Abstract
Background
Porphyromonas gingivalis is a major causative pathogen of chronic periodontitis. Within the inflammatory microenvironment, there exists extreme pH values, elevated temperatures and oxidative stress. Pathogens adapt to these stressful environmental conditions by regulating the transcription of virulence genes, modifying themselves with macromolecules and by aggregating and entering into a biofilm growth phase. Our previous study showed that the P. gingivalis sialidase can help cells obtain sialic acid from the environment, which is used to modify macromolecules on the surface of P. gingivalis cells. In this study, we compared the survival, virulence factors and biofilm formation of a sialidase-deficient strain (ΔPG0352) and the wild-type P. gingivalis W83 strain under various pH values, temperatures and oxidative stress conditions to identify the roles of sialidase in the adaptation of P. gingivalis to stressful conditions.
Results
Compared to the growth of the P. gingivalis W83 strain, the growth of the △PG0352 was more inhibited by oxidative stress (0.25 and 0.5 mM H2O2) and exhibited greater cell structure damage when treated with H2O2 as assessed by transmission electron microscopy. Both Lys-gingipain (Kgp) and Arg-gingipain (Rgp) activities were lower in the ΔPG0352 than those in the P. gingivalis W83 strain under all the assayed culture conditions. The lipopolysaccharide (LPS) activity of the W83 strain was higher than that of the ΔPG0352 under acidic conditions (pH 5.0), but no differences between the strains were observed under other conditions. Compared to the biofilms formed by P. gingivalis W83, those formed by the ΔPG0352 were decreased and discontinuous under acidic, alkaline and oxidative stress conditions.
Conclusion
Compared to the P. gingivalis W83 strain, the survival, virulence and biofilm formation of the ΔPG0352 were decreased under stressful environmental conditions.
Background
Periodontal diseases are complex, multifactorial and polymicrobial inflammatory diseases. They are characterized by the destruction of the supporting tissues around the teeth. The primary pathological changes associated with periodontal diseases, especially chronic periodontitis, are periodontal inflammation, loss of periodontal epithelial attachment, periodontal pocket formation and alveolar bone resorption, which ultimately lead to loosening and exfoliation of the teeth. Data collected by the World Health Organization (WHO) indicate that periodontal diseases affect 10-15% of adults worldwide [1]. It is noteworthy that periodontal diseases have a bidirectional relationship with systemic diseases, such as cardiovascular disease [2], diabetes [3], and rheumatoid arthritis [4], severely affecting the quality of human life.
During the progression of periodontal disease, subgingival microorganisms survive in the inflammatory microenvironment, protecting themselves from the deleterious effects of extreme pH values, elevated temperatures and oxidative stress. Microorganisms must overcome these harsh conditions to colonize or invade the host and can cause inflammation. To survive under these stressful microenvironmental conditions, bacteria will undergo cellular and physiological changes that include regulating the transcription of virulence genes, modifying themselves with macromolecules through reactions such as sialylation and glycosylation and by aggregating and entering into a biofilm growth phase.
Porphyromonas gingivalis is a black-pigmented, gram-negative anaerobe that is the major subgingival etiologic agent contributing to chronic periodontitis [5]. Its pathogenicity is attributed to an array of potential virulence factors, such as cysteine proteinases (gingipains), lipopolysaccharide (LPS), and fimbriae that allow P. gingivalis to attach to host tissues and resist the host innate immune system, ultimately leading to periodontal tissue destruction [6]. Recently, the role of sialidases in the pathogenesis of pathogens has attracted increased attention. Sialidases (neuraminidase) are glycosyl hydrolases that can cleave the connection between glycosylated substrates and sialic acid via a hydrolysis reaction. In a previous study, we confirmed that PG0352 is the sole sialidase-encoding gene in P. gingivalis W83, and we constructed a sialidase-deficient mutant strain (∆PG0352) by homologous recombination. We found that the deletion of PG0352 influenced biofilm formation and capsule biosynthesis and decreased the pathogenicity of P. gingivalis in a mouse subcutaneous abscess model [7].
The survival of P. gingivalis in the inflammatory microenvironment requires that it survive under various stresses, including different pH values, elevated temperatures, and oxidative stress. Some P. gingivalis mutations were observed to influence its survival, especially under these stress conditions. An investigation by Yuan et al. showed that a clpB (encoding a component of the stress response) mutant strain of P. gingivalis W83 demonstrated an increased sensitivity to heat stress, but not to hydrogen peroxide and extreme pH values [8]. McKenzie et al. indicated that the inactivation of pg1372 (encoding a protein with DNA-binding properties) virtually eliminated the ability of P. gingivalis to adapt to oxidative stress [9]. Dou et al. showed that a PG2212 (encoding a zinc finger protein) isogenic mutant strain of P. gingivalis exhibited an increased sensitivity to H2O2 [10]. In this study, we cultured P. gingivalis W83 and its sialidase-deficient mutant strain under stressful environmental conditions, including various pH values, temperatures and oxidative stress conditions, and compared the growth, morphology, tolerance, virulence factors and biofilm formation of these two strains to investigate the roles of sialidase in the adaptation of P. gingivalis to these stressful conditions.
Methods
Bacterial strains and culture conditions
P. gingivalis W83 and △PG0352 were used in this study. The P. gingivalis W83 strain was obtained from the Department of Oral Biology at China Medical University and the △PG0352 was constructed in our previous study [7]. Both strains were cultured anaerobically (10% CO2, 10% H2, and 80% N2) in trypticase soy broth (TSB, BD Diagnostic Systems, Aparks, MD) supplemented with 5 μg/mL hemin, 1 μg/mL menadione and 1 mg/mL yeast extract. For blood agar plates, TSB medium was supplemented with 1.5% agar and 5% sheep blood(Beiruite Bio-technology Co.,Ltd., Beijing, China).
Stress experiments
For all stress-related experiments, the P. gingivalis W83 and △PG0352 strains were incubated until they reached exponential phase (OD600 = 0.8-1.0).
pH-induced stress. The pH of the TSB medium was adjusted to 5.0, 7.2 or 9.0 by the addition of 1 M NaOH or 0.5 M HCl. Bacterial cells were centrifuged at 3000×g for 8 min at 4 °C, then the pellets were washed with phosphate-buffered saline (PBS) and resuspended in TSB medium at different pH values [8].
Temperature-induced stress. First, P. gingivalis W83 and △PG0352 cultures were treated as described above, then cells were resuspended in freshly supplemented TSB. Resuspensions of each strain were divided into three portions that were then cultured at 41, 37 or 34 °C [11].
Hydrogen peroxide-induced oxidative stress. First, P. gingivalis W83 and △PG0352 cultures were treated as described above. Next, hydrogen peroxide (Sigma-Aldrich, St. Louis, MO, USA) was added to the TSB medium and both strains were incubated at final concentrations of 0.1, 0.25, 0.5 or 1 mM H2O2. Final concentrations of hydrogen peroxide (Sigma-Aldrich, St. Louis, MO, USA) were obtained by diluting a 3% solution (8.8 mM) to the appropriate molarity in TSB medium [9]. Cell cultures that were not treated with hydrogen peroxide were used as controls.
Growth curves
The P. gingivalis W83 and ΔPG0352 strains were incubated under different culture conditions for 24 h. The growth curves of both strains under pH- and temperature-induced stress were generated by recording the absorbance at 600 nm (OD600) after 0, 4, 8, 12 and 24 h of culturing using a spectrophotometer (Bio-Rad, USA). The growth curves of the strains grown under oxidative stress were generated by measuring the OD600 after 0, 1, 4, 8 and 12 h of culturing.
Transmission electron microscopy
P. gingivalis W83 and ΔPG0352 cultures were centrifuged and washed three times with PBS. The pellets were fixed with 2.5% glutaraldehyde at 4 °C overnight [12]. Next, cells were washed three times with PBS and suspended in an osmium tetroxide solution for 2 h. The washed cell pellets were then gradually dehydrated in a series of ethanol (30%, 50% and 70%) and acetone (80%, 90% and 100%) solutions for 30 min each. Finally, the pellets were embedded in Epox 812 resin. Thin sections were cut and double-stained with uranyl acetate-lead citrate then were viewed under a transmission electron microscope (TEM) (H7650 Hitachi, Japan) at 70,000× magnification.
Gingipain activity assay
Nα-Benzoyl-DL-arginine p-nitroanilide hydrochloride (BAPNA) and N-(p-Tosyl)-Gly-Pro-Lys 4-nitroanilide acetate salt (both purchased from Sigma-Aldrich, St. Louis, MO, USA) were used as substrates to determine the activities of Arg-gingipain (Rgp) and Lys-gingipain (Kgp), respectively, according to a described previously method [13]. In brief, the P. gingivalis W83 and ΔPG0352 strains were cultured (16 h for pH-induced and temperature-induced stresses and 8 h for hydrogen peroxide-induced oxidative stress); then, cells were harvested by centrifugation at 5000×g for 5 min at 4 °C. Next, the pellets were suspended in 2 mL of reaction buffer, then 100 μL of the prepared pellet suspensions were aliquoted into an ice-cold 96-well microtiter plate. After a 10 min incubation at 37 °C, 100 μL of a 0.5 mM substrate solution was added and the OD405 was measured.
Preparation and activity measurement P. gingivalis LPS
LPS was isolated from 50 mL of bacterial cultures using the TRI-Reagent protocol as previously described [14]. P. gingivalis strains were cultured under varying conditions to mid-log phase, centrifuged at 6500 rpm for 20 min and the pellets were resuspended in TRI-reagent (Takara Bio Inc., Japan). Next, a 1/5 volume chloroform was added and mixed, the mixtures were centrifuged at 12,000 rpm for 10 min, and the top aqueous layers were retained as “crude LPS” extracts. The crude LPS extracts were washed with 1 mL of cold 0.35 M MgCl2 in 95% ethanol and centrifuged at 5000 rpm for 5 min at 4 °C. The pellets were washed twice more with 1 mL of cold 95% ethanol and once with 1 mL of cold 100% ethanol; then, the pellets were air-dried. To remove contaminating phospholipids, LPS extracts of each strain were resuspended to a 1% (w/v) solution in a 2:1 chloroform-methanol solution, centrifuged at 5000 rpm for 5 min at 4 °C, and air-dried. The final products were resuspended in water to attain the same concentrations among the different samples. LPS activity was measured using a Chromogenic End-point Tachypleus Amebocyte Lysate Kit (Chinese Horseshoe Crab Reagent Manufactory, China) according to the manufacturer’s instructions. Briefly, the tachypleus amebocyte lysate solutions, chromogenic substrates and azo reagents were successively added to diluted standards and LPS solutions and incubated for the appropriate time at 37 °C. Next, the mixtures were transferred to a 96-well immunoassay plate. Dilutions of all standards and samples were performed in triplicate. Lastly, a microplate reader (Infinite M200, TACON, Germany) was used to immediately measure the optical densities of the samples at 545 nm. Standard curves were generated by plotting the mean optical density and the activity of each standard dilution and each LPS solution.
Biofilm formation observed by confocal microscopy
Biofilms were grown and microscopically examined in 35 × 10 mm2 polystyrene cell culture plates (Corning, Netherlands) that were coated with artificial saliva (Guangzhou Kodak Adhesives Co. Ltd., China). Cell suspensions (2 mL; 5 × 106 CFU/mL) were aliquoted into polystyrene plates and cultured under varying conditions as described above for 4 days. The plates were washed twice with sterile PBS, and the biofilms were stained with a bacterial viability kit (Live/Dead BacLight bacterial viability kit L-7007; Molecular Probes, US) for 15 min in the dark then were washed twice more with sterile PBS. The polypropylene wells were detached from each other and the slides were air-dried. The biofilms were imaged with a confocal laser scanning microscope (CLSM) (Olympus FV1000, Japan) at an excitation wavelength of 488 nm. The three-dimensional structure of the biofilm was reconstructed from CLSM images using the built-in software (Olympus FV1000 Viewer, Japan). The thickness and density of each biofilm was observed by serially scanning layers from the inside (plaque biofilm and slide surface) to the outside (plaque biofilm-free surface) at a specific interval of 1.5 μm and measurements were subsequently made using ImageJ.
Statistical analysis
All experiments were performed in triplicate for each strain under each condition and were repeated at least three times. The data were analyzed by ANOVA to compare the differences of one strain grown under various conditions. Independent-sample t-tests were used to analyze the differences between the two strains grown in the same culture conditions. The level of significance was set at 0.05.
Results
Compared to P. gingivalis W83, the ΔPG0352 strain exhibited reduced survival under oxidative stress
Both the P. gingivalis W83 and ΔPG0352 strains grew significantly more slowly under acidic conditions (pH 5.0) than in the normal medium (pH 7.2) (P < 0.05), but alkaline conditions (pH 9.0) did not affect the growth of either strain. There were no differences between the growth rates of the P. gingivalis W83 and ΔPG0352 strains under pH-induced stress. (Fig. 1a). For the growth curves at different temperatures, the growth rates of both strains were considerably decreased at 34 °C compared to 37 °C, while both strains barely grew at 41 °C. At a given temperature, the growth rate of the P. gingivalis W83 strain was not significantly different from that of the ΔPG0352 (Fig. 1b). For the growth curves under oxidative stress, a concentration of 0.1 mM H2O2 had no effect on the growth of either the P. gingivalis W83 or ΔPG0352 strains. Final concentrations of 0.25 mM and 0.5 mM H2O2 resulted in a significant decrease in the growth rates of both strains, while both strains were unable to grow after being treated with 1 mM H2O2 (Fig. 1c and d). Compared to the P. gingivalis W83 strain, the ΔPG0352 grew more slowly at final concentrations of 0.25 and 0.5 mM H2O2 (P < 0.05), and there were no significant differences between the two P. gingivalis strains at final concentrations of 0.1 and 1 mM H2O2 (Fig. 1e).
Fig. 1.
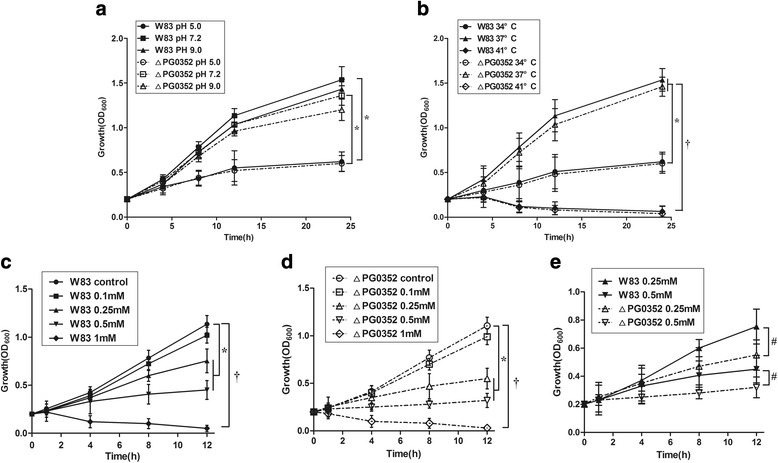
The growth curves of the P. gingivalis W83 and ΔPG0352 strains grown under varying pH values (a), temperatures (b) and oxidative stress conditions (c-e). * P < 0.05;† P < 0.01
Compared to P. gingivalis W83 strain, the cell membrane of the sialidase-deficient mutant was more easily damaged by oxidative stress
Under normal growth conditions, the cell membranes of both the P. gingivalis W83 and ΔPG0352 strains were intact and clearly observable by TEM. The cell structures of both strains showed no significant differences at different pH values (Fig. 2a) or temperatures compared to cells grown under normal culture conditions (Fig. 2b). At a concentration of 0.1 mM H2O2, the cell structure of the P. gingivalis W83 strain was not damaged, while nuclear separation occurred in ΔPG0352 cells. Final concentrations of 0.25, 0.5 and 1 mM H2O2 caused a discontinuity of the cellular membrane, cytoplasm shrinking and plasmolysis in both P. gingivalis W83 and ΔPG0352 cells. As the concentration of H2O2 increased, the cell structures were damaged with increasing severity. For both the P. gingivalis W83 and ΔPG0352 strains, cytoplasm vacuoles were degenerated and intact cells were rarely observed at a final concentration of 1 mM H2O2 (Fig. 2c). The survival rates of the ΔPG0352 mutant in 0.1 mM and 0.25 mM H2O2 were significantly lower than those of P. gingivalis W83. At concentrations of 0.5 mM and 1 mM H2O2, the survival rates of both strains decreased, but there were no significant differences between both P. gingivalis W83 and ΔPG0352 strains under same conditions. (Fig. 2d).
Fig. 2.
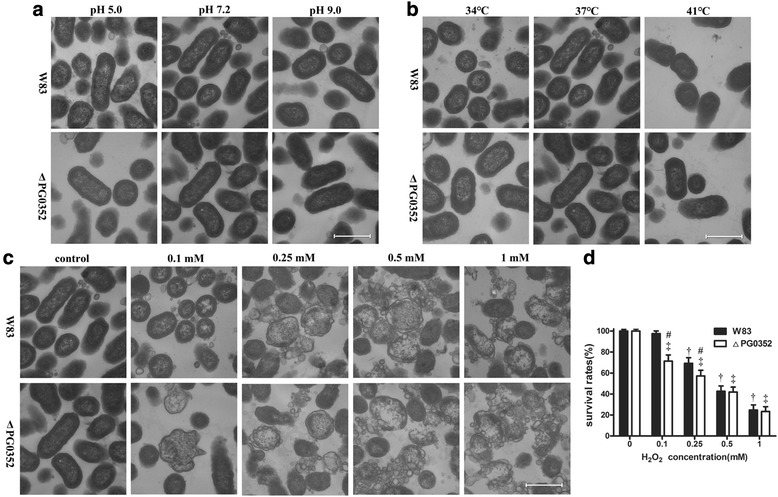
TEM images of P. gingivalis the W83 and ΔPG0352 strains cultured under varying pH values (a), temperatures (b) and oxidative stress conditions (c). Magnification: 70,000×; bar: 500 nm. (d) Survival rates of both strains cultured under oxidative stress; † versus the P. gingivalis W83 strain grown under normal culture conditions, P < 0.05; ‡ versus the ΔPG0352 mutant grown under normal culture conditions, P < 0.05; # versus the P. gingivalis W83 strain under the same culture conditions, P < 0.05
Gingipains and LPS activities in P. gingivalis W83 and ΔPG0352 strains varied under certain stressful culture conditions
Under all stress conditions assayed, including acidic or alkaline conditions (pH 5.0 or 9.0), extreme temperatures and oxidative stress, the activities of Kgp and Rgp in the P. gingivalis W83 and ΔPG0352 strains were lower than those in normal growth conditions. The Kgp activities for the ΔPG0352 were lower than those for the P. gingivalis W83 strain under all stressful conditions except with 0.5 mM and 1 mM H2O2 (Fig. 3a-c). The Rgp activities of the ΔPG0352 were lower than those of the P. gingivalis W83 strain under acidic or alkaline conditions and extreme temperatures, but no significant difference in Rgp activity was observed between these strains under any oxidative stress condition tested (Fig. 3d-f). The LPS activities of both the P. gingivalis W83 and ΔPG0352 strains did not change, and there were no differences between the strains under stressful conditions except for acidic conditions. Under acidic conditions (pH 5.0), the LPS activity of P. gingivalis W83 was significantly higher than that of the ΔPG0352 and was significantly higher than that of the P. gingivalis W83 strain grown under any other culture condition (Fig. 3g-i).
Fig. 3.
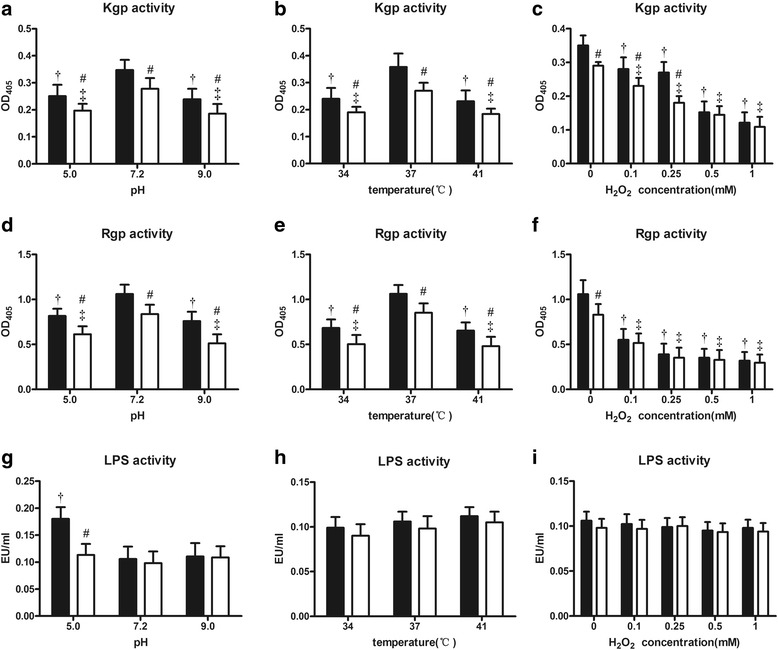
Comparison of virulence factors between the P. gingivalis W83 (■) and ΔPG0352 (□) strains cultured under varying conditions. † the difference in the P. gingivalis W83 strain is significant between normal and stressful conditions, P < 0.05; ‡ the difference in the ΔPG0352 mutant is significant between normal and stressful conditions, P < 0.05; # versus P. gingivalis W83 under the same culture conditions, P < 0.05
Compared to P. gingivalis W83 strain, the biofilm formation of the ΔPG0352 was severely decreased under stressful conditions
Under normal conditions, the biofilms formed by the P. gingivalis W83 and ∆PG0352 strains showed a uniform and dense fluorescence signal, although the biofilm by the ∆PG0352 was thinner than that of the P. gingivalis W83 strain. Biofilm formation by both the P. gingivalis W83 and ∆PG0352 strains decreased under acidic conditions (pH 5.0) and decreased more pronounced under alkaline conditions (pH 9.0). Biofilms of both strains were scattered in bacterial clumps, with the ∆PG0352 mutant forming less biofilm than that of the P. gingivalis W83 strain under both acid and alkaline conditions (Figs. 4a, 5a and d). An increase or decrease in temperature resulted in decreased biofilm formation for both strains, and less biofilm was formed by the ∆PG0352 mutant than by the P. gingivalis W83 strain (Figs. 4b, 5b and e). At a hydrogen peroxide concentration of 0.1 mM, both P. gingivalis strains exhibited continuous biofilm formation, but they were thinner than those generated under normal conditions. As the hydrogen peroxide concentration increased, gaps and pore structures became increasingly prevalent and finally became bacterial mass in the biofilms of both the P. gingivalis W83 and ∆PG0352 strains. Dense biofilm fluorescence signals for the ∆PG0352 were less common than those for the P. gingivalis W83 strain at final concentrations of 0.5 and 1 mM H2O2 (Figs. 4c, 5c and f).
Fig. 4.
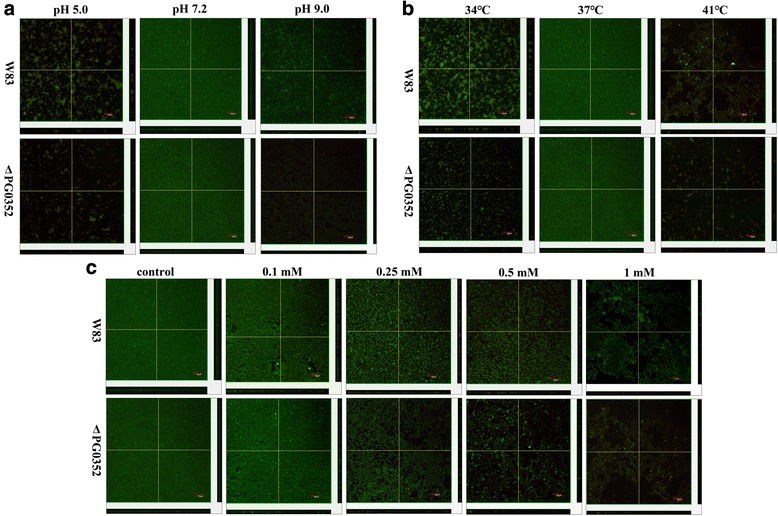
Confocal microscopy images of the P. gingivalis W83 and ΔPG0352 biofilms (stained with live/dead bacterial viability kit) cultured under varying pH values (a), temperatures (b) and oxidative stress conditions (c). Bar: 50 μm
Fig. 5.
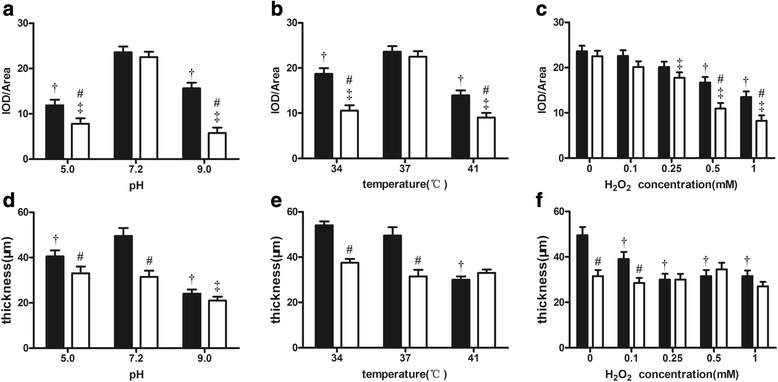
Comparison of the mean thickness (a, b and c) and fluorescence density (d, e and f) of biofilms between the P. gingivalis W83 (■) and ΔPG0352 (□) strains cultured under stressful conditions. † for P. gingivalis W83, the difference is significant between normal and stressful conditions, P < 0.05;. ‡ for the ΔPG0352 mutant, the difference is significant between normal and stressful conditions, P < 0.05; # versus P. gingivalis W83 under the same culture conditions, P < 0.05
Discussion
In this study, the growth curves of the P. gingivalis W83 and ∆PG0352 strains grown in normal and stressful conditions revealed that both strains grew slower at lower temperatures and under oxidative stress; compared to ∆PG0352, the wild-type strain was more sensitive to hydrogen peroxide but not to extreme pH values and temperatures. Bacteria primarily grew slower under sub-optimal temperatures because the activities of enzymes associated with their growth decrease and the enzymatic reactions slow down [15], while P. gingivalis grew slower under oxidative stress because of protein degeneration and cell death. Compared to P. gingivalis W83, the ∆PG0352 was more sensitive to hydrogen peroxide. P. gingivalis is an anaerobic microorganism and enzymes that detoxify oxygen are important in allowing it to inhabit oxygenated environments. Diaz PI et al. found that alkyl hydroperoxide reductase contributed to the tolerance of P. gingivalis to moderate levels of H2O2 [16]. In addition, Lijima R et al. found that sialic acid played a role as a reactive oxygen scavenger, protecting cells against hydrogen peroxide [17]. P. gingivalis cannot produce sialic acid but uses sialidase to cleave sialoglycoconjugates in the environment to produce free sialic acid [18, 19]. Thus, the sialidase-deficient mutant strain cannot release sialic acid to detoxify hydrogen peroxide and is therefore more sensitive to hydrogen peroxide than the W83 strain. Another reason for the oxidative stress sensitivity of the sialidase-deficient mutant is that it cannot synthesize an intact capsule; thus, the cell membrane of the sialidase-deficient mutant is easily damaged under oxidative stress.
Maintaining cell structure is essential for P. gingivalis survival, therefore we performed TEM to assess differences in structure between the P. gingivalis W83 and ∆PG0352 strains that were grown in varying stressful culture conditions. At different pH values or varying temperatures there was no significant difference in the cell structures of either strain compared to those grown under normal culture conditions. As the concentration of H2O2 increased, the balance of osmotic pressure between the inner and outer membranes was gradually lost, damaging the cell structure. The cell structure of the ∆PG0352 mutant was more easily damaged than that of the P. gingivalis W83 strain under oxidative stress. There are two possible reasons for this observation. The first reason is that sialidase deficiency in P. gingivalis influences capsule biosynthesis. Sialidase can cleave sialoglycoconjugates in the environment and produce free sialic acid, which are used for capsule biosynthesis [20]. The capsule contributes to the maintenance of the cell integrity. The ∆PG0352 mutant would produce a defective capsule due to a lack of sialic acid and would have difficulty maintaining cell structure under stressful conditions. The second reason is that the sialylation of macromolecules on the cell surface may play a role in bacteria maintaining their cell structures [21]. In the ∆PG0352, the macromolecules on the cell surface cannot be sialylated because of the sialidase deficiency. Thus, we speculate that the loss of sialidase may influence the sialylation of membrane molecules, limiting cellular resistance to oxidative stress. Taken together, because of the sialidase deficiency, the biosynthesis of capsule and the sialylation of macromolecule on the cell surface are impacted such that the cell structure of the ∆PG0352 is more easily damaged than that of the P. gingivalis W83 strain under oxidative stress.
P. gingivalis produces multiple proteases, with the primary ones being a set of cysteine proteases called gingipains, including an arginine-specific protease (Rgp) and a lysine-specific protease (Kgp). Rgp is encoded by two genes, rgpA and rgpB, and Kgp, which is encoded by a single gene, kgp [22]. The gingipains have multiple functions. They disrupt cell-cell and cell-matrix adhesion, induce apoptosis in several cell types [23–25] and contribute to bacterial survival [26]. Our study found that the gingipain activity of the ∆PG0352 was lower than that of the P. gingivalis W83 strain under either normal or stressful conditions. Vanterpool et al. showed that a complex of proteins could be involved in gingipain biogenesis in P. gingivalis [27]. Because sialidase is involved in the sialylation of gingipains, a sialidase deficiency in P. gingivalis may influence the production, maturation, or secretion of gingipain. We also found that the gingipain activity of both strains was affected by pH, extreme temperatures and oxidative stresses, suggesting that the activities of gingipains require a suitable pH value and a moderate temperature and are weakened under oxidative stress.
LPS is the major constituent of the outer membrane of P. gingivalis and is considered to be a major virulence factor of P. gingivalis. LPS plays an important structural role and mediates interactions with the microenvironment [28]. Its biological activity is very complex, but the most important activity is to stimulate target cells to secrete large amounts of inflammatory cytokines, leading to tissue destruction. LPS consists of lipid A, a distal polysaccharide (or O-antigen) and a non-repeating “core” oligosaccharide [29]. Lipid A, which is embedded in the outer membrane [28], is the biologically active region of LPS that can cause deregulation of the innate immune system through regulation of the toll-like receptor 2 and 4 pathways [30]. The sialylation of lipooligosaccharides occurs in microorganisms, such as Haemophilus influenzae [31], and sialidase may participate in P. gingivalis LPS biosynthesis or the sialylation modification. In our study, we compared the LPS activities of the P. gingivalis W83 and ∆PG0352 strains grown under different stressful conditions and observed no significant differences between the two strains under any assayed condition but acidic one. In the chromogenic end-point TAL assay, Tachypleus amebocyte lysate is primarily agglutinated with lipid A, which is the primary active site of LPS. We speculated that a sialidase deficiency in P. gingivalis may influence LPS biosynthesis or the sialylation modification, but that this may not involve lipid A. Stressful conditions, including alkaline conditions, extreme temperatures and oxidative stress did not influence LPS activity. It is noteworthy that the LPS activity of the P. gingivalis W83 strain in acidic conditions was increased compared to those in other culture conditions and was also higher than in the ∆PG0352. We speculate that P. gingivalis can self-protect against an adverse environment by increasing the activity of LPS to attack the host cells in acidic conditions and that this activity is associated with sialidase. Thus, the activity of LPS in the sialidase-deficient mutant strain cannot increase in acidic conditions, and this mechanism is being further studied in our laboratory.
During biofilm formation, cells must adapt to environmental changes and be able to maintain an appropriate balance between planktonic and biofilm phases. Single-species biofilm formation requires intercellular signaling and cells have a gene transcription profile that is distinct from that of planktonic cells [32]. For P. gingivalis, the level of sialidase gene expression is much higher in biofilms than that in planktonic cells [33], suggesting that sialidase is associated with P. gingivalis biofilm formation. In this study, live/dead bacterial viability fluorescent staining technology and CLSM were carried out to investigate the ability of P. gingivalis to form a biofilm under different conditions and compared the biofilm formation between the P. gingivalis W83 and ∆PG0352 strains. We found that the ∆PG0352 formed less biofilm (in terms of thickness and density) in both normal (in agreement with our previous findings [7]) and also stressful culture conditions. There are three possible reasons for this observation. First, sialic acid is an important component of biofilm [34] and the sialidase-deficient P. gingivalis strain could not obtain enough sialic acid to form an intact biofilm. Second, the sialidase deficiency in P. gingivalis decreased the activity of gingipains, which have been reported to regulate its biofilm formation [35]. Lastly, sialylation of components on the cell surface can help biofilm formation. A previous study showed that sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae [31]; Bloch et al. found that the role of surface glycosylation in the closely related oral pathogen Tannerlla forsythia, as a factor affecting structural aggregation of P. gingivalis in multispecies biofilms [36]. The sialidase-deficient strain could not sialylate the macromolecules on its surface, and its biofilm formation ability was less than that of the wild-type strain. We hypothesized that biofilm formation of P. gingivalis would increase to protect cells under stress conditions, but the results of this study showed that compared to P. gingivalis under normal culture conditions, both P. gingivalis the W83 and ∆PG0352 strains formed less biofilm under stress conditions. There are two possible explanations for this observation. The first is that both strains showed lower growth under these conditions, and the second is that these stressful conditions regulate the expression of P. gingivalis proteins, which associated with biofilm formation. For example, the gingipains and hemagglutinins of P. gingivalis may affect both the amount of monospecies biofilm formation and the composition of multispecies oral biofilms formation [35, 37, 38]. In addition, the biofilms of both strains were dispersed under oxidative stress, primarily because high concentrations of hydrogen peroxide can permeate into the biofilms and destroy them.
Conclusion
In the present study, we observed that the survival, virulence and biofilm formation of a sialidase-deficient mutant of P. gingivalis is decreased under stressful environmental conditions compared to those of the W83 strain. We speculate that the inhibition of P. gingivalis sialidase using a sialidase inhibitor will reduce the survival, virulence and biofilm formation in P. gingivalis, which would be a novel therapy for periodontal disease.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Nature Science Foundation of China (81200785, 81470745), Beijing, China and the National Nature Science Foundation of Liaoning Province (2015010496), Shenyang, Liaoning, China. The authors report no conflicts of interest related to this study.
Availability of data and materials
The data and analyses from the current study are available from the corresponding author upon a reasonable request.
Abbreviations
- CLSM
Confocal laser scanning microscope
- Kgp
Lys-gingipain
- LPS
Lipopolysaccharide
- PBS
Phosphate-buffered saline
- Rgp
Arg-gingipain
- TEM
Transmission electron microscope
- TSB
Trypticase soy broth
- ΔPG0352
Sialidase-deficient porphyromonas gingivalis strain
Authors’ contributions
Conceived and designed the experiments: CL, YP and LL. Performed the experiments: XX, TT and YX. Analyzed the data: XX, TT, YX and LC. Wrote the manuscript: XX, LC and LL. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoyu Xu, Email: xu_910327@163.com.
Tong Tong, Email: 114349079@qq.com.
Xue Yang, Email: yxmiracle@163.com.
Yaping Pan, Email: yppan@cmu.edu.cn.
Li Lin, Email: linli_74k@163.com.
Chen Li, Phone: 86-24-31927706, Email: lichen1980716@163.com.
References
- 1.Petersen PE, Ogawa H, et al. Periodontol. 2012;60(1):15–39. doi: 10.1111/j.1600-0757.2011.00425.x. [DOI] [PubMed] [Google Scholar]
- 2.Lockhart PB, Bolger AF, Papapanou PN, Osinbowale O, Trevisan M, Levison ME, et al. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association?: a scientific statement from the American Heart Association. Circulation. 2012;125(20):2520–2544. doi: 10.1161/CIR.0b013e31825719f3. [DOI] [PubMed] [Google Scholar]
- 3.Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000. 2013;62(1):59-94. [DOI] [PubMed]
- 4.Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, et al. Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD) PLoS Pathog. 2013;9(9):e1003627. doi: 10.1371/journal.ppat.1003627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Hasegawa M, Inohara N. Emerging roles of immunostimulatory oral bacteria in periodontitis development. Trends Microbiol. 2014;22(3):157–163. doi: 10.1016/j.tim.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.How KY, Song KP, Chan KG. Porphyromonas gingivalis: an overview of Periodontopathic pathogen below the gum line. Front Microbiol. 2016;7:53. doi: 10.3389/fmicb.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Kurniyati HB, Bian J, Sun J, Zhang W, et al. Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infect Immun. 2012;80(1):3–13. doi: 10.1128/IAI.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan L, Rodrigues PH, Belanger M, Dunn W, Jr, Progulske-Fox A. The Porphyromonas gingivalis clpB gene is involved in cellular invasion in vitro and virulence in vivo. FEMS Immunol Med Microbiol. 2007;51(2):388–398. doi: 10.1111/j.1574-695X.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 9.McKenzie RM, Johnson NA, Aruni W, Dou Y, Masinde G, Fletcher HM. Differential response of Porphyromonas gingivalis to varying levels and duration of hydrogen peroxide-induced oxidative stress. Microbiol. 2012;158(Pt 10):2465–2479. doi: 10.1099/mic.0.056416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou Y, Aruni W, Luo T, Roy F, Wang C, Fletcher HM. Involvement of PG2212 zinc finger protein in the regulation of oxidative stress resistance in Porphyromonas gingivalis W83. J Bacteriol. 2014;196(23):4057–4070. doi: 10.1128/JB.01907-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami Y, Imai M, Mukai Y, Ichihara S, Nakamura H, Yoshimura F. Effects of various culture environments on expression of major outer membrane proteins from Porphyromonas gingivalis. FEMS Microbiol Lett. 2004;30(2):159–165. doi: 10.1016/S0378-1097(03)00896-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang HY, Cheng JW, Yu HY, Lin L, Chih YH, Pan YP. Efficacy of a novel antimicrobial peptide against periodontal pathogens in both planktonic and polymicrobial biofilm states. Acta Biomater. 2015;25:150–161. doi: 10.1016/j.actbio.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Reyes L, Eiler-McManis E, Rodrigues PH, Chadda AS, Wallet SM, Belanger M, et al. Deletion of lipoprotein PG0717 in Porphyromonas gingivalis W83 reduces gingipain activity and alters trafficking in and response by host cells. PLoS One. 2013;8(9):e74230. doi: 10.1371/journal.pone.0074230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi EC, Hackett M. Rapid isolation method for lipopolysaccharide and lipid a from gram-negative bacteria. Analyst. 2000;125(4):651–656. doi: 10.1039/b000368i. [DOI] [PubMed] [Google Scholar]
- 15.Farewell A, Neidhardt FC. Effect of temperature on in vivo protein synthetic capacity in Escherichia Coli. J Bacteriol. 1998;180(17):4704–4710. doi: 10.1128/jb.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz PI, Zilm PS, Wasinger V, Corthals GL, Rogers AH. Studies on NADH oxidase and alkyl hydroperoxide reductase produced by Porphyromonas gingivalis. Oral Microbiol Immunol. 2004;19(3):137–143. doi: 10.1111/j.0902-0055.2004.00120.x. [DOI] [PubMed] [Google Scholar]
- 17.Iijima R, Takahashi H, Namme R, Ikegami S, Yamazaki M. Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett. 2004;561(1-3):163–166. doi: 10.1016/S0014-5793(04)00164-4. [DOI] [PubMed] [Google Scholar]
- 18.Aruni W, Vanterpool E, Osbourne D, Roy F, Muthiah A, Dou Y, et al. Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect Immun. 2011;79(7):2779–2791. doi: 10.1128/IAI.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cueno ME, Kamio N, Imai K, Ohya M, Tamura M, Ochiai K. Structural significance of the β1K396 residue found in the Porphyromonas gingivalis sialidase beta-propeller domain: a computational study with implications for novel therapeutics against periodontal disease. OMICS. 2014;18(9):591–599. doi: 10.1089/omi.2013.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller A, Severi E, Mulligan C, Watts AG, Kelly DJ, Wilson KS, et al. Conservation of structure and mechanism in primary and secondary transporters exemplified by SiaP, a sialic acid binding virulence factor from Haemophilus influenzae. J Biol Chem. 2006;281(31):22212–22222. doi: 10.1074/jbc.M603463200. [DOI] [PubMed] [Google Scholar]
- 21.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiol. 2007;153(Pt 9):2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 22.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4(6):397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 23.Katz J, Yang QB, Zhang P, Potempa J, Travis J, Michalek SM, et al. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect Immun. 2002;70(5):2512–2518. doi: 10.1128/IAI.70.5.2512-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheets SM, Potempa J, Travis J, Casiano CA, Fletcher HM. Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect Immun. 2005;73(3):1543–1552. doi: 10.1128/IAI.73.3.1543-1552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Casiano CA, Fletcher HM, et al. J Periodontol. 2001;72(5):641–650. doi: 10.1902/jop.2001.72.5.641. [DOI] [PubMed] [Google Scholar]
- 26.Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the mu-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183(1):159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 27.Vanterpool E, Roy F, Zhan W, Sheets SM, Sangberg L, Fletcher HM. VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology. 2006;152(Pt 11):3383–3389. doi: 10.1099/mic.0.29146-0. [DOI] [PubMed] [Google Scholar]
- 28.Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, et al. Temperature-dependent modulation of Porphyromonas gingivalis lipid a structure and interaction with the innate host defenses. Infect Immun. 2011;79(3):1187–1193. doi: 10.1128/IAI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8(2):217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 30.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid a species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;2(9):5041–5051. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swords WE, Moore ML, Godzicki L, Bukofzer G, Mitten MJ, VonCannon J. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun. 2004;72(1):106–113. doi: 10.1128/IAI.72.1.106-113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000;182(10):2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo AW, Seers CA, Boyce JD, Dashper SG, Slakeski N, Lissel JP, et al. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 2009;9:18. doi: 10.1186/1471-2180-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greiner LL, Watanabe H, Phillips NJ, Shao J, Morgan A, Zaleski A, et al. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect Immun. 2004;72(7):4249–4260. doi: 10.1128/IAI.72.7.4249-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuboniwa M, Amano A, Hashino E, Yamamoto Y, Inaba H, Hamada N, et al. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 2009;9:105. doi: 10.1186/1471-2180-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bloch S, Thurnheer T, Murakami Y, Belibasakis GN, Schaffer C, et al. Mol Oral Microbiol. 2017; doi:10.1111/omi.12182. [DOI] [PMC free article] [PubMed]
- 37.Bao K, Belibasakis GN, Thurnheer T, Aduse-Opoku J, Curtis MA, Bostanci N. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 2014;14:258. doi: 10.1186/s12866-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connolly E, Millhouse E, Doyle R, Culshaw S, Ramage G, Moran GP. The Porphyromonas gingivalis hemagglutinins HagB and HagC are major mediators of adhesion and biofilm formation. Mol Oral Microbiol. 2017;32(1):35–47. doi: 10.1111/omi.12151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and analyses from the current study are available from the corresponding author upon a reasonable request.


