Abstract
Huntington's disease (HD) is a fatal, dominant neurogenetic disorder. HD results from polyglutamine repeat expansion (CAG codon, Q) in exon 1 of HD, conferring a toxic gain of function on the protein huntingtin (htt). Currently, no preventative treatment exists for HD. RNA interference (RNAi) has emerged as a potential therapeutic tool for treating dominant diseases by directly reducing disease gene expression. Here, we show that RNAi directed against mutant human htt reduced htt mRNA and protein expression in cell culture and in HD mouse brain. Importantly, htt gene silencing improved behavioral and neuropathological abnormalities associated with HD. Our data provide support for the further development of RNAi for HD therapy.
Keywords: short hairpin RNAs, triplet repeat diseases, gene therapy, nanomedicine
Huntington's disease (HD) is one of nine dominant neurodegenerative diseases resulting from polyglutamine repeat expansions, leading to a toxic gain of function (1, 2). Hallmark HD characteristics include cognitive and behavioral disturbance, involuntary movements (chorea), neuronal inclusions, and striatal and cortical neurodegeneration (2). Huntingtin (htt) alleles containing >35 CAG repeats generally cause HD, with age at onset correlating inversely with expansion length, a common characteristic of the polyglutamine repeat disorders. The disease usually develops in midlife, but juvenile-onset cases can occur with CAG repeat lengths >60. Death typically occurs 10–15 years after symptom onset.
Therapies aimed at delaying disease progression have been tested in HD animal models. For example, beneficial effects have been reported in animals treated with substances that increase transcription of neuroprotective genes (histone deacetylase) (3), prevent apoptosis (caspase inhibitors) (4), enhance energy metabolism (coenzyme Q/remacemide and creatine) (5, 6), and inhibit the formation of polyglutamine aggregates (trehalose, Congo red, and cystamine) (7–9). These approaches target downstream and possibly indirect effects of disease allele expression. In contrast, no therapies have been described that directly reduce mutant htt gene expression, thereby targeting the fundamental, underlying pathological insult.
The therapeutic promise of silencing mutant htt expression was demonstrated in a tetracycline-regulated mouse model of HD (10). When mutant htt was inducibly expressed, pathological and behavioral features of the disease developed, including the characteristic neuronal inclusions and abnormal motor behavior. Upon repression of transgene expression in affected mice, pathological and behavioral features resolved. Thus, reduction of htt expression by using RNA interference (RNAi) may allow protein clearance mechanisms within neurons to normalize mutant htt-induced changes. We hypothesize that directly inhibiting the expression of mutant htt will slow or prevent HD-associated symptom onset in a relevant animal model.
Screening of putative therapies for HD has benefited from the existence of several HD mouse models (11, 12). HD-like phenotypes are displayed in knock-in mice (13, 14), drug-induced models (15), and transgenic mice expressing full-length mutant htt (e.g., yeast artificial chromosome-transgenic mice) (16–18) or an N-terminal fragment of htt (10, 19, 20). Mice expressing truncated N-terminal fragments of htt have been valuable for proof-of-principle evaluation of therapies because they show rapidly progressive motor abnormalities and striatal neuropathology, phenotypes that do not develop or develop very late in knock-in or yeast artificial chromosome-transgenic mice. Mice expressing truncated forms of htt thus replicate more severe forms of the disease. In this study, we tested whether RNAi induced by short hairpin RNAs (shRNAs) (21) could reduce expression of mutant htt and improve HD-associated abnormalities in a transgenic mouse model of HD.
Materials and Methods
Plasmids and Adenoassociated Virus (AAV) Construction. Myc-tagged HD-N171-82Q was expressed from a pCMV-HD-N171-82Q plasmid (20). PCR (Pfu DNA polymerase, Stratagene) was used to amplify the U6 promoter along with shRNAs targeting human htt (shHD2.1; Fig. 1A), eGFP (shGFP) (22), or Escherichia coli β-galactosidase (base pairs 1152–1172; shLacZ). PCR products were cloned, sequenced, and inserted into pAAV.CMV. hrGFP, which contains AAV serotype 2 inverted terminal repeats, a CMV-humanized Renilla GFP (hrGFP)-simian virus 40 poly(A) reporter cassette, and sequences used for homologous recombination into baculovirus (23). Recombinant AAV serotype 1 vectors were generated as described in ref. 23. AAV titers were determined by using quantitative PCR and/or DNA slot blot analysis and were 5 × 1012 vector genomes per ml.
Fig. 1.
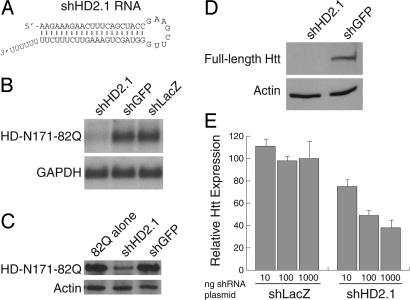
RNAi reduces human htt expression in vitro. (A) RNA sequence of shHD2.1. The 21-nt antisense strand is cognate to nucleotides 416–436 of human htt mRNA (GenBank accession no. NM_002111). (B and C) Northern and Western blots demonstrate shHD2.1-mediated reduction of HD-N171-82Q mRNA and protein expression 48 h after transfection of target- and shRNA-expressing plasmids. GAPDH and actin serve as loading controls. (D) Western blots show that shHD2.1 inhibits expression of full-length human htt protein, 48 h after transfection. (E) shHD2.1 induces dose-dependent reduction of human htt mRNA. Cells were transfected with either shLacZ- or shHD2.1-expressing plasmids in the indicated amounts. Relative htt expression was determined by using quantitative PCR 24 h later.
Animals. All animal studies were approved by the University of Iowa Animal Care and Use Committee. HD-N171-82Q mice were purchased from The Jackson Laboratory (20, 24) and maintained on a B6C3F1/J background. Hemizygous and age-matched WT littermates were used for the experiments, as indicated.
Northern Blot Analysis. HEK293 cells were transfected (Lipofectamine 2000, Invitrogen) with pCMV-HD-N171-82Q and plasmids expressing shHD2.1, shGFP, or shLacZ at shRNA-to-target ratios of 8:1. Forty-eight hours after transfection, RNA was harvested by using TRIzol reagent (Invitrogen), and 10 μg was assessed by Northern blot analysis (NorthernMax, Ambion) using probes to human htt or human GAPDH. Band intensities were quantified by using a Storm 860 phosphorimager and imagequant 1.2 software, both from Molecular Dynamics.
For in vivo studies, total RNA was isolated from hrGFP-positive striata. Thirty micrograms of RNA was run on 15% polyacrylamide/urea gels, transferred to Hybond-N+ membranes (Amersham Pharmacia), and then probed with 32P-labeled sense oligonucleotides at 36°C for 3 h, washed in 2× SSC at 36°C, and exposed to film.
Western Blot Analysis. HEK293 cells were transfected as described with shHD2.1 or shGFP singly or in combination with pCMV-HD-N171-82Q. Forty-eight hours later, cells were lysed to recover total protein. Western blots were incubated with anti-myc (1:5,000; Invitrogen), anti-full-length human htt (1:5,000; MAB2166, Chemicon), or anti-human β-actin (1:5,000; Clone AC-15, Sigma) followed by horseradish peroxidase-coupled goat anti-mouse or goat anti-rabbit secondary antibodies (1:20,000 and 1:100,000, respectively; Jackson Immunochemicals, West Grove, PA). Blots were developed by using ECL-Plus reagents (Amersham Pharmacia). For evaluation of transduced brain, 3-week-old mice were injected as described, and protein was harvested from striata 2 weeks later. Twenty-five micrograms of protein was run on SDS/PAGE as described, transferred to nitrocellulose, and then probed with antibodies to detect human htt (1:500, mEM48; a gift from X. J. Li, Emory University School of Medicine, Atlanta) and mouse prion protein (1:40,000, Chemicon). Secondary antibody incubations were performed as described above.
Quantitative RT-PCR. In vitro shRNA dose–response. HEK293 cells were transfected with 0 (mock), 10, 100, or 1,000 ng of either shLacZ or shHD2.1, and RNA was harvested 24 h later. After DNase treatment (DNA-Free, Ambion), random-primed, first-strand cDNA was generated from 500 ng of total RNA by using TaqMan reverse transcription reagents (Applied Biosystems) according to the manufacturer's protocol. Assays were performed on a sequence detection system by using primers and probe sets specific for human htt and mammalian rRNA (Prism 7000 and TaqMan 2X Universal Master Mix; Applied Biosystems). Relative gene expression was determined by using the relative standard curve method.
In vivo htt mRNA expression. Striata were dissected from 5.5-month-old mice, snap-frozen in liquid nitrogen, and pulverized. cDNA was generated as described above. Relative gene expression was assayed by using TaqMan primer/probe sets specific for human htt and mammalian rRNA or TaqMan Assays-by-Design primers/probes specific for mouse htt (mHdh, Applied Biosystems). All values were calibrated to contralateral, uninjected striata. Human htt detection, shHD2.1 samples, n = 8 striata; shLacZ, n = 7; uninjected, n = 4. Mouse htt detection, injected HD samples, n = 4; uninjected samples n = 2.
AAV Injections. AAV injections were performed in 4-week-old mice by using the following parameters (coordinates are reported with respect to the bregma): striatal, 0.5 mm anterior, 2.5 mm lateral, 2.5-mm depth, 5 μl per site, 250 nl/min infusion rate; cerebellar, 0.1-mm depth, 1 μl per site, 250 nl/min infusion rate.
Behavioral Analysis. Stride length measurements. Mice injected bilaterally at 4 weeks of age were analyzed at 4 months of age. Analyses were performed as described in ref. 25, with some modifications. Specifically, mice were allowed to walk across a paper-lined chamber (100 cm × 10 cm with 10-cm walls) and into an enclosed box. Mice were given one practice run and were then tested three times to produce three separate footprint tracings, totaling 42 measurements each for front and rear footprints per mouse. Measurements were averaged, and data were presented as box plots. ANOVA with Scheffé's post hoc test was performed to determine statistical significance. Uninjected mice, n = 4; injected WT, n = 3; injected HD-N171-82Q, n = 6 mice per group.
Rotarod performance test. Two separate experimental cohorts of mice were injected at 4 weeks of age and tested on the rotarod (model 7650, Ugo Basile, Varese, Italy) at 10 and 18 weeks of age as described in ref. 26. Data from trials 2–4 for each day are presented as means ± SEM. Uninjected WT, n = 6; shLacZ WT, n = 5, shHD2.1 WT, n = 6; uninjected HD-N171-82Q, n = 5; shLacZ HD-N171-82Q, n = 10; shHD2.1 HD-N171-82Q, n = 11.
Immunofluorescence. Forty-micrometer free-floating coronal sections from mice (n = 5 per group) were stained with mEM48 antibody (1:500; 24 h, 4°C), followed by AlexaFluor 568-labeled goat anti-mouse secondary antibody (1:200; 4 h, room temperature; Molecular Probes). Sections were mounted onto slides and covered in Gel/Mount (Biomeda, Foster City, CA), and images were captured by using fluorescent microscopy with either a DM RBE (Leica) or confocal microscope (Zeiss) equipped with a charge-coupled device camera (SPOT RT, Diagnostic Instruments, Sterling Heights, MI).
Results
shHD2.1 Reduces Human htt Expression in Vitro. In vitro screening was used to identify effective shRNAs directed against a CMV promoter-transcribed HD-N171-82Q mRNA, which is identical to the pathogenic truncated htt fragment transgene present in HD-N171-82Q mice (20). Hairpin constructs targeting sequences in human exons 1–3 were evaluated by cotransfection. One htt-targeted shRNA, shHD2.1 (Fig. 1 A), reduced HD-N171-82Q mRNA and protein levels by ≈85% and ≈55% respectively, compared with control shRNA-treated samples (Fig. 1 B and C).
To test whether shHD2.1 could silence endogenous full-length human htt expression, HEK 293 cells were transfected with plasmids expressing shHD2.1 or shGFP. ShHD2.1, but not control shRNAs, directed gene silencing of endogenous htt mRNA and protein (Fig. 1 D and E).
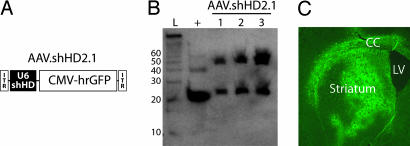
Expression of shRNA in Mouse Brain. We next tested for U6 promoter-transcribed shHD2.1 expression in vivo and its effects on HD-associated symptoms in mice. This RNA polymerase III-dependent promoter has not previously been evaluated in striata for sustained expression in vivo, although shRNAs have been expressed in brain by using either the RNA polymerase II-dependent CMV promoter in striatum (22) or the H1 promoter in cerebellar degeneration models (26). U6 promoter-driven shHD2.1 and the control hairpin shLacZ were cloned into AAV shuttle plasmids that contained a separate CMV-hrGFP reporter cassette (Fig. 2A). High-titer AAV1 particles (AAV.shHD2.1 and AAV.shLacZ), which have broad neuronal tropism, were generated (23), and hairpin expression was assessed after injection into mouse striatum. The N171-82Q mouse model was used because shHD2.1 targets sequences in exon 2, precluding the use of the R6/2-transgenic model, which expresses only exon 1 of the HD gene. As shown in Fig. 2B, precursor and processed shRNAs (≈50 and 21 nt, respectively) were expressed 3 weeks after transduction, indicating sustained expression and appropriate processing of shRNAs in the striatum. Analysis of coronal brain sections from injected mice showed widespread transduction (Fig. 2C; hrGFP fluorescence) up to 5 months after injection.
Fig. 2.
AAV.shHD2.1 delivers widespread RNAi expression to mouse striatum. (A) AAV.shHD2.1 viral vector. ITR, inverted terminal repeat. (B) Northern blot showing shHD2.1 transcripts are expressed in vivo. Processed antisense (lower band) and unprocessed (upper band) shHD2.1 transcripts in three different AAV.shHD2.1-injected mice are shown. L, ladder; +, positive control oligonucleotide. The blot was probed with radiolabeled sense probe. (C) Typical AAV1 transduction pattern (hrGFP) in mouse brain. CC, corpus callosum; LV, lateral ventricle.
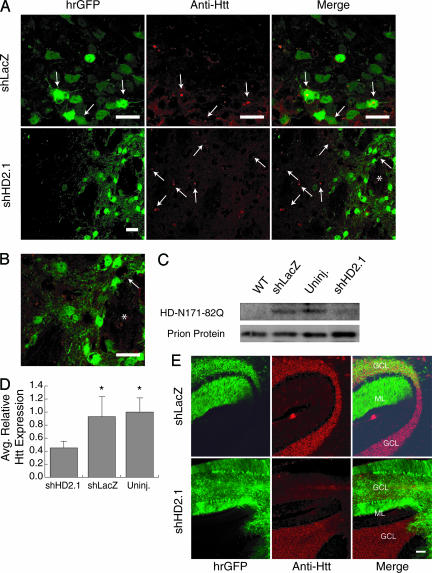
AAV.shHD2.1 Reduces HD-N171-82Q Expression in Vivo. We next investigated the effects of RNAi on the characteristic HD-associated neuronal inclusions and HD-N171-82Q mRNA levels in vivo. Tissues were harvested from end-stage HD-N171-82Q mice (≈5.5 months of age) because striatal inclusions are less robust at earlier ages in this model. In striata from HD-N171-82Q mice injected with AAV.shHD2.1, htt-reactive inclusions were absent in transduced cells, compared with untransduced regions (Fig. 3A Lower and Fig. 3B). Conversely, abundant inclusions were detected in transduced regions from AAV.shLacZ-injected HD mice (Fig. 3A Upper). No inclusions were observed in WT mice (data not shown). Western blot analysis performed on striatal extracts harvested before inclusion formation (2 weeks after injection; 5 weeks of age) also showed that soluble HD-N171-82Q monomer was decreased in mouse striata transduced with AAV.shHD2.1, compared with uninjected or AAV.shLacZ-injected controls (Fig. 3C). In addition, HD-N171-82Q mRNA was reduced 51–55% in A AV.shHD2.1-injected HD mice, compared w ith AAV.shLacZ-treated or untreated HD mice (Fig. 3D). AAV.shHD2.1 and AAV.shLacZ had no effect on endogenous mouse htt mRNA expression. (Average htt expression: uninjected HD, 1.00 ± 0.09; uninjected WT, 1.13 ± 0.04; AAV.shLacZ-injected HD, 1.10 ± 0.08; AAV.shHD2.1-injected HD, 1.08 ± 0.05.)
Fig. 3.
AAV.shHD2.1 eliminates the accumulation of htt-reactive neuronal inclusions and reduces HD-N171-82Q mRNA in vivo.(A) Representative photomicrographs show htt-reactive inclusions (arrows) in HD striatal cells transduced with AAV.shLacZ but not AAV.shHD2.1. (Scale bar, 20 μm.) (B) Higher-magnification photomicrograph from A (Lower Right) showing lack of htt-reactive inclusions in cells transduced by AAV.shHD2.1. * serves as a marker for orientation. (Scale bar, 20 μm.) (C) Representative Western blot demonstrates decreased HD-N171-82Q expression in mouse striata transduced with AAV.shHD2.1, compared with uninjected or AAV.shLacZ-injected striata. Prion protein was used as a loading control to normalize for tissues expressing the HD-N171-82Q transgene. (D) AAV.shHD2.1-treated HD mice showed a 55% average reduction in HD-N171-82Q mRNA, compared with AAV.shLacZ or uninjected HD mice. Data are means + SEM relative to uninjected HD samples. *, Difference from AAV.shHD2.1 samples, P < 0.05; ANOVA. (E) Mice were injected with either AAV.shHD2.1 or AAV.shLacZ directly into the cerebellum. Cerebellar sections confirm that AAV.shHD2.1, but not AAV.shLacZ, reduces htt immunoreactivity. GCL, granule cell layer; ML, molecular layer. (Scale bar, 100 μm.)
Neuronal inclusions in HD-N171-82Q striata are variable. Inclusions may be present in 10–50% of all striatal neurons in different end-stage HD-N171-82Q mice (20). In contrast, robust and widespread EM48-positive inclusions are present in cerebellar granule cells by ≈3 months of age (see ref. 20 and Fig. 3), and cerebellar HD-N171-82Q mRNA levels are ≈8-fold higher, compared with striatum (quantitative real-time RT-PCR, data not shown). This high-level cerebellar expression is partially attributable to the transcriptional profile of the prion promoter driving HD-N171-82Q transgene expression (20). Cerebellar inclusions are not typically found in the brains of adult-onset HD patients. However, cerebellar pathology has been reported in juvenile-onset HD cases, which are the most severe forms of the disease, and interestingly, in Hdh140 knock-in mice as early as 4 months of age (14, 27–30). The abundant inclusions in HD-N171-82Q cerebellar neurons provide a second target for assessing the effects of AAV.shHD2.1 on target protein levels. Direct cerebellar injections were performed in a separate cohort of mice, and HD-N171-82Q expression was examined by immunofluorescence. The data in Fig. 3 show that shHD2.1 reduces HD-N171-82Q expression as measured by EM48 immunoreactivity in transduced (hrGFP-positive) cells, whereas AAV.shLacZ does not. Together, the data show that AAV.shHD2.1, but not control AAV.shLacZ, reduces mutant htt expression and prevents the formation of the disease-associated neuronal inclusions.
Striatal Delivery of AAV.shHD2.1 Improves Established Behavioral Phenotypes. The effects of shRNA treatment on established behavioral deficits and animal weight were tested. RNAi directed to striatum did not normalize the notable weight differences between HD-N171-82Q and WT mice (shHD2.1-injected, 22.7 ± 3.8 g; shLacZ, 22.6 ± 2.8 g), compared with age-matched WT mice (shHD2.1, 26.3 ± 0.4; shLacZ, 27.3 ± 5.8), confirming that intracerebral injection confines RNAi therapy to the site of application (20, 26). However, significant improvements in stride length measurements and rotarod deficits were noted.
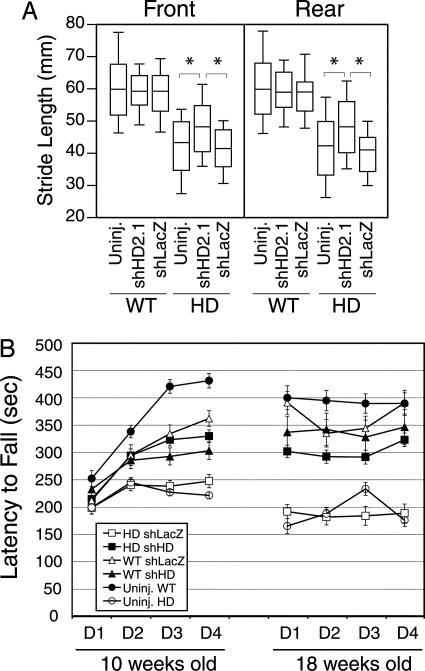
Stride length and rotarod tests were performed on uninjected mice, and mice were injected bilaterally with either AAVshHD2.1 or AAVshLacZ into the striatum. As shown in Fig. 4A, HD-N171-82Q mice display significantly shorter stride lengths than WT mice, consistent with prior work (14, 25, 31). Gait deficits in AAV.shHD2.1-treated HD-N171-82Q mice were significantly improved, compared with AAV.shLacZ-treated (improvements for front and rear strides, 13% and 15%, respectively; P < 0.0001) and uninjected HD-N171-82Q mice (front and rear strides, 14% and 18%, respectively; P < 0.0001). Gait improvements did not fully resolve; all HD-N171-82Q groups remained significantly different from their age-matched WT littermates. There was no effect of A AV.shLacZ or AAV.shHD2.1 expression on the stride lengths of WT mice.
Fig. 4.
AAV.shHD2.1 improves behavioral deficits in HD-N171-82Q mice. (A) The box plot shows that the bilateral striatal delivery of AAV.shHD2.1 improved stride length in HD-N171-82Q mice. HD mice had significantly shorter stride lengths, compared with WT mice. AAV.shHD2.1 mediated significant gait improvement, compared with control-treated HD mice. *, P < 0.0001; ANOVA with Scheffé's post hoc test. (B) Bilateral striatal delivery of AAV.shHD2.1 significantly improved rotarod performance in HD-N171-82Q mice. Only AAV.shLacZ-injected and uninjected HD-N171-82Q mice declined significantly with time. Data are means ± SEM.
The accelerating rotarod test was used to confirm the beneficial behavioral effects of RNAi targeted to the mutant human HD allele (20). Mice were left uninjected or were injected bilaterally into the striatum with either AAV.shLacZ or AAV.shHD2.1 at 4 weeks of age, followed by rotarod analyses at 10 and 18 weeks of age (Fig. 4B). By 10 weeks, uninjected and AAV.shLacZ-injected HD mice showed impaired performance, compared with all other groups, and continued to demonstrate significantly reduced performance over the course of the study (P < 0.05, relative to all other groups). Importantly, HD mice treated with AAVshHD2.1 showed dramatic behavioral improvements, compared with control-treated HD mice (P < 0.0008) (Fig. 4B). AAV.shLacZ-treated HD mice showed a 22% decline (P < 0.005; ANOVA), whereas AAV.shHD2.1-treated HD mice displayed a modest, nonsignificant 3% drop in rotarod performance between 10 and 18 weeks of age. The partial normalization of rotarod deficits in HD mice injected with AAV.shHD2.1, compared with WT mice, was consistent with the gait analyses.
We found no significant difference in stride length or decline in rotarod performances over time, in WT mice left untreated or injected with shRNA-expressing AAVs (Fig. 4). We did note differences in rotarod performance between uninjected and injected WT mice at the 10-week time point, however. Importantly, this difference in rotarod performance resolved by 18 weeks of age. These data suggest that there was some detrimental effect of direct brain injection on rotarod performance from which the mice recovered over time. Although these data do not rule out the possibility that RNAi expression may cause potentially detrimental off-target effects on a cellular or molecular level, they do suggest that RNAi expression in mammalian brain had no overt negative impact on motor behavior (Fig. 4).
Discussion
We show that motor and neuropathological abnormalities in a relevant HD mouse model are significantly improved by reducing striatal expression of a pathogenic htt allele by using AAV1-delivered shRNA. Our laboratory previously showed that RNAi can improve neuropathology and behavioral deficits in a mouse model of spinocerebellar ataxia type 1 (26), a dominant neurodegenerative disorder that affects a population of neurons distinct from those degenerating in HD.
The shHD2.1 hairpin sequence was developed before studies describing optimal shRNA design (32–35). Review of the hairpin sequence according to currently understood rules suggests that our intended guide strand may not be preferentially loaded into the RNAi-inducing silencing complex. However, shHD2.1 did reduce htt expression in vitro and in vivo, and importantly, our Northern blot analysis data suggest that the processed active guide strand was protected by the RNAi-inducing silencing complex in vivo. It is possible that shRNAs that more closely follow currently understood guidelines for processing in vitro may further improve correction of HD-associated phenotypes in vivo.
Striatal delivery of AAV.shHD2.1 had no effect on weight loss in HD-N171-82Q mice. The underlying cause of weight loss in mice and humans is unclear but may be an indicator of the systemic nature of the disease irrespective of, or indirectly related to, brain involvement (36). It has been suggested that expanded htt may cause hypothalamic dysfunction leading to weight loss in HD patients and HD-N171-82Q mice (37, 38). The inability to normalize weight differences in this study may be attributed to the focal striatal delivery of AAV.shHD2.1. It is possible that normalization of weight differences and greater improvements in motor function could be accomplished by a broader injection regimen.
Prior work demonstrated an essential role for htt in embryogenesis and postnatal neurogenesis (39–42). However, the effect of partial reduction of normal htt expression in adult, postmitotic neurons in vivo is unknown. In the current study, shHD2.1 reduced expression of a mutant, disease-causing human htt transgene but had no effect on normal mouse htt expression because of sequence differences between mouse and human genes. In HD patients, shHD2.1 would be expected to reduce the expression of both the mutant and normal htt alleles. Thus, RNAi as a therapy for HD will require addressing two important issues. First, can adult striatal neurons tolerate and benefit from partial nonselective knockdown of both the normal and disease alleles? Our data show that HD-like symptoms can be improved by partial reduction of mutant htt expression, suggesting that complete elimination of mutant allele expression may not be required. Second, disease allele-specific silencing will require identification and testing of disease-linked polymorphisms, one of which has been identified in exon 58 (43). This proof-of-principle work in the HD-N171-82Q-transgenic model provides evidence that disease allele-specific silencing can be accomplished in vivo.
In summary, we show that RNAi can dramatically improve HD-associated abnormalities, including pathological and behavioral deficits, in a mouse model of HD. Our data suggest the feasibility of treating HD by directly reducing mutant htt gene expression by using RNAi and support its general applicability to treating other dominant neurodegenerative disorders.
Acknowledgments
We thank C. McLennan for manuscript preparation, T. Taylor for excellent technical assistance, Jianqiang Shao for microscopy help, X. J. Li for providing mEM48 antibody, and D. Borchelt (Johns Hopkins School of Medicine, Baltimore) for furnishing the HD-N171-82Q plasmid. In addition, we thank P. B. McCray, Jr., for comments on the manuscript. This work was supported by grants from the National Institutes of Health and the Cure HD Initiative (to S.Q.H. and B.L.D.).
Author contributions: S.Q.H. and B.L.D. designed research; S.Q.H., P.D.S., X.H., S.L.E., I.H.M., and Q.M. performed research; L.Y. and R.M.K. contributed new reagents/analytic tools; S.Q.H., P.D.S., H.L.P., and B.L.D. analyzed data; S.Q.H. and B.L.D. wrote the paper; H.L.P. was a consultant and collaborator; and B.L.D. was the principal investigator.
Abbreviations: HD, Huntington's disease; htt, huntingtin; RNAi, RNA interference; shRNA, short hairpin RNA; AAV, adenoassociated virus; hrGFP, humanized Renilla GFP.
References
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2.Gusella, J. F. & MacDonald, M. E. (2000) Nat. Rev. Neurosci. 1, 109–115. [DOI] [PubMed] [Google Scholar]
- 3.Ferrante, R. J., Kubilus, J. K., Lee, J., Ryu, H., Beesen, A., Zucker, B., Smith, K., Kowall, N. W., Ratan, R. R., Luthi-Carter, R. & Hersch, S. M. (2003) J. Neurosci. 23, 9418–9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ona, V. O., Li, M., Vonsattel, J. P., Andrews, L. J., Khan, S. Q., Chung, W. M., Frey, A. S., Menon, A. S., Li, X. J., Stieg, P. E., et al. (1999) Nature 399, 263–267. [DOI] [PubMed] [Google Scholar]
- 5.Ferrante, R. J., Andreassen, O. A., Dedeoglu, A., Ferrante, K. L., Jenkins, B. G., Hersch, S. M. & Beal, M. F. (2002) J. Neurosci. 22, 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreassen, O. A., Dedeoglu, A., Ferrante, R. J., Jenkins, B. G., Ferrante, K. L., Thomas, M., Friedlich, A., Browne, S. E., Schilling, G., Borchelt, D. R., et al. (2001) Neurobiol. Dis. 8, 479–491. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka, M., Machida, Y., Niu, S., Ikeda, T., Jana, N. R., Doi, H., Kurosawa, M., Nekooki, M. & Nukina, N. (2004) Nat. Med. 10, 148–154. [DOI] [PubMed] [Google Scholar]
- 8.Karpuj, M. V., Becher, M. W., Springer, J. E., Chabas, D., Youssef, S., Pedotti, R., Mitchell, D. & Steinman, L. (2002) Nat. Med. 8, 143–149. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez, I., Mahlke, C. & Yuan, J. (2003) Nature 421, 373–379. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto, A., Lucas, J. J. & Hen, R. (2000) Cell 101, 57–66. [DOI] [PubMed] [Google Scholar]
- 11.Beal, M. F. & Ferrante, R. J. (2004) Nat. Rev. Neurosci. 5, 373–384. [DOI] [PubMed] [Google Scholar]
- 12.Levine, M. S., Cepeda, C., Hickey, M. A., Fleming, S. M. & Chesselet, M. F. (2004) Trends Neurosci. 27, 691–697. [DOI] [PubMed] [Google Scholar]
- 13.Lin, C. H., Tallaksen-Greene, S., Chien, W. M., Cearley, J. A., Jackson, W. S., Crouse, A. B., Ren, S., Li, X. J., Albin, R. L. & Detloff, P. J. (2001) Hum. Mol. Genet. 10, 137–144. [DOI] [PubMed] [Google Scholar]
- 14.Menalled, L. B., Sison, J. D., Dragatsis, I., Zeitlin, S. & Chesselet, M. F. (2003) J. Comp. Neurol. 465, 11–26. [DOI] [PubMed] [Google Scholar]
- 15.McBride, J. L., Behrstock, S. P., Chen, E. Y., Jakel, R. J., Siegel, I., Svendsen, C. N. & Kordower, J. H. (2004) J. Comp. Neurol. 475, 211–219. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson, J. G., Agopyan, N., Gutekunst, C. A., Leavitt, B. R., LePiane, F., Singaraja, R., Smith, D. J., Bissada, N., McCutcheon, K., Nasir, J., et al. (1999) Neuron 23, 181–192. [DOI] [PubMed] [Google Scholar]
- 17.Slow, E. J., van Raamsdonk, J., Rogers, D., Coleman, S. H., Graham, R. K., Deng, Y., Oh, R., Bissada, N., Hossain, S. M., Yang, Y. Z., et al. (2003) Hum. Mol. Genet. 12, 1555–1567. [DOI] [PubMed] [Google Scholar]
- 18.Reddy, P. H., Williams, M., Charles, V., Garrett, L., Pike-Buchanan, L., Whetsell, W. O., Jr., Miller, G. & Tagle, D. A. (1998) Nat. Genet. 20, 198–202. [DOI] [PubMed] [Google Scholar]
- 19.Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W. & Bates, G. P. (1996) Cell 87, 493–506. [DOI] [PubMed] [Google Scholar]
- 20.Schilling, G., Becher, M. W., Sharp, A. H., Jinnah, H. A., Duan, K., Kotzuk, J. A., Slunt, H. H., Ratovitski, T., Cooper, J. K., Jenkins, N. A., et al. (1999) Hum. Mol. Genet. 8, 397–407. [DOI] [PubMed] [Google Scholar]
- 21.Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457–467. [DOI] [PubMed] [Google Scholar]
- 22.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 23.Urabe, M., Ding, C. & Kotin, R. M. (2002) Hum. Gene Ther. 13, 1935–1943. [DOI] [PubMed] [Google Scholar]
- 24.Schilling, G., Jinnah, H. A., Gonzales, V., Coonfield, M. L., Kim, Y., Wood, J. D., Price, D. L., Li, X. J., Jenkins, N., Copeland, N., et al. (2001) Neurobiol. Dis. 8, 405–418. [DOI] [PubMed] [Google Scholar]
- 25.Carter, R. J., Lione, L. A., Humby, T., Mangiarini, L., Mahal, A., Bates, G. P., Dunnett, S. B. & Morton, A. J. (1999) J. Neurosci. 19, 3248–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia, H., Mao, Q., Eliason, S. L., Harper, S. Q., Martins, I. H., Orr, H. T., Paulson, H. L., Yang, L., Kotin, R. M. & Davidson, B. L. (2004) Nat. Med. 10, 816–820. [DOI] [PubMed] [Google Scholar]
- 27.Nance, M. A. & Myers, R. H. (2001) Ment. Retard. Dev. Disabil. Res. Rev. 7, 153–157. [DOI] [PubMed] [Google Scholar]
- 28.Fennema-Notestine, C., Archibald, S. L., Jacobson, M. W., Corey-Bloom, J., Paulsen, J. S., Peavy, G. M., Gamst, A. C., Hamilton, J. M., Salmon, D. P. & Jernigan, T. L. (2004) Neurology 63, 989–995. [DOI] [PubMed] [Google Scholar]
- 29.Seneca, S., Fagnart, D., Keymolen, K., Lissens, W., Hasaerts, D., Debulpaep, S., Desprechins, B., Liebaers, I. & De Meirleir, L. (2004) Eur. J. Pediatr. 163, 717–721. [DOI] [PubMed] [Google Scholar]
- 30.Byers, R. K., Gilles, F. H. & Fung, C. (1973) Neurology 23, 561–569. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler, V. C., Gutekunst, C. A., Vrbanac, V., Lebel, L. A., Schilling, G., Hersch, S., Friedlander, R. M., Gusella, J. F., Vonsattel, J. P., Borchelt, D. R. & MacDonald, M. E. (2002) Hum. Mol. Genet. 11, 633–640. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S. & Khvorova, A. (2004) Nat. Biotechnol. 22, 326–330. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N. & Zamore, P. D. (2003) Cell 115, 199–208. [DOI] [PubMed] [Google Scholar]
- 34.Khvorova, A., Reynolds, A. & Jayasena, S. D. (2003) Cell 115, 209–216. [DOI] [PubMed] [Google Scholar]
- 35.Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., Ueda, R. & Saigo, K. (2004) Nucleic Acids Res. 32, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djousse, L., Knowlton, B., Cupples, L. A., Marder, K., Shoulson, I. & Myers, R. H. (2002) Neurology 59, 1325–1330. [DOI] [PubMed] [Google Scholar]
- 37.Li, S. H., Yu, Z. X., Li, C. L., Nguyen, H. P., Zhou, Y. X., Deng, C. & Li, X. J. (2003) J. Neurosci. 23, 6956–6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kremer, H. P. & Roos, R. A. (1992) Arch. Neurol. (Chicago) 49, 349. [DOI] [PubMed] [Google Scholar]
- 39.Nasir, J., Floresco, S. B., O'Kusky, J. R., Diewert, V. M., Richman, J. M., Zeisler, J., Borowski, A., Marth, J. D., Phillips, A. G. & Hayden, M. R. (1995) Cell 81, 811–823. [DOI] [PubMed] [Google Scholar]
- 40.Duyao, M. P., Auerbach, A. B., Ryan, A., Persichetti, F., Barnes, G. T., McNeil, S. M., Ge, P., Vonsattel, J. P., Gusella, J. F., Joyner, A. L., et al. (1995) Science 269, 407–410. [DOI] [PubMed] [Google Scholar]
- 41.White, J. K., Auerbach, W., Duyao, M. P., Vonsattel, J. P., Gusella, J. F., Joyner, A. L. & MacDonald, M. E. (1997) Nat. Genet. 17, 404–410. [DOI] [PubMed] [Google Scholar]
- 42.Dragatsis, I., Levine, M. S. & Zeitlin, S. (2000) Nat. Genet. 26, 300–306. [DOI] [PubMed] [Google Scholar]
- 43.Ambrose, C. M., Duyao, M. P., Barnes, G., Bates, G. P., Lin, C. S., Srinidhi, J., Baxendale, S., Hummerich, H., Lehrach, H., Altherr, M., et al. (1994) Somatic Cell Mol. Genet. 20, 27–38. [DOI] [PubMed] [Google Scholar]