Abstract
High-molecular-mass polyethylenimines (PEIs) are widely used vectors for nucleic acid delivery. We found that removal of the residual N-acyl moieties from commercial linear 25-kDa PEI enhanced its plasmid DNA delivery efficiency 21 times in vitro, as well as 10,000 times in mice with a concomitant 1,500-fold enhancement in lung specificity. Several additional linear PEIs were synthesized by acid-catalyzed hydrolysis of poly(2-ethyl-2-oxazoline), yielding the pure polycations. PEI87 and PEI217 exhibited the highest efficiency in vitro: 115-fold and 6-fold above those of the commercial and deacylated PEI25s, respectively; moreover, PEI87 delivered DNA to mouse lung as efficiently as the pure PEI25 but at a lower concentration and with a 200-fold lung specificity. These improvements stem from an increase in the number of protonatable nitrogens, which presumably results in a tighter condensation of plasmid DNA and a better endosomal escape of the PEI/DNA complexes. As a validation of the potential of such linear, fully deacylated PEIs in gene therapy for lung diseases, systemic delivery in mice of the complexes of a short interfering RNA (siRNA) against a model gene, firefly luciferase, and PEI25 or PEI87 afforded a 77% and 93% suppression of the gene expression in the lungs, respectively. Furthermore, a polyplex of a siRNA against the influenza viral nucleocapsid protein gene and PEI87 resulted in a 94% drop of virus titers in the lungs of influenza-infected animals.
Keywords: influenza, linear polyethylenimine, short interfering RNA, flu therapy, transfection
Nucleic acid-based medical intervention promises revolutionary advances in the treatment of human diseases. Originally aimed to correct genetic disorders, such as hemophilia (1) and cystic fibrosis (2), and therefore termed gene therapy, this pharmaceutical strategy has broadened its scope significantly as both DNA and RNA have emerged as potential therapeutic agents against many nongenetic diseases. Not only can defective genes, in principle, be corrected by functional ones, but also unwanted gene expression can be inhibited to provide therapeutic benefits, as exemplified by the use of RNA interference (3). Consequently, the nucleic acid-based treatments are being developed for such acquired illnesses as cancer (4), heart disease (5), neurodegenerative disorders (6), and infectious diseases (7).
Nucleic acids typically exert their therapeutic effect inside the cell. Consequently, delivery issues, such as crossing the plasma membrane and endosomal escape, are obstacles in nucleic acid-based therapies (8). Biological vectors, namely viruses, are used in most of the ongoing clinical trials (www.wiley.co.uk/genetherapy/clinical). However, safety concerns associated with viral vectors, such as induction of immune reactions (9, 10) [some leading to death (10)], cancers due to insertional mutagenesis (11), and the appearance of viruses in the semen of the treated patients (12), threaten their clinical utility. Therefore, nonviral vectors, in particular, cationic liposomes (13) and polycations (4, 8), are being explored as alternatives. However, these synthetic vectors, including even the best ones, high-molecular-mass polyethylenimines (PEIs) (14, 15), are inferior to their viral counterparts in the efficiency and specificity of nucleic acid delivery.
In the present study, we show that commercial preparations of linear PEI (molecular mass = 25 kDa) contain residual N-acyl groups, which severely handicap nucleic acid transfection. Removal of the residual N-acyl groups dramatically enhances DNA transfection efficiency of this PEI. Consistent with these findings, three additional linear PEIs, synthesized ab initio in the nonacylated form, exhibited remarkable DNA and short interfering RNA (siRNA) delivery efficiency and specificity. As a demonstration of their therapeutic potential, we show that influenza virus infection in the lungs of mice is inhibited 94% by siRNA delivered with PEI lacking N-acyl groups.
Materials and Methods
Chemicals and General Methods. Branched PEI25 and 50-kDa and 500-kDa poly(2-ethyl-2-oxazoline)s (PEOZs) were from Sigma–Aldrich. Commercial linear PEI25 and 200-kDa PEOZ were from Polysciences. NMR spectra were recorded by using a Varian Mercury 300-MHz NMR spectrometer with chemical shifts expressed with reference to the water peak in 2H2O (4.80 ppm). Elemental analyses were performed by MHW Laboratories (Phoenix, AZ).
Plasmids and siRNA. The plasmids gWiz β-gal and gWiz Luc containing the β-gal and luciferase genes, respectively, driven by the human CMV promoter, were obtained from Aldevron (Fargo, ND) as 5.0-mg/ml stock solutions in water. The plasmid pd1GL3-RL containing two Luc genes, firefly and Renilla Luc (denoted as GL3 and RL, respectively, in the plasmid acronyms), driven by two CMV promoters, was prepared as follows. D1GFP from the parental vector pd1EGFP-N1 (BD Biosciences) was replaced by the GL3 gene cut from pGL3-Basic (Promega). This modified pd1 vector was then cut at the AseI site upstream of the CMV promoter and blunted. Separately, the RL gene, together with its own CMV promoter (CMV-RL), was cut from pRL-CMV vector (Promega) and blunted, followed by insertion into the pd1 vector mentioned above to obtain pd1GL3-RL. This plasmid was propagated in Escherchia coli DH5α (Kan/Neo selection).
siRNAs, from Dharmacon (Lafayette, CO), had the following sequences. GFP-siRNA: sense, 5′-GGCUACGUCCAGGA GCGCAUU-3′; complementary strand, 5′-UGCGCUCCUGGACGUAGCCUU. NP-siRNA (in which NP is influenza nucleocapsid protein): sense, 5′-GGAUCUUAUUUCUUC GAGdTdT-3′; complementar y strand, 5′-CUCCGA AGAAAUAAGAUCCdTdT-3′. fLuc-siRNA (in which fLuc is firefly luciferase): sense, 5′-CUUACGCUGAGUACUUCGATT-3′; complementary strand, 5′-UCGAAGUACUCAGCGUAAGTT-3′.
Synthesis. Fully deacylated linear PEI25 was prepared by the acid-catalyzed hydrolyses of the commercial, incompletely deacylated PEI25. Separately, 22-, 87-, and 217-kDa linear PEIs without N-acyl groups were synthesized from commercial PEOZs. Typically, 1.2 g of the PEI25 (89% deacylated based on NMR) or 3.0 g of the PEOZs was added to 120 ml of 24% (wt/vol) HCl, followed by refluxing for 96 h. The first reaction mixture contained a white precipitate throughout the reaction. The POEZ crystals dissolved completely in ≈2 h, but, 3 h later, a white precipitate appeared. The precipitate in each case was isolated by filtration, air-dried, dissolved in water, and lyophilized. The resultant white powders were confirmed by a combination of NMR (see Results and Discussion) and elemental analysis to be pure PEI hydrochlorides. Elemental analysis results of different PEIs were as follows. Commercial PEI25: C, 49.92; H, 10.31; N, 25.70. PEI25: C, 27.64; H, 6.82; N, 15.26; Cl, 49.53. PEI22: C, 28.34; H, 7.12; N, 15.44; Cl, 37.90. PEI87: C, 29.02; H, 7.09; N, 15.70; Cl, 47.95. PEI217: C, 29.24; H, 7.31; N, 16.51; Cl, 44.50.
The starting PEOZs were also subjected to elemental analyses, and the C/N ratios obtained were consistent with their molecular formula (C5H9NO; note that for PEI, it is C2H5N). Because hydrolysis of PEOZ merely removes propionyl groups, the molecular masses of the resultant PEIs could be calculated from those of the precursors; thus, 50-kDa, 200-kDa, and 500-kDa PEOZs yielded 22-kDa, 87-kDa, and 217-kDa PEIs, respectively.
Stock solutions of all of the PEIs were prepared in water and stored at 4°C (150 mM in -CH2CH2NH-units, pH 2), unless stated otherwise. Appropriate dilutions were made immediately before each experiment.
pH Titration. Acid titrations were carried out by using a 2-ml solution of each PEI (113 mM in -CH2CH2NH-units) adjusted to pH 11.5 with NaOH. Sequential 20-μl additions of 1 M HCl were performed, and the pH after each addition was measured; a 113 mM solution of NaCl was titrated similarly as a control.
Ethidium Bromide (EtdBr) Displacement Assay. To a 2-ml solution of gWizLuc DNA (33 μg) and EtdBr (5 μg) in 10 mM PBS, 5-μl aliquots of 5 mM PEI stock solutions were added sequentially. Fluorescence of free EtdBr (FI), fluorescence of EtdBr/DNA (FD), and fluorescence of EtdBr/DNA after each addition of PEI (FC) were recorded (λex = 523 nm and λem = 587 nm, with excitation and emission band widths of 5 and 10 nm, respectively). Relative fluorescence values were calculated by using [(FC – FI)/(FD – FI)] × 100%.
Cell Culture and Transfection. A549 cells (human lung carcinoma) from the American Type Culture Collection were cultured in Ham's F12K medium (American Type Culture Collection), following the American Type Culture Collection protocol. Twenty-four hours before transfection, 1.5 × 105 cells per well were plated on Costar six-well tissue culture clusters.
The plasmid DNA (gWizβ-gal), 1.25 μg per well, was used for transfection. The polyplexes, prepared as described in ref. 16, were diluted to 3 ml with Ham's F12K medium containing 10% FBS and antibiotics immediately before their addition to the cells. Beforehand, the growth medium was removed from each well, and 1.0 ml per well of the aforementioned transfection medium was added, followed by incubation at 37°C in a humidified-air (5% CO2) atmosphere for 24 h. Thereafter, the cells were assayed for β-gal expression (14). Total protein was estimated from the bicinchoninic acid (Sigma–Aldrich) assay (14). The results were expressed as mean ± SD (n = 6).
Cytotoxicity Measurements. Cells were cultured as described above. PEI solutions, prepared as above except that DNA was omitted, were added at 1 ml per well. Control cells were treated with medium alone. After incubation at 37°C in a humidified-air atmosphere (5% CO2) for 24 h, the medium was removed, and the cells were treated with 1.0 ml of Ham's F12K medium containing 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma–Aldrich) and assayed (14). The results were expressed as mean ± SD (n = 6).
Gene Delivery in Mice. All animal experiments adhered to the Principles of Laboratory Animal Care (National Institutes of Health publication no. 85-23, revised in 1985). To obtain the desired N/P ratios (those of PEI nitrogen to DNA phosphate), appropriate volumes of PEI stock solutions were diluted to 600 μl with 5% aqueous glucose and added to equal volume of the glucose solutions containing 420 μg of the plasmid DNA (gWiz Luc), followed by pipette mixing. The resulting polyplexes were incubated at room temperature for 10 min. Then, 6- to 8-week-old C57BL/6 male mice (Taconic Farms) that were under anesthesia were injected retroorbitally with 200 μl of the polyplexes containing 70 μg of DNA. After 24 h, the mice were euthenized by CO2 inhalation; their lungs, kidneys, livers, hearts, and spleens were collected, washed with PBS, and suspended in 400 μl of lysis buffer prepared by mixing 4 ml of 5× passive lysis buffer (Promega), 800 μl of 8.7 mg/ml PMSF (Sigma–Aldrich) in methanol, 400 μl of the protease inhibitor mixture (Sigma–Aldrich), and 14.8 ml of water. The samples were freeze-thawed, homogenized by probe-sonication for 15 s, and centrifuged. The supernatants (10 μl, either as such or after dilution with the buffer) were mixed with 50 μl of the luciferase assay reagent (Promega), and the luminescence was measured by using an Optocomp I luminometer (MGM Instruments, Hamden, CT). Protein concentrations were determined by using the bicinchoninic acid assay. The results were expressed as mean ± SD (n = 5).
siRNA Delivery in Mice. Polyplexes were prepared at different N/P ratios as described above, except that gWizLuc plasmid was replaced by either pd1GL3-RL plasmid (420 μg) or fLuc-siRNA (360 μg), and the volume of the polyplex solutions was 600 μl each. DNA/PEI polyplexes were mixed with either the siRNA/PEI polyplex or an equal volume of the blank glucose solution. Two hundred μl of the aforementioned polyplexes containing either 70 μg of DNA or 70 μg of DNA plus 60 μg of siRNA was injected retroorbitally into anesthetized mice. After 24 h, the mice were killed; their lungs, kidneys, livers, hearts, and spleens were collected and processed as described above by using 400 μl of the cell lysis buffer (Marker Gene Technologies, Eugene, OR). To 10 μl of the supernatants (either as such or after dilution with the buffer) were added sequentially 50 μl of Firefly Luciferase Assay Reagent II and Stop and Glo Reagent (Dual Luciferase Assay System, Promega), and the luminescence from the samples was measured after each of the two additions. The ratio of the luminescence due to firefly and Renilla Luc in the absence of siRNA was used as a control for evaluating the suppression of fLuc expression by fLuc-siRNA. The results were expressed as mean ± SD (n = 5).
Influenza Virus Infection of Mice and Assay of Lung Virus Titers. Influenza A/Puerto Rico/8/34 (PR8, subtype H1N1) was provided by Dr. Peter Palese (Mount Sinai School of Medicine, New York). Three hours before infection, 200 μl of PEI87/siRNA polyplexes containing either 120 μg of NP- or GFP-siRNA (N/P = 5) in 5% aqueous glucose was injected retroorbitally into each mouse. For infection, 30 μl of PBS with 0.3% BSA and 1× penicillin/streptomycin containing 12,000 plaque-forming units of the virus was instilled into the nostrils of anesthetized mice. After 24 h, their lungs were harvested, suspended in 1 ml of PBS with 0.3% BSA and 1× penicillin/streptomycin, freeze-thawed twice, and homogenized by sonication for 30 s to release the virus. This procedure was followed by centrifugation and storing the supernatants at –80°C.
Madin–Darby Canine Kidney (MDCK) (American Type Culture Collection)/hemagglutinin assay was used to estimate the virus titers. Briefly, MDCK cells (2 × 104 cells per well; 96-well plates) were cultured in DMEM containing 10% FBS, antibiotics, and glutamine (GIBCO–Invitrogen) according to American Type Culture Collection protocols. After 12 h, the medium was removed, and 25 μl of lung homogenates, either undiluted or 10-fold serially diluted (seven steps), was added into wells in triplicate. After incubation for 1 h at room temperature, 175 μl of infection medium (culture medium containing 4 μg/ml trypsin) was added to each well, and incubation was continued under cell culture conditions for 48 h. The presence of the virus in the culture supernatants was determined by hemagglutination of chicken RBCs (Charles River Laboratories). The virus titers are presented as a logarithm of the tissue culture 50% infective dose ± SD (n = 5). Student's t test was performed to obtain the P values.
Results and Discussion
PEI, particularly the branched and linear forms of the 25-kDa polymer, is the premier polycationic vector for nucleic acid delivery (14, 16–23). Branched and linear 25-kDa PEIs are synthesized by different routes. The former is prepared by an acid-catalyzed, ring-opening homopolymerization of aziridine (Fig. 1A) (24) that leads to a multidirectional chain growth. In contrast, the linear PEI is synthesized in two steps. First, PEOZ is obtained by a ring-opening isomerization polymerization of 2-ethyl-2-oxazoline (Fig. 1B) in the presence of initiators (25). Then, PEOZ (N-propionyl-PEI) is acid-hydrolyzed to cleave off the N-propionyl groups to yield PEI (26).
Fig. 1.
Compound structures and syntheses in this study. (A and B) Chemical structures of aziridine and 2-ethyl-2-oxazoline, respectively. (C) Synthesis of fully deacylated linear PEI25 by exhaustive acid hydrolysis of its commercial predecessor. (D) Synthesis of fully deacylated linear PEI22, PEI87, and PEI217 by the acid hydrolysis of PEOZs. Conditions: (i) 24% (wt/vol) HCl, 110°C, 96 h; k = 517, l = 64, m = 581, PEI25; n = 504 for 50-kDa PEOZ, 2,018 for 200-kDa PEOZ, and 5,044 for 500-kDa PEOZ.
Because N-deacylations are sluggish, we suspected that the removal of the N-propionyl groups in the commercial PEIs might be incomplete. Indeed, the 1H NMR spectrum of commercial linear PEI25 exhibited a triplet at 1.05 ppm and a doublet at 2.43 ppm corresponding to the -CH3 and -CH2-protons of the propionyl moiety, respectively. Based on the ratio of the intensity of the -CH3 signal of propionyl group and the -CH2-CH2-signal of the ethylenimine units (2.75 ppm), the extent of removal of the N-propionyl groups in commercial PEI25 was ≈89%. Because the amide (as opposed to amine) nitrogens are nonbasic due to the delocalization of their lone electron pairs with the carbonyl group, DNA binding and proton–sponge capacity of the commercial linear PEI25 might be compromised. Therefore, exhaustive hydrolysis of the commercial linear 25-kDa PEI, widely used for gene delivery, was carried out by using concentrated HCl at 110°C, as outlined in Materials and Methods (Fig. 1C). The NMR spectrum of the resultant polymer revealed no residual N-propionyl moieties. In addition, three more linear PEIs were prepared by the same exhaustive acid hydrolysis of their commercial precursor PEOZs (Fig. 1D). The fully deacylated PEI25 and the PEI22, PEI87, and PEI217 thus produced exhibited a singlet at 3.52 ppm by NMR corresponding to -CH2-CH2-NH2+-but no signal corresponding to the N-propionyl moieties, confirming their complete removal.
Removal of the residual N-propionyl groups markedly affected the acid titration curve of PEI. For instance, at a HCl concentration where the pH of the commercial polycation had already reached 5, that of the fully N-deacylated PEI25 was still around pH 8 (Fig. 2A), indicating its much higher buffer capacity in this pH range. Similarly, the fully deacylated PEI87 also exhibited a much enhanced buffering capacity compared with the commercial PEI25 (Fig. 2 A). A greater buffer capacity should ensure a more efficient endosomal escape of the polyplexes and hence a greater transfection efficiency.
Fig. 2.
Full deacylation increases the buffer capacity and DNA binding efficiency of linear PEI. (A) Acid titration profiles of aqueous solutions of linear commercial PEI25 (squares), fully deacylated PEI25 (open circles), linear PEI87 (filled circles), and NaCl as a control (triangles). Solutions (113 mM) were adjusted to pH 11.5 at room temperature and then titrated with 1 M HCl. (B) Displacement of the intercalated fluorophore EtdBr from plasmid DNA by linear commercial PEI25 (squares), fully deacylated linear PEI25 (open circles), and PEI87 (filled circles).
Although the difference in the number of protonatable nitrogens between the fully deacylated PEI25 and its commercial counterpart is only 11%, it can have a profound effect on the polycation's behavior, as illustrated by the following calculation. Given the 43-Da molecular mass of the -CH2CH2NH-unit, the average number of the -CH2CH2NH-monomers in PEI25 is 581, which is also the length of the contiguous stretch of potentially protonatable nitrogens. Assuming a uniform distribution of the N-propionyl moieties in the commercial PEI25, its contiguous stretch of protonatable nitrogens is only 64. This 9-fold difference may substantially affect the stability of the polyplexes as they traverse through the various extracellular and intracellular barriers involved in gene delivery (8). Supporting this notion, the branched 2-kDa PEI (47 contiguous nitrogens) is some 100-fold less efficient in DNA transfection than the 25-kDa counterpart (14, 16). Furthermore, functionalization of 10% of the nitrogens of branched PEI25 with lactose, which sterically interferes with DNA binding, reduces its DNA transfection efficiency in vitro 10-fold (27). Thus, interference of intervening nitrogens in PEI in forming salt bridges with the contiguous phosphates of DNA can adversely affect DNA condensation (see the next paragraph) and reduce the transfection efficiency.
That the higher content of the protonatable nitrogens would increase PEI's DNA condensation capacity was verified by fluorescence spectroscopy using an EtdBr displacement assay. EtdBr, a DNA intercalator, exhibits ≈10-fold greater fluorescence emission upon binding to DNA. Condensation of DNA by PEI distorts the double helix and consequently displaces EtdBr, thus quenching its fluorescence. This method has been successfully used to compare the relative DNA condensation efficiencies of polycations and cationic lipids (28, 29). We found that fully deacylated PEI25 indeed displaced EtdBr from DNA far more efficiently (and PEI87 even more so) than its commercial predecessor (Fig. 2B), hence supporting our underlying hypothesis.
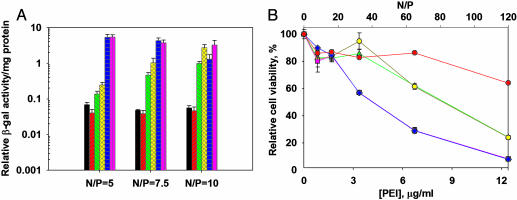
To compare the transfection efficiency of fully deacylated PEIs and their commercial counterparts, polyplexes were tested at the N/P ratios of 5, 7.5, and 10. The results are shown in Fig. 3A. Both branched and linear commercial PEI25s exhibited similarly low efficiencies at all of the N/P ratios tested. Complete deacylation of linear PEI25 increased its optimal transfection efficiency 21-fold. The fully deacylated PEI22, PEI87, and PEI217 were even more efficient compared with the commercial linear PEI25; their Luc activity rose 58-, 115-, and 116-fold, respectively (Fig. 3A). The fully deacylated PEI22, similar in size to PEI25, exhibited optimal efficiency at N/P = 10. In contrast, the fully deacylated PEI87 and PEI217 exhibited optimal efficiencies already at N/P = 5. These results suggest that the DNA transfection efficiency of linear PEIs in vitro is markedly affected by their extent of deacylation, and the optimal N/Ps vary with their molecular mass.
Fig. 3.
The effect of full deacylation of PEIs on their transfection efficiency and toxicity in vitro. (A) Expression of β-gal in A549 cells after transfection with branched (black bars), commercial linear (red bars), and fully deacylated (green bars) PEI25s, as well as linear, hydrolytically pure PEI22 (yellow bars), PEI87 (blue bars), and PEI217 (pink bars). (B) Cytotoxicities induced by linear polycations: commercial (red circles) and fully deacylated (green triangles) PEI25s, as well as hydrolytically pure PEI22 (yellow circles), PEI87 (blue squares), and PEI217 (pink diamonds) in A549 cells.
Due to their positive charge, PEIs and other polycations can interact electrostatically with such cellular anionic macromolecules as proteins, various types of RNA, and genomic DNA. This binding impairs the normal cellular functions of these polyanions, resulting in toxicity. Thus, we assessed the cytotoxicity of the fully deacylated PEIs by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The cytotoxicity of PEIs is known to increase with their molecular mass (16). Indeed, PEI87 and PEI217 were more toxic than PEI22 and PEI25 at high concentrations (Fig. 3B). Interestingly, the commercial linear PEI25 was less toxic than its fully deacylated successor, suggesting that a higher charge density contributes to the toxic effect, presumably due to stronger electrostatic interactions with cellular anionic macromolecules. However, none of the PEIs exhibited appreciable toxicity at concentrations corresponding to the N/P ratios of up to 20 in our in vitro transfection experiments (Fig. 3B, top x axis). Moreover, unlike PEI25 and PEI22, PEI87 and PEI217 exhibited optimal efficiencies at N/P ratios as low as 5 and, therefore, can be used at half the concentration to minimize toxicity while still achieving optimal transfection efficiency.
To test the effect of the full deacylation of PEIs on their in vivo gene delivery, we used a mouse model of systemic delivery. Linear commercial PEI25, fully deacylated PEI25 and PEI87 (whose in vitro efficiencies resemble those of PEI22 and PEI217, respectively), as well as branched PEI25, were selected for these experiments. The polyplexes were prepared at the N/P ratios of 5, 7.5, and 10 for all PEI25s. Because PEI87 exhibited similar efficiencies in this N/P range in vitro, in addition to the 5 and 7.5, an N/P ratio of 3.75 was also used for in vivo experiments. Thus, DNA-encoding fLuc gene was complexed with various PEIs and administered into mice, and Luc activities in various organs were assayed 24 h after delivery.
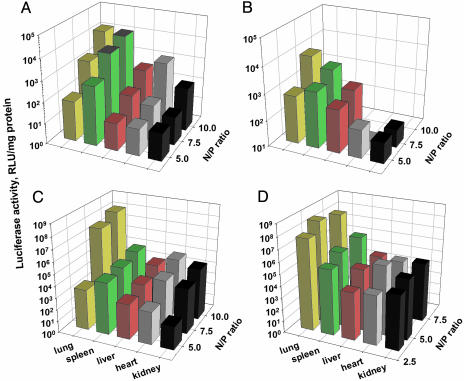
As seen in Fig. 4A, at the N/P ratios of 5 and 7.5, the polyplexes of commercial linear PEI25 exhibited the highest gene expression in the spleen; at N/P = 10, similar expression was observed in the lung. Raising the N/P ratio from 5 to 7.5 and to 10 enhanced the gene expression in the spleen 9 and 18 times, respectively, and enhanced the gene expression in the lung 21 and 176 times, respectively (Fig. 4A). Note that the Luc expression levels were low, <104 relative light units/mg of protein (Fig. 4A). As in vitro, branched PEI25 exhibited low efficiency in gene delivery in vivo, comparable to that of its linear commercial counterpart (Fig. 4B). The organ specificity of branched PEI25 was also poor, with the polyplexes formed at the N/P ratios of 5 and 7.5 having similar expressions in the lung and the spleen. Moreover, the polyplexes formed by this polycation were highly toxic at N/P = 10, causing the death of all of the mice injected with them.
Fig. 4.
Comparison of the delivery efficiencies to different organs in mice of a plasmid containing the luciferase gene mediated by different PEIs. Commercial linear (A) and branched (B) PEI25s, as well as hydrolytically pure linear PEI25 (C) and PEI87 (D), were used. Only the mean values are shown.
The fully deacylated PEI25 exhibited dramatically higher transfection efficiency than its commercial counterpart, with maximum Luc activity soaring to 108 relative light units/mg of protein (Fig. 4C). At N/P = 5, the highest expression was observed in the spleen, followed by the lung, with the difference between the two being an order of magnitude. However, when the N/P ratio was raised to 7.5 and 10, the gene expression in the lung rose 4 orders of magnitude, whereas that in the spleen merely doubled (Fig. 4C). Thus, upon the complete deacylation of commercial linear PEI25, its ability to deliver DNA to the mouse lung jumped 10,000-fold, and the lung-versus-spleen specificity increased 1,500-fold (at the optimal N/P ratio of 10; Fig. 4 A and C).
PEI87, as in vitro, exhibited optimal transfection efficiency at N/P = 5 (Fig. 4D). The highest gene expression was observed in the lung, followed by the spleen. The level of expression for PEI87 in the lung was 3 times the PEI25's but the organ specificity (lung vs. spleen) was lower, although still 200-fold.
A plausible reason for the preferential gene expression in the lung is that more polyplexes are taken up by it than by other organs. Because lung is highly vascularized and has a larger surface area (30), the uptake of the polyplexes should be facilitated. Also, the lung contains the first capillary beds traversed by intravenously injected materials.
The foregoing results suggest that fully deacylated linear PEIs may be used for the targeted nucleic acid delivery to the lung. We explored this possibility with the fully deacylated linear PEIs to deliver siRNAs to the mouse lung. We first used a siRNA specific for a model reporter gene, fLuc. The plasmid used (pd1GL3-RL) contained two Luc genes, firefly and Renilla Luc, and hence allowed examination of the selective suppression of fLuc expression by a siRNA specific for this gene (fLuc-siRNA) alone. Mice were administered PEI/pd1GL3-RL polyplexes (N/P = 10 and 5, respectively, for PEI25 and PEI87) alone or in the presence of PEI/fLuc-siRNA polyplexes at different N/P ratios. The ratios of fLuc and Renilla Luc activities in the lung was used to estimate the siRNA delivery efficiency. The polyplexes of PEI25 with fLuc-siRNA at the N/P ratios of 5 and 7.5 afforded a 76 ± 5% and 77 ± 3% suppression of the fLuc activity, respectively. The PEI87/fLuc-siRNA polyplexes at the N/P ratios of 3.75 and 5 were more potent, resulting in a 92 ± 2% and 93 ± 3% suppression of the fLuc activity, respectively. These results demonstrate that fully deacylated PEI25 and PEI87 are effective in delivering not only plasmid DNA, but also siRNA, in vivo; hence, the latter strategy may be exploited for combating infectious diseases affecting the lung, such as influenza.
Influenza virus production can be inhibited by siRNAs specific for different viral genes, especially those encoding the NP (31). Therefore, polyplexes of PEI87 with siRNA specific for NP (NP-siRNA) were administered retroorbitally (N/P = 5, 120 μg siRNA). Mice were then infected with influenza virus intranasally, and, 24 h later, the virus titers in lungs were measured. When PEI/NP-siRNA polyplexes were given, the virus titer in the lungs of influenza-infected mice plunged by as much as 17 times after 24 h (P = 0.002) (Table 1). In contrast, the mice treated with the PEI87/GFP-siRNA polyplexes or with the free PEI87 under otherwise the same conditions produced no significant decrease in virus titers (1.2 times reduction; P = 0.64 in both cases). Note that the observed 94% drop in the virus titer in the lung is highly significant therapeutically, because previous vaccine development studies have shown that even a 90% reduction in the virus titer leads to the survival from a lethal challenge (32).
Table 1. Inhibition of virus production in the lungs of influenza-infected mice by delivering influenza nucleoprotein siRNA mediated by linear, hydrolytically pure PEI87.
| Challenge | Mean virus titer in the lung* | Decrease in viral titer | P value |
|---|---|---|---|
| 5% glucose (control) | 3.08 ± 0.4 | — | — |
| PEI87/GFP-siRNA | 3.00 ± 0.0 | 1.2 | 0.64 |
| PEI87 alone | 3.00 ± 0.0 | 1.2 | 0.64 |
| PEI87/NP-siRNA | 1.86 ± 0.5 | 16.6 | 0.002 |
—, not applicable.
Expressed as logarithm of the tissue culture 50% infective dose; see Materials and Methods for details.
In closing, we have demonstrated that by removing the residual N-acyl moieties and hence maximizing the number of protonatable nitrogens of linear PEIs, the polycations' in vitro gene delivery efficiency, as well as in vivo efficiency and specificity, could be enhanced by several orders of magnitude. Moreover, effective suppression of the influenza virus titers in the mouse lung by delivering the resultant polyplexes with a therapeutically relevant siRNA was demonstrated. Because our methodology for the preparation of hydrolytically pure linear PEIs is simple and straightforward, the findings reported here should facilitate both basic and clinical research employing polycation-mediated gene and siRNA delivery.
Acknowledgments
This work was supported by National Institutes of Health Grants EB000244 (to A.M.K.) and AI56267 (to J.C.) and by the National Science Foundation-funded Biotechnology Process Engineering Center at Massachusetts Institute of Technology (A.M.K.). J.J.L. thanks the National Science and Engineering Research Council of Canada for a Postdoctoral Fellowship.
Author contributions: M.T., J.C., and A.M.K. designed research; M.T., J.J.L., Q.G., and C.Z. performed research; M.T. and Q.G. contributed new reagents/analytic tools; M.T. analyzed data; and M.T., J.C., and A.M.K. wrote the paper.
Abbreviations: PEI, polyethylenimine; PEIn, PEI with molecular mass n (in kDa); PEOZ, poly(2-ethyl-2-oxazoline); siRNA, short interfering RNA; NP, nucleocapsid protein; fLuc, firefly luciferase; EtdBr, ethidium bromide.
References
- 1.Nathwani, A. C., Davidoff, A. M. & Tuddenham, E. G. (2004) Haemophilia 10, 309–318. [DOI] [PubMed] [Google Scholar]
- 2.Lee, T. W., Matthews, D. A. & Blair, G. E. (January 19, 2005) Biochem. J., 10.1042/BJ20041923. [DOI] [PMC free article] [PubMed]
- 3.Izquierdo, M. (2005) Cancer Gene Ther. 12, 217–227. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. G., Peng, W., Akinc, A., Hossain, N., Kohn, A., Padera, R., Langer, R. & Sawicki, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 16028–16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quarck, R. & Holvoet, P. (2004) Curr. Gene Ther. 4, 207–223. [DOI] [PubMed] [Google Scholar]
- 6.Lowenstein, P. R. & Castro, M. G. (2004) Curr. Opin. Pharmacol. 4, 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fanning, G., Amado, R. & Symonds, G. (2003) J. Gene Med. 5, 645–653. [DOI] [PubMed] [Google Scholar]
- 8.Thomas, M. & Klibanov, A. M. (2003) Appl. Microbiol. Biotechnol. 62, 27–34. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser, J. (2004) Science 304, 1423–1425. [DOI] [PubMed] [Google Scholar]
- 10.Marshall, E. (1999) Science 286, 2244–2245. [DOI] [PubMed] [Google Scholar]
- 11.Check, E. (2005) Nature 433, 561. [Google Scholar]
- 12.Marshall, E. (2001) Science 294, 2268–2269. [DOI] [PubMed] [Google Scholar]
- 13.Liu, Y., Fong, S. & Debs, R. J. (2003) Methods Enzymol. 373, 536–550. [DOI] [PubMed] [Google Scholar]
- 14.Thomas, M. & Klibanov, A. M. (2002) Proc. Natl. Acad. Sci. USA 99, 14640–14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diebold, S. S., Kursa, M., Wagner, E., Cotten, M. & Zenke, M. (1999) J. Biol. Chem. 274, 19087–19094. [DOI] [PubMed] [Google Scholar]
- 16.Thomas, M., Ge, Q., Lu, J. J., Chen, J. & Klibanov, A. M. (2005) Pharm. Res. 22, in press. [DOI] [PMC free article] [PubMed]
- 17.Forrest, M. L., Meister, G. E., Koerber, J. T. & Pack, D. W. (2004) Pharm. Res. 21, 365–371. [DOI] [PubMed] [Google Scholar]
- 18.Thomas, M. & Klibanov, A. M. (2003) Proc. Natl. Acad. Sci. USA 100, 9138–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akinc, A., Thomas, M., Klibanov, A. M. & Langer, R. (November 15, 2004) J. Gene. Med., 10.1002/jgm.696. [DOI] [PubMed]
- 20.Sonawane, N. D., Szoka, F. C. J. & Verkman, A. S. (2003) J. Biol. Chem. 278, 44826–44831. [DOI] [PubMed] [Google Scholar]
- 21.Pham, P. L., Perret, S., Doan, H. C., Cass, B., St-Laurent, G., Kamen, A. & Durocher, Y. (2003) Biotechnol. Bioeng. 84, 332–342. [DOI] [PubMed] [Google Scholar]
- 22.Derouazi, M., Girard, P., Van Tilborgh, F., Iglesias, K., Muller, N., Bertschinger, M. & Wurm, F. M. (2004) Biotechnol. Bioeng. 87, 537–545. [DOI] [PubMed] [Google Scholar]
- 23.Choosakoonkriang, S., Lobo, B. A., Koe, G. S., Koe, J. G. & Middaugh, C. R. (2003) J. Pharm. Sci. 92, 1710–1722. [DOI] [PubMed] [Google Scholar]
- 24.Fischer, D., von Harpe, A., Kunath, K., Petersen, H., Li, Y. & Kissel, T. (2002) Bioconjugate Chem. 13, 1124–1133. [DOI] [PubMed] [Google Scholar]
- 25.Hoogenboom, R., Fijten, M. W. M., Brandli, C., Schroer, J. & Schubert, U. S. (2003) Macromol. Rapid Commun. 24, 98–103. [Google Scholar]
- 26.Jeong, J. H., Song, S. H., Lim, D. W., Lee, H. & Park, T. G. (2001) J. Controlled Release 73, 391–399. [DOI] [PubMed] [Google Scholar]
- 27.Kunath, K., von Harpe, A., Fischer, D. & Kissel, T. (2003) J. Controlled Release 88, 159–172. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. H., Lim, Y. B., Choi, J. S., Lee, Y., Kim, T. I., Kim, H. J., Yoon, J. K., Kim, K. & Park, J. S. (2003) Bioconjugate Chem. 14, 1214–1221. [DOI] [PubMed] [Google Scholar]
- 29.Prata, C. A., Zhao, Y., Barthelemy, P., Li, Y., Luo, D., McIntosh, T. J., Lee, S. J. & Grinstaff, M. W. (2004) J. Am. Chem. Soc. 126, 12196–12197. [DOI] [PubMed] [Google Scholar]
- 30.Agu, R. M., Ugwoke, M. I., Armand, M., Kinget, R. & Verbeke, N. (2001) Respir. Res. 2, 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge, Q., McManus, M. T., Nguyen, T., Shen, C. H., Sharp, P. A., Eisen, H. N. & Chen, J. (2003) Proc. Natl. Acad. Sci. USA 100, 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein, S. L., Tumpey, T. M., Misplon, J. A., Lo, C. Y., Cooper, L. A., Subbarao, K., Renshaw, M., Sambhara, S. & Katz, J. M. (2002) Emerging Infect. Dis. 8, 796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]