fold increase) ± SD.
Detection of the Cell Surface EGFR Levels. The procedure was adapted from published protocols in refs. 11 and 12. Briefly, HEK293 cells were rinsed with ice cold PBS (pH 8.0) and reacted with 1 ml of ice-cold sulfo-NHS-LC-biotin reagent [Pierce, no. 21335, freshly dissolved in cold PBS (pH 8.0) to 1 mg/ml]. After 30 min at 4°C, the reagent was removed, and the reaction was quenched by the addition of cold 100 mM glycine in PBS. The labeled and collected cells were lysed by brief sonication in RIPA buffer, supplemented with protease and phosphatase inhibitors. The supernatant fluid was clarified by centrifugation in a Sorvall biofuge pico microcentrifuge at 16,000 × g for 20 min at 4°C, and protein concentration of each sample was assessed by spectrometry. Three hundred micrograms of cell lysate was reacted with 2 μg of anti-EGFR antibody (Upstate Biotechnology, no. 06-129) at 4°C overnight, and 40 μl of protein glutathione-agarose beads (50% slurry) were used to pull down the immune complexes. Each sample was resuspended in 100 μl of SDS loading buffer, boiled, and subjected to electrophoresis in a denaturing gel. The separated proteins were transferred to nitrocellulose membrane, blocked with 5% nonfat milk at room temperature for 3 h, and probed for 1 h at room temperature with horseradish peroxidase-streptavidin (Pierce, 1 mg/ml) diluted in 1% BSA-PBS (1:3,000). The probed blot was reacted with enhanced chemiluminescence plus reagent (Amersham Pharmacia). The reactive protein bands were quantified by using the Storm 860 phosphorimager (General Dynamics, Falls Church, VA) or exposed to x-ray film.
Results
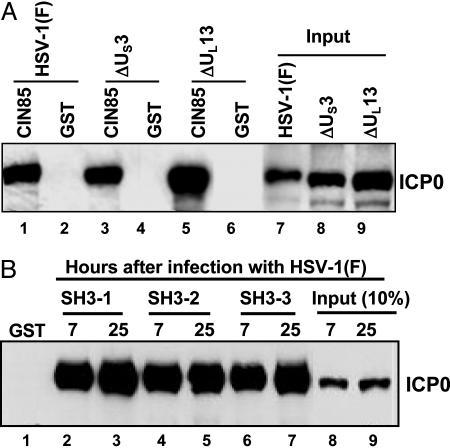
ICP0 Residues Encoded by Exon 3 Interact in a Reciprocal Manner with CIN85. As shown in Fig. 1, ICP0 contains several putative SH3 domain binding sites located in sequences encoded in exons 1 and 3, respectively (Fig. 1). These putative binding sites conform to the recognition consensus motif PX (P/A) XXR of CIN85. The experiments described below indicate that exon 3 encodes sequences that specifically bind in a reciprocal manner with the SH3 domains of CIN85 protein. In the first series of experiments, GST alone or GST-tagged full-length CIN85 protein bound to glutathione-agarose beads was reacted overnight at 4°C with lysates of cells harvested 16 h after exposure of cells to 10 pfu per cell of wild-type virus HSV-1(F) or mutant viruses lacking the US3 (ΔUS3) or UL13 (ΔUL13) protein kinases. After reaction, the beads were collected and rinsed, and the bound proteins were solubilized, electrophoretically separated on a denaturing gel, transferred to a nitrocellulose sheet, and reacted with anti-ICP0 monoclonal antibody. As shown in Fig. 2A, GST-CIN85 but not GST alone pulled down ICP0 from lysates of infected cells. The interaction of ICP0 with CIN85 does not require the posttranslational modification of ICP0 by the viral protein kinases encoded by US3 or UL13 genes (Fig. 2 A, lanes 3 and 5.)
Fig. 1.
Amino acid sequence of ICP0 showing putative binding sites of CIN85 SH3 domains.
Fig. 2.
CIN85 interacts with ICP0 through its SH3 domains. (A) CIN85 pulls down ICP0. GST-tagged full-length CIN85 or GST bound to beads were reacted overnight with equal amount of lysates of HEp-2 cells mock infected or exposed to 10 pfu per cell of HSV-1(F), ΔUL13, or ΔUs3 mutant for 16 h. The pulled-down proteins and the corresponding input lysates were resolved by electrophoresis in a denaturing gel and reacted with anti-ICP0 monoclonal antibody. (B) SH3 domains of CIN85 pull down ICP0. HEp-2 cells were harvested 7 or 25 h after infection with wild-type virus. Equal amounts of infected cell lysates were reacted for 4 h at 4°C with GST-tagged SH3 domains of CIN85 (designated as GST-SH3–1, GST-SH3–2, or GST-SH3–3, respectively) absorbed to glutathione-agarose beads. The proteins adhering to the beads were electrophoretically separated in denaturing gels and reacted with anti-ICP0 monoclonal antibody as described in Materials and Methods.
The sequence of CIN85 contains three SH3 domains predicted to bind to ICP0. To test this prediction, GST-tagged SH3 domains 1, 2, or 3 of CIN85 described in Materials and Methods bound to glutathione-agarose beads were reacted with lysates of cells exposed to 10 pfu of HSV-1(F) per cell and harvested 7 or 25 h after infection. The beads were processed as described above. As shown in Fig. 2B, all three SH3 domains of CIN85 pulled down ICP0, whereas GST alone did not.
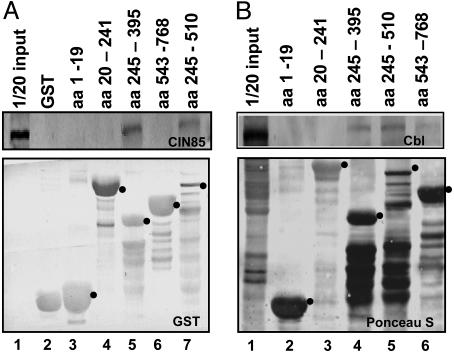
The objective of the third series of experiments was to verify whether the location of the binding site in ICP0 corresponds to the location of the sites predicted to bind CIN85. In these experiments, specific fragments of ICP0 fused to GST were reacted with lysates of HEK293 cells overnight at 4°C. The beads were then rinsed, and the bound proteins were solubilized, electrophoretically separated in denaturing polyacrylamide gels, transferred to a nitrocrellulose sheet, and reacted with antibody to CIN85 (Fig. 3A Upper), or GST (Lower). The dots identify the bands containing chimeric proteins consisting of GST fused to the appropriate sequence of ICP0. The results shown in Fig. 3A Upper indicated that the residues binding CIN85 were located between residues 245 to 395 (Fig. 3, lane 5), although we cannot exclude the possibility that additional binding sites are located between residues 395 to 510 (Fig. 3, lane 7).
Fig. 3.
CIN85 (A) and Cbl (B) are pulled down by domains of ICP0 containing SH3 domain binding sites. GST-ICP0 chimeric proteins bound to agarose beads as described in Materials and Methods were reacted with equal amounts of uninfected HEK293 cell lysates. The proteins bound to the beads were solubilized, separated in denaturing gels, transferred to a nitrocellulose membrane, and reacted with antibodies against CIN85 (A Upper) or GST (A Lower), Cbl (B Upper) or stained with Ponceau S (B Lower). The dots identify the bands that contain full-size GST-ICP0 chimeric proteins.
As indicated in the introduction, CIN85 is an adapter protein that interacts with Cbl. The question arose as to whether the complex containing ICP0 and CIN85 also contained Cbl. To test this hypothesis, the experiment described above was repeated, except that the electrophoretically separated proteins bound to the glutathione-agarose beads were probed with the anti-Cbl antibody (Fig. 3B Upper) as described in Materials and Methods. The nitrocellulose sheet was also stained with Ponceau S (Fig. 3B Lower). The results shown in Fig. 3B Upper were that Cbl was pulled down by the residues 245–395 and 245–510. Trace amounts of Cbl were also pulled down by the residues 543–768.
We conclude that ICP0 and CIN85 interact in a reciprocal manner in pull down experiments. As predicted, the interactive sites in CIN85 included the three SH3 domains. The fragments of ICP0 that interact with CIN85 in pull down experiments contain the sequences predicted to bind the CIN85 SH3 domains. Finally, the Cbl ubiquitin ligase was also present in the protein complexes precipitated by the ICP0 fragments that pulled down CIN85.
CIN85 and Cbl Are Relatively Stable for at Least 8 h After Infection. In this series of experiments, replicate cultures of HEp-2 cells were mock-infected or exposed to 10 pfu of HSV-1(F) per cell and harvested at 0, 2, 4, 8, or 12 h after infection. A replicate set of cultures was exposed to 0.5 mg/ml of MG132 (Calbiochem) for 4 h at 4 or 8 h after infection. Aliquots of the harvested cell lysates were resolved electrophoretically on a denaturing gel, electrically transferred to a nitrocellulose membrane, and reacted with anti-CIN85 antibody. We did not observe an increase in either Cbl or CIN85 in cells treated with MG132 or a significant decrease in either protein for at least 8 h after infection. A slight decrease in CIN85 was noted in cells harvested 12 h after infection (data not shown).
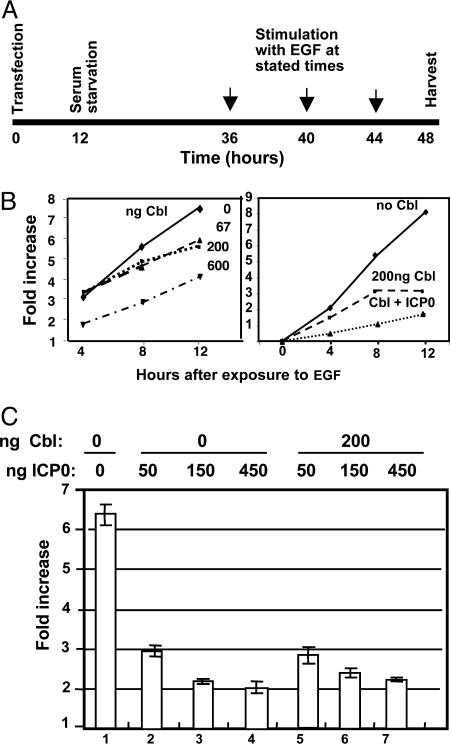
The Accumulation of EGF-Induced Reporter Protein Is Affected by Cbl and ICP0. A central question arising from the experiments described above concerned the role of ICP0 in the negative regulation of receptor tyrosine kinases by CIN85 and Cbl. The design of the experiments described in this section was as follows (Fig. 4A): HEK293 cells grown on 12-well plates were transfected with a mixture of plasmids consisting of an EGF-inducible reporter gene construct SRE-Luc (10 ng per well), the construct CMV-LacZ (0.4 ng per well) that served as an internal control for normalization of transfection efficiency, the EGFR expression vector pRK-EGFR (20 ng per well), and either the effector plasmids encoding Cbl or ICP0 or an equal amount of the empty carrier plasmid MTS1. The transfected cells were incubated in regular medium for 12 h at 37°C and then for an additional 24 h in serum-free medium. At the end of the second incubation period, the cells were exposed to EGF (100 ng/ml) for 4, 8, or 12 h, then harvested, lysed, and analyzed for luciferase and β-galactosidase activities as described in Materials and Methods. Luciferase activity was normalized against that of β-galactosidase for each transfection, and EGF-induced increase in luciferase activity was quantified in triplicate for every pair and was expressed as the average fold increase ± SD.
Fig. 4.
EGF-induced reporter gene expression is decreased in a dose-dependent manner by cotransfection with Cbl or ICP0. (A) Experimental design: HEK293 cells grown in 12-well plates were transfected with SRE-luciferase reporter gene construct, CMV-LacZ, expression vector pRK-EGFR, and variable amounts of plasmids expressing Cbl or ICP0. The empty expression vector MTS1 was added to the transfection mixture as needed to maintain a constant amount of transfected DNA. After 12 h, the cells were replenished with serum-free medium for 24 h and then incubated in medium containing 100 ng/ml EGF for 4, 8, or 12 h. The cells were then harvested, lysed, and the luciferase and β-galactosidase activities were assayed as described in Materials and Methods. Luciferase activity was normalized against β-galactosidase levels for each transfection, and EGF-induced fold increase in luciferase activity was quantified for every pair in the triplicate and was expressed as the average induction (fold increase) ± SD. (B) Cbl and ICP0 down-regulate EGF-induced reporter gene. EGF-induced luciferase activity in cells transfected with Cbl alone or with ICP0 is shown. (C) ICP0 down-regulates EGF-induced reporter gene. EGF-induced luciferase activity in cells transfected with increasing amounts of ICP0 alone or in combination with Cbl is shown.
In the first series of experiments, the transfection mixtures contained different amounts of the plasmid encoding the effector gene Cbl to test the efficacy of the reporter gene cotransfection assay. As shown in Fig. 4B Left, the decrease in the accumulation of EGF-induced luciferase activity in transfected cells was time- and Cbl dosage-dependent. In the second series of experiments, we measured the effect of addition of the plasmid-encoding ICP0 to the transfection mixture containing Cbl. ICP0 plus 200 ng of Cbl reduced the accumulation of EGF-induced luciferase to a greater extent than Cbl alone.
In the third series of experiments, we compared the EGF-induced luciferase activity in HEK293 cells transfected with various amounts of ICP0 alone or ICP0 plus 200 ng of Cbl. The results shown in Fig. 4C indicate ICP0 alone or ICP0 plus 200 ng of Cbl were equivalent with respect to their ability to mediate a decrease in the amounts of EGF-induced luciferase activity.
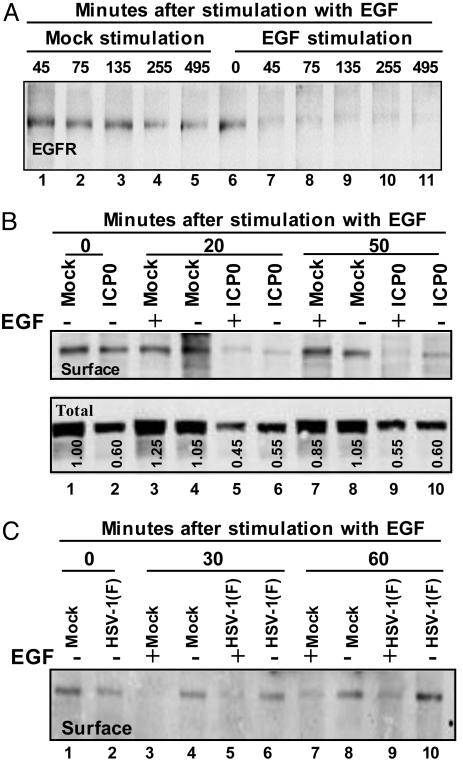
The Effect of HSV-1 or Transfection with ICP0 on the Stability of EGFR. We report three series of experiments. The first series of experiments was designed to test the time course of degradation of EGFR on cell surfaces after exposure to EGF. The design of this experiment was an abridged version of the protocol described in Fig. 4A. Briefly, T-25 flask cultures of HEK293 cells were serum-starved for 24 h and then either mock-treated or exposed to 100 ng/ml EGF for time intervals ranging from 45 to 495 min. The cells were then rinsed, and the cell surface proteins were labeled with a water soluble but membrane-impermeable biotinylating reagent sulfo-NHS-LC-biotin. The cell surface level of EGFR was detected as described in Materials and Methods. The results shown in Fig. 5A indicate that EGFR was stable in mock-treated cells but disappeared entirely from the cell surface by 45 min after exposure of the cells to EGF.
Fig. 5.
Cell surface and total EGFR levels are affected by ICP0 or by wild-type virus. (A) Effect of EGF on EGFR on cell surface. Replicate HEK293 cells grown in T-25 flasks were stimulated or mock-stimulated with 100 ng/ml EGF at time intervals shown. EGFR on cell surfaces was biotinylated and detected as described in Materials and Methods. (B) EGFR in cells transfected with ICP0. Replicate cultures of HEK293 cell flasks were transfected with EGFR and MTS1-ICP0 or the same amount of empty vector MTS1. At 12 h after transfection, the cultures were replenished with serum-free medium. At 36 h after transfection, the cells were mock-stimulated or exposed to EGF for 20 or 50 min. The cell surface levels of EGFR were assayed as described above (Upper). An aliquot of the total cell lysate removed before immunoprecipitation served to measure the level of EGFR in the total cell lysate (Lower). (C) EGFR in cells infected with HSV-1(F). Replicate cultures of HEp-2 cells were incubated in serum-free medium for 24 h, then mock-infected or infected with HSV-1(F). At 4 h after infection, the cells were mock-stimulated or exposed to EGF for 30 or 60 min. The cell surface levels of EGFR were assayed as detected above.
The objective of the second series of experiments was to test the effects of ICP0 cotransfected with EGFR. These experiments were done according to the protocol described in Fig. 4A. Briefly, T-25 flask cultures of HEK293 were transfected with 125 ng of a plasmid encoding EGFR and 2,500 ng of empty MTS1 plasmid or the same amount of MTS1-ICP0 plasmid. At 12 h after transfection, the cultures were replenished with serum-free medium. After 24 h of incubation, the cells were rinsed and stimulated with EGF as described above. Fig. 5B Upper shows the amounts of EGFR on the cell surface. Fig. 5B Lower shows 1/10 of the total amount of EGFR in the mock-stimulated or stimulated cells. The relevant findings (Fig. 5B) were as follows.
The total amount of EGFR reproducibly decreased in cells transfected with ICP0 (Fig. 5B Lower compared with mock-transfected cells). Compare lanes 2, 5, 6, 9, and 10 with corresponding lanes 1, 3, 4, 7, and 8. Exposure of mock-infected cells to EGF had and no significant effect on total EGFR.
In cells transfected with ICP0, the amount of EGFR at the cell surface at time 0 (Fig. 5B Upper, lane 2) was lower than in mock-transfected cells (lane 1). Cells transfected with ICP0 showed decreased surface levels of EGFR at the times after exposure to EGF (Fig. 5B Upper). In both EGF-treated and untreated cells, the levels of EGFR decreased to background levels after 20 min of incubation (lanes 5 and 6), and we therefore could not measure an additive effect of EGF on the decrease in surface level of EGFR mediated by ICP0.
In the last series of experiments, we examined the effects of HSV-1 infection on the accumulation of EGFR. Replicate T-25 flask cultures of HEp-2 cells were mock infected or exposed to 10 pfu of HSV-1(F) per cell for 4 h. The cells were then mock stimulated or stimulated with 100 ng/ml EGF for 30 or 60 min. The cell surface levels of EGFR were assayed as described above. As illustrated in Fig. 5C, the cell surface levels of EGFR in infected cells stimulated with EGF at 4 h after infection were lower than in cells that were not stimulated (Fig. 5C, compare lanes 5 and 6 and 9 and 10). As expected, at all times after infection, the levels of EGFR in mock-infected stimulated cells decreased rapidly relative to the amount of EGFR in unstimulated, mock-infected cells. It is noteworthy at the instant of addition of EGF, the cell surface EGFR in infected HEp-2 cell (lane 2) was already significantly lower that in mock-infected cells (lane 1). The results indicate that HSV-1 caused a decrease in cell surface EGFR even in the absence of EGF stimulation.
Discussion
In this report we have shown the following:
A previously uncharted domain representing ≈30% of the coding sequence of ICP0 contains multiple SH3 domain binding sites embedded with the CIN85 binding motif. CIN85 pulled down ICP0. Moreover, ICP0 was bound by GST fused to each of the three SH3 domains of CIN85. ICP0 fragments containing the putative CIN85 SH3 domain binding sites pulled down CIN85 or Cbl, whereas ICP0 fragments lacking the consensus sequences (e.g., sequences encoded by exon 2) did not.
EGF-dependent reporter gene expression was down-regulated in a dose-dependent manner in cells transfected with Cbl or a combination of Cbl and ICP0. EGF induced gene expression was also reduced in cells transfected with ICP0. It is noteworthy that the level of EGFR was reduced in cells contransfected with EGFR and ICP0 as compared with EGFR alone. This finding is particularly striking in light of the reports that ICP0 up-regulates the expression of genes introduced into cells by transfection (1).
Cell surface levels of EGFR were reduced in mock-infected cells exposed to EGF but not in untreated cells. In cells transfected with ICP0, cell surface and total cell levels of EGFR were reduced in the absence of EGF stimulation.
The significance of these finding stems from the following considerations: Cbl is a 982-residue, zinc-binding RING finger multifunctional protein involved in numerous regulatory activities that include attenuation of signaling through receptor tyrosine kinases and the formation of complexes that mediate insulin-dependent glucose transport and osteoclast function. Cbl contains numerous motifs that enable it to interact with >40 known proteins (reviewed in refs. 3 and 13). The most extensively studied functions of Cbl are those involved in attenuation of signaling through receptor tyrosine kinases (13). In the performance of these functions, Cbl expresses two activities: it promotes endocytosis by linking receptor tyrosine kinases to the endocytic machinery and by promoting the ubiquitylation of the receptors. Specifically, ligand-induced dimerization and autophosphorylation of the receptor enables Cbl to bind to the phosphotyrosine motifs at the carboxyl terminus of the receptor. The phosphorylation of Cbl by the activated receptor enables the binding of CIN85-endophilin to the complex containing the activated receptor tyrosine kinase and ultimately the internalization of the receptor in clathrin-coated vesicles. Depending on the receptor or the metabolic status of the cell, the receptor may be monoubiquitylated, polyubiquitylated, or monoubiquitylated at multiple sites. The ubiquitin ligation activities occur primarily at the RING finger domain (residues 365–428), although the location of the substrate binding sites may vary.
In this report, we used EGFR as the prototypic receptor tyrosine kinase to evaluate the significance of the interaction of ICP0 with CIN85. However, Cbl regulates a very wide range of cell surface receptors. The reported receptors reviewed in detail by Dikic et al. (13) include growth factor receptors (EGFR, platelet-derived growth factor receptor, hepatocyte growth factor receptor, and nerve growth factor receptor), a death receptor (Fas R), cytokine receptors (IL-2, IL-3, IL-4, stem cell factor receptor TNF-α, and IFN-γ) and immune receptors. Signaling by some of these receptors may be deleterious to viral replication, and it is not surprising that HSV would evolve functions designed to block signaling by reduction of surface receptors. Although we have not identified the specific target of HSV, preliminary studies have shown that IFN-γ receptor is also impaired by a process facilitated by ICP0 as described in this article (Y.L. and B.R., unpublished data). The studies reported here indicate that in addition to blocking presentation of antigenic peptides on cell surfaces (reviewed in ref. 14), HSV-1 also blocks signaling to the cell through receptor tyrosine kinases. In effect, HSV isolates the infected cells from its environment.
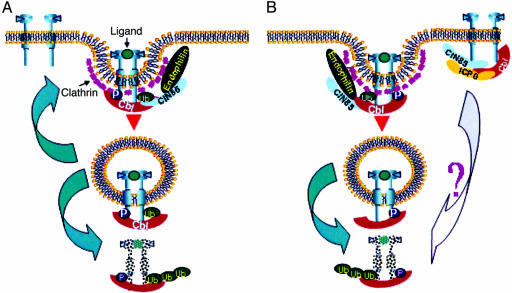
ICP0 is a multifunctional protein engaged in two fundamental activities: it enables efficient expression of viral genes most likely by precluding the silencing of the viral genome; it also blocks host response to infection by dispersal of ND10 nuclear structures, thereby precluding activation of antiviral genes (15). To perform its multiple functions, ICP0 interacts with numerous proteins and is extensively posttranslationally modified; it is localized in the nucleus early in infection and shuttles between the nucleus and cytoplasm at later stages in infection. ICP0 has also been shown to act as a ubiquitin ligase involved in the degradation of promyelocytic leukemia protein, Sp100, the DNA-dependent protein kinase, and cdc34 (1). The mechanism by which ICP0 mediates the decrease in both surface receptor tyrosine kinase and activation of genes induced by the ligand is at this stage unknown. As illustrated schematically in Fig. 6, ICP0 could block the recycling to the cell surface or promote active degradation of nonactivated receptor tyrosine kinases.
Fig. 6.
Schematic representation of the endocytosis and degradation or recycling of receptor tyrosine kinases in mock-infected (A) or infected (B) cells. In mock-infected cells, binding of the ligand results in dimerization and autophosphorylation of the receptor, recruitment and phosphorylation of Cbl, interaction with the adaptor protein CIN85 and endophilin, endocytosis, and either ultimate recycling or degradation of the receptor. In infected cells, ICP0, CIN85, and Cbl may form a complex that promotes the degradation of receptor tyrosine kinases in a ligand-independent fashion. In addition, ICP0 may preclude recycling of ligand-activated receptors.
Acknowledgments
We thank Iwona Szymkiewicz and Ivan Dikic (University of Frankfurt, Frankfurt) for invaluable reagents and Dale E. Bredesen (Buck Institute) for advice. These studies were aided by National Cancer Institute Grants CA87661, CA83939, CA71933, CA78766, and CA88860 to the University of Chicago and by U.S. Public Health Service Grants NS45093 and NS33376 to the Buck Institute for Age Research.
Author contributions: B.R. designed research; Y.L. performed research; A.K. contributed new reagents/analytic tools; A.K. analyzed data; and B.R. wrote the paper.
Abbreviations: EGFR, EGF receptor; HSV-1, herpes simplex virus 1; ICP0, infected cell protein 0.
Note Added in Proof. Two articles published after the submission of this report indicated that EGFR is degraded in a clathrin-dependent pathway in cells exposed to low levels of EGF and by both clathrin-dependent and -independent pathway in cells exposed to high concentration of EGF (16, 17). Our dose–response studies indicated the enhancement of degradation of EGFR from cell surfaces by ICP0 is greater at high concentrations (20 ng) than at low concentrations (1.5 ng) of EGF (data not shown). It is not clear at this time whether the enhancement of EGFR degradation in cells exposed to higher concentrations of EGF is ICP0-dependent or reflects degradation of EGFR independent of ICP0.
References
- 1.Hagglund, R. & Roizman, B. (2004) J. Virol. 78, 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurakin, A. V., Wu, S. & Bredesen, D. E. (2003) J. Biol. Chem. 278, 34102–34109. [DOI] [PubMed] [Google Scholar]
- 3.Thien, C. B. & Langdon, W. Y. (2001) Nat. Rev. Mol. Cell Biol. 2, 294–307. [DOI] [PubMed] [Google Scholar]
- 4.Dikic, I. & Giordano, S. (2003) Curr. Opin. Cell Biol. 15, 128–135. [DOI] [PubMed] [Google Scholar]
- 5.Soubeyran, P., Kowanetz, K., Szymkiewicz, I., Langdon, W. Y. & Dikic, I. (2002) Nature 416, 183–187. [DOI] [PubMed] [Google Scholar]
- 6.Petrelli, A., Gilestro, G. F., Lanzardo, S., Comoglio, P. M., Migone, N. & Giordano, S. (2002) Nature 416, 187–190. [DOI] [PubMed] [Google Scholar]
- 7.Purves, F. C., Ogle, W. O. & Roizman, B. (1993) Proc. Natl. Acad. Sci. USA 90, 6701–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purves, F. C., Longnecker, R. M., Leader, D. P. & Roizman, B. (1987) J. Virol. 61, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez, P., Van Sant, C. & Roizman, B. (2001) J. Virol. 75, 3832–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sciortino, M. T., Taddeo, B., Poon, A. P. W., Mastino, A. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 8318–8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravid, T., Sweeney, C., Gee, P., Carraway, K. L., III, & Goldkorn, T. J. (2002) J. Biol. Chem. 277, 31214–31219. [DOI] [PubMed] [Google Scholar]
- 12.Ohnishi, T., Muroi, M. & Tanamoto, K. (2001) J. Immunol. 167, 3354–3359. [DOI] [PubMed] [Google Scholar]
- 13.Dikic, I., Szymkiewicz, I. & Soubeyran, P. (2003) Cell. Mol. Life Sci. 60, 1805–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roizman, B. & Knipe, D. K. (2001) in Fields Virology eds. Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B. & Straus, S. E. (Lippincott-Raven, Philadelphia) 4th Ed., 2399–2459.
- 15.Chee, A. V., Lopez, P. Pandolfi, P. P. & Roizman, B. (2003) J. Virol. 77, 7101–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigismund, S., Woelk, T., Puri, C., Maspero, E., Tacchetti, C., Trnsiico, O., Di Fiore P. P. & Polo S. (2005) Proc. Natl. Acad. Sci. USA 102, 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, H. & De Camilli, P. (2005) Proc. Natl. Acad. Sci. USA 102, 2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]