Abstract
Background
Environmental sampling based on boot swabs and/or liquid manure samples is an upcoming strategy for the identification of paratuberculosis (paraTB) positive herds, but only limited data are available regarding the diagnostic performance of molecular detection methods (qPCR) versus faecal culture (FC) for this purpose. In the present study, the test characteristics of two different qPCR protocols (A and B) and a standardized FC protocol, for the detection of Mycobacterium avium subsp. paratuberculosis in boot swabs and liquid manure samples were evaluated.
Results
In 19 paraTB unsuspicious and 58 paraTB positive herds boot swabs and liquid manure were sampled simultaneously and analyzed in three different diagnostic laboratories. Using boot swabs and liquid manure, a substantial to excellent accordance was found between both qPCRs, for boot swabs also with culture, while for liquid manure the detection rate of culture was decreased after prolonged storage at −20 °C. The quantitative results of both qPCR methods correlated well for the same sample and also for boot swabs and liquid manure from the same herd. When cut-off threshold cycle (CT-)-values were applied as recommended by the manufacturers, herd level specificity (Sp) of qPCR B was below 100% for boot swabs and for both qPCRs for liquid manure. A decreased herd level sensitivity was encountered after adjustment of Sp to 100% and re-calculation of the cut-off CT-values.
Conclusions
qPCR is equally suitable as bacterial culture for the detection of Mycobacterium avium subsp. paratuberculosis in boot swabs and liquid manure samples. Both matrices represent easily accessible composite environmental samples which can be tested with reliable results. The data encourage qPCR testing of composite environmental samples for paraTB herd diagnosis.
Keywords: Cultural isolation, Diagnostic performance, DNA extraction, Environmental samples, qPCR
Background
Environmental sampling constitutes an upcoming strategy for the identification of paratuberculosis (paraTB)-positive dairy herds or for the classification of the paraTB herd status because of reasonable expenses and the simple sampling technique [1–3]. Sampling six composite environmental faecal samples from manure concentration areas and manure storage areas is one option for herd classification within the Voluntary Bovine Johne’s Disease Control Program (VBJDCP) in the US [4]. Using this number of samples per holding, the sensitivity of the technique is not significantly influenced by the sampled locations, while reduction of the sample number without loss of sensitivity demands proper selection of the sampled areas, preferably alleyways and manure lagoons, where manure from numerous cows accumulates and is well mixed [5]. Recently, the boot swab (BS) sampling technique which is established for Salmonella testing in poultry flocks was adapted for environmental sampling of Mycobacterium avium subsp. paratuberculosis (MAP) in dairy herds, an approach overcoming the issue of sampling location and further reducing effort and cost [6]. The detection limit of this approach in terms of within-herd prevalence (WHP) is low [7], but depends on the laboratory methods used to substantiate MAP in the samples. Faecal culture (FC), still considered the most sensitive method for MAP detection is time-consuming and prone to microbial contamination [8]. Molecular techniques utilizing nucleic acid amplification are time saving alternatives. Meanwhile, several end-point and more recently Real-Time PCR (qPCR) protocols with excellent analytical sensitivity based on different targets have been developed for the identification of MAP [9–15].
However, diagnostic sensitivity of molecular methods relative to cultural isolation depends on the extraction protocol used to isolate MAP DNA from the samples. Several factors influence the efficiency of DNA extraction: homogeneity of MAP distribution in the samples, adequate disruption of the mycobacterial cell wall, and removal of DNA amplification inhibitors [14, 16]. Different silica membrane-based and magnetic bead-based extraction protocols for MAP from faeces have been proposed [17–20]. Significant differences in the purity and yield of the obtained DNA have been reported. However, similar extraction efficiency can be achieved with optimized silica membrane-based as well as magnetic bead-based procedures [21]. The detection rate can be improved by including a microfiltration step in the sample pre-treatment [22].
Conflicting data exist regarding test performance of direct qPCR relative to cultural isolation for the detection of MAP in individual and pooled faecal samples. While some authors report a higher diagnostic sensitivity of direct qPCR [23–26], others found similar [27, 28], or even lower [29] detection rates of qPCR in comparison to culture.
A good agreement of quantitative results between both diagnostic methods was found when pooled faecal and environmental samples were examined, reflected in a good to excellent correlation between threshold cycle (CT-) value and colony forming unit (CFU) counts [30].
Because comparative data for BS and liquid manure (LM) are lacking the aim of the present study was to determine the diagnostic performance of two different direct qPCR protocols (one silica membrane-based, one magnetic bead-based) and faecal culture for these matrices. We hypothesize
that direct qPCR protocols are equally suitable for the detection of MAP in BS and LM samples as cultural isolation, and
that BS and LM samples are equally suitable for the detection of MAP in the barn environment of MAP infected cattle.
Methods
Study population
Seventy-seven dairy herds from Thuringia, Hesse and Saxony, all federal states of Germany, with different within herd prevalence of paraTB were selected for this study. Nineteen herds were certified as ‘paraTB-unsuspicious’ that is comparable to the ‘herd classification no. 6’ of the uniform program standards of the VBJDCP [4]. The other 58 herds were classified as MAP positive based on recent results of FC of individual faecal samples obtained for whole herd testing. All herds were housed in free stalls.
BS and LM sampling
The BS samples were collected as described by Eisenberg et al. [6] by a veterinarian of the animal health service from each federal state. Briefly, three BS were taken in parallel mainly in localizations with high cow traffic involving manure concentration areas which had been proven to be suitable for environmental sampling [31].
Additionally, samples from LM storage areas (tanks, lagoons, pits or pre-flooders) were obtained as described recently by Donat et al. [7].
Three laboratories working under quality management standards were involved in the study (LI – LIII). After sampling, BS and LM were immediately transported to either LI or LII. LM was divided into three aliquots of equal volume. All samples were frozen and stored at −20 °C for 1 to 2 months except LM for qPCR B. For technical reasons these latter samples were stored for 8 to 10 months at −20 °C. One BS and one aliquot of LM of each herd were shipped frozen to the two other laboratories.
FC of all samples was performed in the three laboratories. Furthermore, the samples were tested by two different qPCR methods, qPCR A (LII) and qPCR B (LIII).
Boot swab handling
BS were placed in 125 mL plastic cups with screw cap and cut into small pieces with sterile scissors. In LI and LII, 100 mL of sterile physiologic salt solution (Merck, Darmstadt, Germany) were added and faecal material was rinsed off the boot swab pieces by automatically shaking at 200 rpm for 30 min. The eluate was transferred into a sterile 50 mL centrifuge tube (Sarstedt, Nümbrecht-Elsenroth, Germany). The supernatant was discarded after centrifugation for 15 min at 2000 x g.
In LIII, 3 g of cut BS were directly processed for bacterial culture and qPCR as described below.
Bacterial culture
FC of processed BS as well as LM was done according to the official manual of diagnostic procedures published by the Friedrich-Loeffler-Institut, the German Federal Research Institute of Animal Health [32].
First, 3 g of faecal material or 5 g of cut BS (LIII) were mixed with 30 mL of a 0.75% hexadecylpyridinium chloride solution (Acros Organics, Fisher Scientific GmbH, Nidderau, Germany) for decontamination. After sedimentation of coarse material for 5 min, 20 mL of the supernatant were decanted, horizontally shaken for 30 min at 200 rpm and subsequently incubated for 48 h at room temperature in the dark. Then, the supernatant was discarded and 0.2 mL of the sediment was inoculated onto each of 3 tubes of commercial Herrold’s Egg Yolk Medium (HEYM) with mycobactin J, amphotericin B, nalidixic acid and vancomycin (ANV; Becton Dickinson, Heidelberg, Germany). After cultivation under aerobic conditions (37 °C ± 2 °C) for 7 days in a slanting position, the tubes were locked airtight and incubated vertically for another 11 weeks. Differentiation of suspected colonies was carried out by IS900 PCR [33]. Samples were characterized as MAP-positive (MAP+), MAP-negative (MAP-) or non-assessable (n.a.) due to microbial contamination. The number of cultivable bacteria was recorded semi-quantitatively as a score; score 1: up to 10 colonies/tube, score 2: 11–50 colonies/tube, score 3: 51–100 colonies/tube, score 4: >100 colonies/tube. The average score of the three tubes per sample was calculated.
DNA preparation and PCR
qPCR A, utilizing nucleic acid purification with magnetic beads
Nucleic acids were extracted from faeces using the MagMax™ Total NucleicAcid isolation kit (Life Technologies, Darmstadt, Germany) according to the instruction of the manufacturer, which uses zirconia beads for the mechanical disruption and magnetic beads for nucleic acid purification.
The samples were thawed, homogenized and 0.3 g were transferred into 1 mL phosphate buffered saline. After homogenization by vortexing for 3 min and centrifugation at 100 x g for 60 s, 175 μl of the supernatant were added to the zirconia bead tubes previously filled with 235 μl lysis/binding solution. The samples were homogenized three times for 30 s at 6800 rpm (Precellys® 24 homogenizer, Bertin Technologies, Montigny-le-Bretonneux, France) and centrifuged at 16,000 x g for 3 min.
To clarify the lysate, 300 μl of the sample were transferred into a new 1.5 mL tube and centrifuged for a second time at 16,000 x g for 6 min.
The MagMax™ Express 96 instrument (Life Technologies) was applied for nucleic acid purification. After four washing steps the purified DNA was directly used for the qPCR.
For the detection of MAP DNA, the VetMAX™ MAP Real-Time PCR screening kit (Life Technologies), performed on a 7500 Fast qPCR cycler (Life Technologies), was used according to the instruction of the manufacturer.
A sample was detected to be positive at CT-values ≤37.0, inconclusive at CT-values >37.0 and <40.0. Samples with atypical amplification curves or with undetermined CT-values for target and internal control (PCR inhibition) were considered n.a.
qPCR B, utilizing sample enrichment and silica membrane-based nucleic acid purification
5.0 g of cut BS or 3.0 g of LM, respectively, were suspended in 20 mL sterile distilled water and left for 10 to 20 min at room temperature for sedimentation of coarse material. To concentrate bacteria and remove PCR inhibiting substances, 10 mL of the supernatant were centrifiltrated using Adiafilter (ADIFIL 100, Adiagene, Saint Brieux, France) and the pellet was re-dissolved in 500 μL sterile distilled water. Bacterial cells were mechanically disrupted by bead-beating with 300 mg glas beads (Peqlab, Erlangen, Germany) for 10 min at 30 Hz using the TissueLyser (Qiagen, Hilden, Germany). After an initial centrifugation step (5 min, 15,000 x g) the nucleic acid extraction was carried out using the QIAamp® DNA Mini kit (Qiagen) according to the instruction of the manufacturer. The qPCR was performed with the Adiavet™ Paratb Real time kit (Adiagene) using the 7500 Real Time PCR system (Applied Biosystems, Foster City, CA, USA) as recommended. Samples were tested in duplicate and the mean CT-value of both replicates was calculated. A sample was detected to be positive at mean CT-values ≤38.0, inconclusive at mean CT-values >38.0 and ≤40.0 and n.a. when one replicate yielded a measurable and the other an undetermined CT-value for the target.
Statistical data analysis
Data recording and descriptive statistics were performed using a Microsoft Excel® spreadsheet (Microsoft Office 2007, Microsoft Corporation, Redmond, USA). Further statistical examinations were done by means of the statistical programme packages BMDP [34] and StatXact 9.0 [35]. The figures were created with the statistical software package PASW Statistics 17 (SPSS, Quarry Bay, Hongkong).
For each matrix, BS and LM samples, the Spearman’s rank correlation coefficient (programme BMDP3D) was calculated in order to analyse the relationship between the semi-quantitative colony growth score of FC and the CT-values of qPCR.
Kappa coefficients were calculated with the programme BMDP4F to describe the degree of agreement between the dichotomized outcomes (negative or positive) of the FC and the qPCR test systems. The following levels of agreement were considered for the interpretation of the kappa coefficient (ĸ) [36]: < 0.20: slight; 0.21–0.40: low; 0.41–0.60: moderate; 0.61–0.80: substantial; 0.81–1.00: excellent. Furthermore, both calculations were performed for the description of the accordance between qPCR A and B for each matrix, and for the accordance between BS and LM using either PCR A or B. Herd level Se and Sp for both qPCR test systems applied either on BS or LM samples were estimated by a frequency-based approach classifying herds according to their certification status (MAP+, MAP-) using the cut-off CT-values as recommended by the manufacturers for faecal samples. The corresponding exact 95% confidence intervals of the binomial distribution were calculated according to Clopper and Pearson [37]. Cut-offs for both qPCR tests were estimated by TG-ROC-analysis [38, 39]. Herd level Se and the cut-offs of the qPCRs were re-calculated after adjusting the values for herd level Sp to 100%. These calculations were performed using R software [40], version 2.15.2 (2012–10-26), with package DiagnosisMed [41]. In general, the statistical significance level was set at α = 0.05.
Results
A total of 77 pairs of BS and LM samples were tested in parallel by FC in the three laboratories. Additionally, all samples were analyzed by one of two different direct qPCR methods. The proportion of culture positive samples was comparable between the three laboratories and between BS and LM, except for LM in LIII, where a markedly lower number of samples were tested positive (see Table 1). LM tended to a higher number of n.a. samples (due to fungal overgrowth) than BS, although the proportion was low in both matrices (BS 0–1.3%, LM 2.6%). Compared to cultural isolation, with qPCR the number of positive BS and LM was slightly lower and the proportion of n.a. samples was higher, especially when LM were analyzed (Table 1). The reasons for n.a. results differed between the two qPCR protocols. When qPCR A was applied they were mainly due to inhibition. With qPCR B the samples contained only very low amounts of target DNA leading to inconsistent results of the two replicates. A substantial to excellent accordance was observed between the results (positive or negative) of FC and qPCR for BS within the two laboratories and in LII also for LM (Table 2). In LIII, due to the reduced number of culture-positive LM samples (Table 1), only a moderate accordance between the results of both methods could be achieved (Table 2). Semi-quantitative FC results and qPCR CT-values were significantly negative correlated with concordant Spearman rank correlation coefficients (rs) for BS (qPCR A − 0.878; qPCR B − 0.889) and LM (qPCR A − 0.886; qPCR B − 0.730) (Table 2).
Table 1.
Results of the analysis of boot swabs and liquid manure samples (n/ %) from 77 herds by faecal culture in three labs (I to III) and by two different qPCR methods (qPCR A and qPCR B) for Mycobacterium avium ssp. paratuberculosis (MAP)
| Methode of MAP detection | Laba | Boot swab | Liquid manure | ||||
|---|---|---|---|---|---|---|---|
| posb | negc | n.a.d | pos | neg | n.a. | ||
| Faecal culture | I | 40/ 51.9 | 37 /48.0 | 0 /0.0 | 39 /50.6 | 36 /46.8 | 2 /2.6 |
| Faecal culture | II | 41/ 53.2 | 36 /46.8 | 0 /0.0 | 38 /49.4 | 37 /48.0 | 2 /2.6 |
| Faecal culture | III | 41 /53.2 | 35 /45.5 | 1 /1.3 | 21 /27.3 | 54 /70.1 | 2 /2.6 |
| qPCR A | II | 35 /45.5 | 39 /50.6 | 3 /3.9 | 33 /42.9 | 35 /45.5 | 9 /11.7 |
| qPCR B | III | 37 /48.0 | 36 /46.8 | 4 /5.2 | 38 /49.4 | 32 /41.6 | 7 /9.1 |
aLaboratory
bMAP-positive
cMAP-negative
dNon-assessable faecal culture due to contamination or non-assessable qPCR results
Table 2.
Accordance between the results of faecal culture and the two qPCRs for boot swabs and liquid manure samples
| Matrix | qPCR | rs a,b | ĸ ± SEapp c |
|---|---|---|---|
| Boot swab | A | - 0.878 | 0.757 ± 0.076 |
| B | - 0.889 | 0.808 ± 0.069 | |
| Liquid manure | A | - 0.866 | 0.794 ± 0.074 |
| B | - 0.730 | 0.514 ± 0.102 |
aSpearman rank correlation coefficient
bp-value for all Spearman rank correlation coefficients: P < 0.0001
cCohen’s kappa coefficient ± approximate standard error
The test performance of the two qPCRs in terms of positive and negative outcome was comparable as reflected in Cohen’s kappa values which demonstrate a substantial accordance for BS (κ: 0.629 ± 0.092) and LM (κ: 0.688 ± 0.090). The quantitative results of qPCR A and B correlated well, with rs of 0.886 (BS) and 0.885 (LM) (Fig. 1a and b).
Fig. 1.
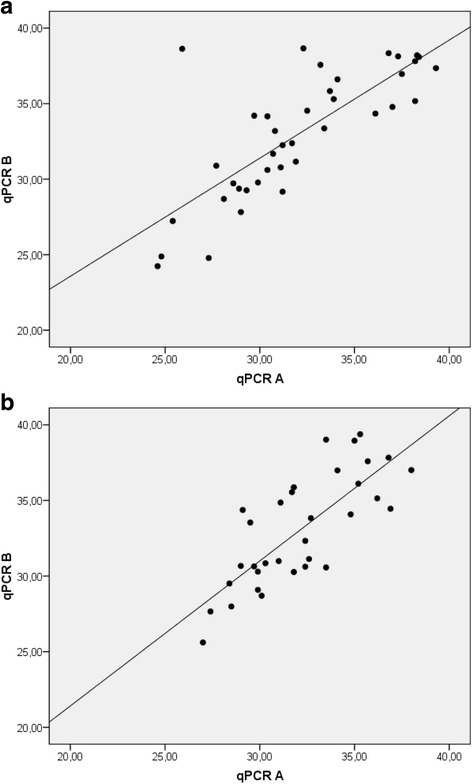
Correlation diagram of the CT-values of qPCR A and B for test positive samples. a using boot swabs and b using liquid manure samples. Included are only samples with positive qPCR results in both tests based on the cut-off values as recommended by the manufacturers’
Using qPCR A test results (positive or negative) for both specimens, BS and LM, from one herd corresponded clearly (κ: 0.784 ± 0.077), while the accordance between the results of qPCR B for both matrices was less pronounced (κ: 0.543 ± 0,104) (Table 3). However, the quantitative qPCR results (CT-values) of BS and LM from the same herd correlated markedly for qPCR A (rs: 0.845) and qPCR B (rs: 0.779).
Table 3.
Accordance between matrices, boot swab and liquid manure, using qPCR A or qPCR B
| qPCR | rs a,b | ĸ ± SEapp c |
|---|---|---|
| A | 0.845 | 0.784 ± 0.077 |
| B | 0.779 | 0.543 ± 0.104 |
aSpearman rank correlation coefficient
bp-value for all Spearman rank correlation coefficients: P < 0.0001
cCohen’s kappa coefficient ± approximate standard error
Herd level Se of qPCR B was slightly higher than that of qPCR A for both matrices, when cut-off CT-values were applied as recommended by the manufacturers. However, under these conditions, the herd level Sp of qPCR B for BS and of qPCR A and B for LM was below 100% (Table 4), leading to false positive test results. Re-calculation after adjustment of Sp to approximately 100% resulted in decreased cut-off CT-values and, consequentially, in a reduced Se for these test-specimen combinations (Table 5).
Table 4.
Test characteristics of qPCR A and B for analysis of boot swabs and liquid manure samples using the cut-off CT-values as recommended by the manufacturers’ for faecal samples
| Matrix | qPCR | Cut-off CT a | Test + /Status +b | Se [%] (95% CI) |
Test - /Status -c | Sp [%] (95% CI) |
|---|---|---|---|---|---|---|
| Boot swab | A | 37.00 | 35/57 | 61.40 (47.57–74.00) |
17/17 | 100.00 (80.49–100.00) |
| B | 38.00 | 36/55 | 65.45 (51.42–77.76) |
16/17 | 94.44 (72.71–99.86) |
|
| Liquid manure | A | 37.00 | 32/51 | 62.75 (48.08–75.87) |
16/17 | 94.12 (71.31–99.85) |
| B | 38.00 | 36/53 | 67.92 (53.68–80.08) |
15/17 | 88.24 (63.56–98.54) |
aCT-values ≤37.00 (qPCR A) and ≤38.00 (qPCR B) were considered positive and CT-values >37.00 (qPCR A) and >38.00 (qPCR B) were considered negative.
bNumber of test positive samples per total number of assessable samples from MAP-positive herds (n/n).
cNumber of test negative samples per total number of assessable samples from MAP-negative herds (n/n)
Table 5.
Results of the TG-ROC analysis: Cut-off CT-values and Se of qPCR A and B when Sp was adjusted to 100.0%
| Matrix | qPCR | Cut-off CT | Se [%] (95% CI) |
Sp [%]a
(95% CI) |
|---|---|---|---|---|
| Boot swab | A | 39.30 | 73.68 (60.34–84.46) |
100.00 (80.49–100.00) |
| B | 36.12 | 50.91 (37.07–64.65) |
100.00 (81.47–100.00) |
|
| Liquid manure | A | 35.10 | 50.98 (36.60–65.25) |
100.00 (80.49–100.00) |
| B | 35.99 | 50.94 (36.84–64.94) |
100.00 (80.94–100.00) |
aSp was fixed at 100.0%, the corresponding Se and cut-offs were calculated
Discussion
The present data support the concept that qPCR is equally suitable for the detection of MAP in composite environmental samples as bacterial culture on solid media. Recently, it has been shown that use of qPCR can yield rapid, quantitative estimates of MAP load in pooled faecal and composite environmental samples that were collected according to the recommendations of the USDA National Animal Health Monitoring System (NAHMS) [30]. However, in this study, qPCR was applied to BS for the first time, and test performance on samples from paraTB-unsuspicious herds has not been studied so far.
The herd level Sp of both qPCR methods was below 100% when cut-off CT values were applied as to the recommendations of the manufacturers. Under these conditions, positive PCR results in supposedly paraTB-unsuspicious herds could have different reasons: (1) a very low WHP of MAP-infected animals which was not detected before or, (2) cross-contamination of samples during processing from sample collection in the herd to DNA extraction and PCR set up in the laboratory. It cannot be excluded that single very low shedders have been overlooked in herds with a history of complete negative results of bacterial culture on the individual animal level. Moreover, cross contamination during PCR workflow which has only rarely been addressed in the literature [42–44], has to be considered, particularly when samples are collected in the typical environment of dairy cattle herds and when sample preparation protocols comprise several manual handling steps. Semi-automated sample preparation seems to be less prone to cross contamination, because qPCR A, for which such a DNA extraction protocol was applied, had a higher Sp than qPCR B. However, the results have to be interpreted with caution because the number of paraTB-unsuspicious herds in this study (n = 19) was rather low.
Both qPCR methods are equally suitable for the examination of environmental samples as bacterial culture as reflected by the substantial to excellent accordance between both methods for BS (both qPCRs) and LM (qPCR A). Storage of LM samples at −20 °C for several months resulted in a decreased detection rate of bacterial culture (Table 1), leading to a lower Cohen’s κ value (0.514 ± 0.102) and reduced correlation of the quantitative results (rs: - 0.730, Table 2). This confirms data from Aly et al. [30] and points to the fact that MAP detection by qPCR is less affected by prolonged frozen sample storage than detection of viable bacteria by culture. However, the proportion of n.a. test results of both qPCR methods was higher than for faecal culture. Reasons were, on the one hand, samples with divergent results because two replicates per sample were analyzed (qPCR B). We assume that even a very low content of target DNA which is irregularly distributed in the DNA extract is detected because of the high analytical Se of the method. Re-sampling and testing of the respective herds may result in a reliable herd classification. On the other hand, n.a. qPCR results were due to complete or incomplete inhibition of amplification (qPCR A), reflected by no or non-typical amplification curves. It has been shown that inhibition is relieved by dilution of the DNA extract [45], which is also recommended by the qPCR manufacturer but was not done in the present study. Generally, LM were more affected by n.a. results than BS. It seems that in LM i) dilution of MAP positive faeces is more probable and ii) the mixture of faecal material with urine and other effluents increases the risk of qPCR inhibition.
The excellent (qPCR A) or substantial (qPCR B) accordance between the results of BS and LM indicates that both matrices can be reliably examined by qPCR. Combination of BS and LM leads to an even higher detection rate than testing only one matrix [7]. Cohen’s κ value of qPCR B (0.543, Table 3) was lower than that of qPCR A (0.784). This is due to the fact that qPCR B detected a higher proportion of samples with low bacterial load and therefore, frequently only one of the two matrices was positive, leading to lower accordance and a lower κ value. In contrast, the excellent accordance between BS and LM when tested with qPCR A is due to a lower sensitivity of the method causing a higher proportion of negative qPCR results in samples with low bacterial load. This applies predominantly to samples from herds with low WHP [7]. A good correlation between the CT-values of BS and LM was noted for both qPCRs. This shows that the MAP load in LM is related to the MAP load in the herd environment which in turn is dependent on WHP [7].
Despite a slightly higher Se of qPCR B both PCRs showed comparable results. Cohen’s κ values point to a moderate accordance. qPCR B encompasses a DNA extraction protocol that includes a microfiltration step in combination with silica membrane-based mini-columns which has been shown to be one of the most sensitive protocols [22].
Using the cut-off CT-values as recommended by the manufacturers, Sp and Se of both qPCR’s for both matrices are comparable to each other (Table 4). As the definition of the herd status MAP+ based on herd history including also herds without actual shedders at the day of environmental sampling, the estimates for sensitivity are rather conservative in our study. Similar values have been achieved when a set of composite environmental samples was examined by bacterial culture [3] as recommended by the USDA for herd classification [4].
Only a low number of paraTB-unsuspicious herds was included in the present study (n = 19). Therefore, only a few false positive test results account for a considerable drop of Sp of the methods. As already discussed above, several reasons for false positive test results have to be considered. Altogether, a larger proportion of paraTB-unsuspicious herds is necessary to verify the Sp results of both qPCRs for BS and LM. Adjustment of Sp to about 100% and re-calculation of the cut-off CT-values resulted in diminished Se, because samples with high CT-values were classified negative, except for qPCR A when boot swabs were examined. For this test configuration, re-calculation resulted in a higher cut-off CT-value, and Se was not altered, because no false positive samples existed. As discussed for Sp, a few false positive test results account for marked shifts of the re-calculated cut-off CT-values and, therefore, these cut-offs cannot be recommended for practical use.
Conclusions
qPCR is equally suitable as bacterial culture for the detection of MAP in composite environmental samples. Boot swab and LM samples represent easily accessible composite environmental samples which can be tested with reliable results. Examination of larger numbers of herds is necessary to fix valid cut-off CT-values for qPCR methods which ensure a high herd level Sp in combination with a high diagnostic Se.
Acknowledgements
The authors thank Sandy Werner, Annett Held, Kerstin Trübger and Ulrike Kling for excellent technical assistance, all involved veterinarians for sample collection and the dairy farmers for allowing access to their herds.
Funding
The study was funded by the Hessian Animal Diseases Fund, the Saxon Animal Diseases Fund, the Thuringian Animal Diseases Fund and the Thuringian Dairy Industry Association (Landesvereinigung Thüringer Milch e. V.).
Availability of data and materials
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ANV
Amphotericin B, nalidixic acid, vancomycin
- BS
Boot swab(s)
- CFU
Colony forming units
- CT
Threshold cycle
- FC
Faecal culture
- HEYM
Herrold’s Egg Yolk medium with mycobactin and ANV
- LM
Liquid manure
- MAP
Mycobacterium avium subsp. paratuberculosis
- n.a.
Non-assessable
- paraTB
Paratuberculosis
- qPCR
Quantitative polymerase chain reaction
- rs
Spearman rank correlation coefficient
- Se
Sensitivity
- Sp
Specificity
- WHP
Within-herd prevalence
- κ
kappa
Authors’ contributions
KF, TE, PMZ, KD, EE and HK conceived of the study, participated in its design and coordination. NH contributed to sample collection. TE, KS, EE and HK coordinated sample processing, bacterial culture and PCR in the different laboratories. KF performed the statistical analyses. All authors contributed to the analysis and interpretation of the data. NH, EE, KD and HK drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nathalie Hahn, Email: leaha_2001@web.de.
Klaus Failing, Email: Klaus.Failing@vetmed.uni-giessen.de.
Tobias Eisenberg, Email: Tobias.Eisenberg@lhl.hessen.de.
Karen Schlez, Email: Karen.Schlez@lhl.hessen.de.
Peter-Michael Zschöck, Email: Peter-Michael.Zschoeck@lhl.hessen.de.
Karsten Donat, Email: kdonat@thueringertierseuchenkasse.de.
Esra Einax, Email: eeinax@thueringertierseuchenkasse.de.
Heike Köhler, Email: heike.koehler@fli.bund.de.
References
- 1.Pillars RB, Grooms DL, Woltanski JA, Blair E. Prevalence of Michigan dairy herds infected with Mycobacterium avium subspecies paratuberculosis as determined by environmental sampling. Prev Vet Med. 2009;89:191–196. doi: 10.1016/j.prevetmed.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Aly SS, Anderson RJ, Whitlock RH, Fyock TL, McAdams SC, Byrem TM, et al. Cost-effectiveness of diagnostic strategies to identify Mycobacterium avium subspecies paratuberculosis super-shedder cows in a large dairy herd using antibody enzyme-linked immunosorbent assays, quantitative real-time polymerase chain reaction, and bacterial culture. J Vet Diagn Investig. 2012;24:821–832. doi: 10.1177/1040638712452107. [DOI] [PubMed] [Google Scholar]
- 3.Lavers CJ, McKenna SLB, Dohoo IR, Barkema HW, Keefe GP. Evaluation of environmental fecal culture for Mycobacterium avium subspecies paratuberculosis detection in dairy herds and association with apparent within-herd prevalence. Can Vet J. 2013;54:1053–1060. [PMC free article] [PubMed] [Google Scholar]
- 4.Uniform Program Standards for the Voluntary Bovine Johne’s Disease Control Program, (2010).
- 5.Wolf R, Barkema HW, De Buck J, Orsel K. Sampling location, herd size, and season influence Mycobacterium avium ssp paratuberculosis environmental culture results. J Dairy Sci. 2015;98:275–287. doi: 10.3168/jds.2014-8676. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberg T, Wolter W, Lenz M, Schlez K, Zschöck M. Boot swabs to collect environmental samples from common locations in dairy herds for Mycobacterium avium ssp paratuberculosis (MAP) detection. J Dairy Res. 2013;80:485–489. doi: 10.1017/S002202991300040X. [DOI] [PubMed] [Google Scholar]
- 7.Donat K, Hahn N, Eisenberg T, Schlez K, Köhler H, Wolter W, et al. Within-herd prevalence thresholds for the detection of Mycobacterium avium subspecies paratuberculosis-positive dairy herds using boot swabs and liquid manure samples. Epidemiol Infect. 2016;144:413–424. doi: 10.1017/S0950268815000977. [DOI] [PubMed] [Google Scholar]
- 8.Kim YG, Bechnielsen S, Gordon JC, Slemons RD, Spangler E. Comparison of two methods for isolation of Mycobacterium paratuberculosis from bovine fecal samples. Am J Vet Res. 1989;50:1110–1113. [PubMed] [Google Scholar]
- 9.Ravva SV, Stanker LH. Real-time quantitative PCR detection of Mycobacterium avium subsp paratuberculosis and differentiation from other mycobacteria using SYBR Green and TaqMan assays. J Microbiol Methods. 2005;63:305–317. doi: 10.1016/j.mimet.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Herthnek D, Bölske G. New PCR systems to confirm real-time PCR detection of Mycobacterium avium subsp paratuberculosis. BMC Microbiol. 2006;6 [DOI] [PMC free article] [PubMed]
- 11.Schönenbrücher H, Abdulmawjood A, Bülte M. Real time-PCR-assay for the detection of Mycobacterium avium ssp paratubercuiosis - Development and validation. Fleischwirtschaft. 2006;86:123–125. [Google Scholar]
- 12.Schönenbrücher H, Abdulmawjood A, Failing K, Bülte M. New Triplex Real-Time PCR Assay for Detection of Mycobacterium avium subsp. paratuberculosis in bovine feces. Appl Environ Microbiol. 2008;74:2751–2758. doi: 10.1128/AEM.02534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irenge LM, Walravens K, Govaerts M, Godfroid J, Rosseels V, Huygen K, et al. Development and validation of a triplex real-time PCR for rapid detection and specific identification of M. avium subsp. paratuberculosis in faecal samples. Vet Microbiol. 2009;136:166–172. doi: 10.1016/j.vetmic.2008.09.087. [DOI] [PubMed] [Google Scholar]
- 14.Sevilla IA, Garrido JM, Molina E, Geijo MV, Elguezabal N, Vazquez P, et al. Development and evaluation of a novel multicopy-element-targeting triplex PCR for detection of Mycobacterium avium subsp paratuberculosis in feces. Appl Environ Microbiol. 2014;80:3757–3768. doi: 10.1128/AEM.01026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fawzy A, Eisenberg T, El-Sayed A, Zschöck M. Improvement of sensitivity for Mycobacterium avium subsp. paratuberculosis (MAP) detection in bovine fecal samples by specific duplex F57/IC real-time and conventional IS900 PCRs after solid culture enrichment. Trop Anim Health Prod. 2015;47:721–726. doi: 10.1007/s11250-015-0784-9. [DOI] [PubMed] [Google Scholar]
- 16.Amaro A, Duarte E, Amado A, Ferronha H, Botelho A. Comparison of three DNA extraction methods for Mycobacterium bovis, Mycobacterium tuberculosis and Mycobacterium avium subsp avium. Lett Appl Microbiol. 2008;47:8–11. doi: 10.1111/j.1472-765X.2008.02372.x. [DOI] [PubMed] [Google Scholar]
- 17.Cook KL, Britt JS. Optimization of methods for detecting Mycobacterium avium subsp paratuberculosis in environmental samples using quantitative, real-time PCR. J Microbiol Methods. 2007;69:154–160. doi: 10.1016/j.mimet.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Herthnek D, Nielsen SS, Lindberg A, Bölske G. A robust method for bacterial lysis and DNA purification to be used with real-time PCR for detection of Mycobacterium avium subsp paratuberculosis in milk. J Microbiol Methods. 2008;75:335–340. doi: 10.1016/j.mimet.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhang MZ, Zhang SP. An efficient DNA extraction method for polymerase chain reaction-based detection of Mycobacterium avium subspecies paratuberculosis in bovine fecal samples. J Vet Diagn Investig. 2011;23:41–48. doi: 10.1177/104063871102300106. [DOI] [PubMed] [Google Scholar]
- 20.Salgado M, Steuer P, Troncoso E, Collins MT. Evaluation of PMS-PCR technology for detection of Mycobacterium avium subsp paratuberculosis directly fromty bovine fecal specimens. Vet Microbiol. 2013;167:725–728. doi: 10.1016/j.vetmic.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Leite FL, Stokes KD, Robbe-Austerman S, Stabel JR. Comparison of fecal DNA extraction kits for the detection of Mycobacterium avium subsp. paratuberculosis by polymerase chain reaction. J Vet Diagn Investig. 2013;25:27–34. doi: 10.1177/1040638712466395. [DOI] [PubMed] [Google Scholar]
- 22.Sting R, Hrubenja M, Mandl J, Seemann G, Salditt A, Waibel S. Detection of Mycobacterium avium subsp paratuberculosis in faeces using different procedures of pre-treatment for real-time PCR in comparison to culture. Vet J. 2014;199:138–142. doi: 10.1016/j.tvjl.2013.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Bögli-Stuber K, Kohler C, Seitert G, Glanemann B, Antognoli MC, Salman MD, et al. Detection of Mycobacterium avium subspecies paratuberculosis in Swiss dairy cattle by real-time PCR and culture: a comparison of the two assays. J Appl Microbiol. 2005;99:587–597. doi: 10.1111/j.1365-2672.2005.02645.x. [DOI] [PubMed] [Google Scholar]
- 24.Scott HM, Fosgate GT, Libal MC, Sneed LW, Erol E, Angulo AB, et al. Field testing of an enhanced direct-fecal polymerase chain reaction procedure, bacterial culture of feces, and a serum enzyme-linked immunosorbent assay for detecting Mycobacterium avium subsp paratuberculosis infection in adult dairy cattle. Am J Vet Res. 2007;68:236–245. doi: 10.2460/ajvr.68.3.236. [DOI] [PubMed] [Google Scholar]
- 25.Logar K, Kopinc R, Bandelj P, Staric J, Lapanje A, Ocepek M. Evaluation of combined high-efficiency DNA extraction and real-time PCR for detection of Mycobacterium avium subsp paratuberculosis in subclinically infected dairy cattle: comparison with faecal culture, milk real-time PCR and milk ELISA. BMC Vet Res. 2012;8 [DOI] [PMC free article] [PubMed]
- 26.Hanifian S, Khani S, Barzegari A, Shayegh J. Quantitative real-time PCR and culture examination of Mycobacterium avium subsp paratuberculosis at farm level. Vet Microbiol. 2013;162:160–165. doi: 10.1016/j.vetmic.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Alinovi CA, Ward MP, Lin TL, Moore GE, Wu CC. Real-time PCR, compared to liquid and solid culture media and ELISA, for the detection of Mycobacterium avium ssp paratuberculosis. Vet Microbiol. 2009;136:177–179. doi: 10.1016/j.vetmic.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Plain KM, Marsh IB, Waldron AM, Galea F, Whittington AM, Saunders VF, et al. High-throughput direct fecal PCR assay for detection of Mycobacterium avium subsp paratuberculosis in sheep and cattle. J Clin Microbiol. 2014;52:745–757. doi: 10.1128/JCM.03233-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller SM, Stephan R, Künzler R, Meylan M, Wittenbrink MM. Comparison of fecal culture and F57 real-time polymerase chain reaction for the detection of Mycobacterium avium subspecies paratuberculosis in Swiss cattle herds with a history of paratuberculosis. Acta Vet Scand. 2014;56 [DOI] [PMC free article] [PubMed]
- 30.Aly SS, Mangold BL, Whitlock RH, Sweeney RW, Anderson RJ, Jiang JM, et al. Correlation between Herrold’s egg yolk medium culture and real-time quantitative polymerase chain reaction results for Mycobacterium avium subspecies paratuberculosis in pooled fecal and environmental samples. J Vet Diagn Investig. 2010;22:677–683. doi: 10.1177/104063871002200501. [DOI] [PubMed] [Google Scholar]
- 31.Donat K, Kube J, Dressel J, Einax E, Pfeffer M, Failing K. Detection of Mycobacterium avium subspecies paratuberculosis in environmental samples by faecal culture and real-time PCR in relation to apparent within-herd prevalence as determined by individual faecal culture. Epidemiol Infect. 2015;143:975–985. doi: 10.1017/S0950268814002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anonymus. Paratuberculose. Amtliche Methodensammlung: Meldepflichtige Tierkrankheiten. Friedrich-Loeffler-Institut, Bundesforschungsinstitut für Tiergesundheit. 2014.
- 33.Englund S, Ballagi-Pordany A, Bölske G, Johansson KE. Single PCR and nested PCR with a mimic molecule for detection of Mycobacterium avium subsp. paratuberculosis. Diagn Microbiol Infect Dis. 1999;33:163–171. doi: 10.1016/S0732-8893(98)00098-4. [DOI] [PubMed] [Google Scholar]
- 34.Dixon WJ. BMDP Statistical Software Manual. Berkeley, Los Angeles, London: University of California Press; 1993. [Google Scholar]
- 35.Anonymus. Cytel Studio StatXact Vers 9.0.0. Statistical Software for Exact Nonparametric Inference. 9.0.0 ed. Cambridge, MA 02139, U.S.A.: CYTEL Inc.; 2010.
- 36.Grouven U, Benden R, Ziegler A, Lange S. The kappa coefficient. Dt Med Wschr. 2007;132(Suppl 2):65–68. doi: 10.1055/s-2007-959046. [DOI] [PubMed] [Google Scholar]
- 37.Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. doi: 10.1093/biomet/26.4.404. [DOI] [Google Scholar]
- 38.Greiner M, Sohr D, Göbel PA. Modified ROC analysis for the selection of cut-off values and the definition of intermediate results of serodiagnostic tests. J Immunol Methods. 1995;185:123–132. doi: 10.1016/0022-1759(95)00121-P. [DOI] [PubMed] [Google Scholar]
- 39.Greiner M. Two-graph receiver operating characteristic (TG-ROC): update version supports optimization of cut-off values that minimize overall misclassification costs. J Immunol Methods. 1996;191:93–94. doi: 10.1016/0022-1759(96)00013-0. [DOI] [PubMed] [Google Scholar]
- 40.Team RDC . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 41.Brasil P. DiagnosisMed: Diagnostic test accuracy evaluation for medical professionals. R package. 0.2.3. ed2010.
- 42.Kox LFF, Rhienthong D, Miranda AM, Udomsantisuk N, Ellis K, Vanleeuwen J, et al. More reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1994;32:672–678. doi: 10.1128/jcm.32.3.672-678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marsh I, Whittington R, Millar D. Quality control and optimized procedure of hybridization capture-PCR for the identification of Mycobacterium avium subsp paratuberculosis in faeces. Mol Cell Probes. 2000;14:219–232. doi: 10.1006/mcpr.2000.0309. [DOI] [PubMed] [Google Scholar]
- 44.Liu HB, Gan Y, Yang B, Weng H, Huang CM, Yang DF, et al. Performance evaluation of the Maxwell 16 System for extraction of influenza virus RNA from diverse samples. PloS one. 2012;7(10): e48094. doi:10.1371/journal.pone.0048094. [DOI] [PMC free article] [PubMed]
- 45.Acharya KR, Dhand NK, Whittington RJ, Plain KM. PCR Inhibition of a quantitative PCR for detection of Mycobacterium avium subspecies paratuberculosis DNA in feces: Diagnostic implications and potential solutions. Front Microbiol. 2017;8:115. doi:10.3389/fmicb.2017.00115. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


