Abstract
Background
Aeromonas spp. are gram-negative bacteria that can cause a variety of infections in both humans and animals and play a controversial role in diarrhea outbreaks. Our aim was to identify clinical and environmental Aeromonas isolates associated with a cholera outbreak in a northeast county of Brazil at the species level. We also aimed to determine the genetic structure of the bacterial population and the virulence potential of the Aeromonas isolates.
Methods and results
Analysis based on concatenated sequences of the 16S rRNA and gyrB genes suggested the classification of the 119 isolates studied into the following species: A. caviae (66.9%), A. veronii (15.3%), A. aquariorum (9.3%), A. trota (3.4%), A. hydrophila (3.4%) and A. jandaei (1.7%). One isolate did not fit any Aeromonas species assessed, which might indicate a new species. The haplotype network based on 16S rRNA gene sequences identified 59 groups among the 119 isolates and 26 reference strains, and it clustered almost all A. caviae isolates into the same group. The analysis of the frequency patterns of seven virulence-associated genes (alt, ast, hlyA, aerA, exu, lip, flaA/B) revealed 29 virulence patterns composed of one to seven genes. All the isolates harbored at least one gene, and three of them harbored all seven virulence genes.
Conclusion
The results emphasize the need to improve local water supply and maintain close monitoring of possible bacterial contamination in the drinking water.
Keywords: Aeromonas, Taxonomy, Virulence, Diarrhea, Outbreaks
Background
The genus Aeromonas comprises many species of gram-negative bacteria, and it harbors common inhabitants of aquatic environments with some species involved in human and animal infections. In humans, gastroenteritis is the most frequent clinical manifestation of an Aeromonas infection. Aeromonas-associated diarrhea occurs worldwide in both industrialized and developing countries, affecting all age groups and both healthy and immunocompromised individuals. However, a role for Aeromonas in diarrhea outbreaks remains controversial [1, 2].
The Aeromonas taxonomy is complex and ever changing due to the introduction of new identification criteria leading to the description of new species and reclassification of known ones [3]. Although analyses based on 16S rRNA gene sequencing are universally accepted to establish phylogenetic relationships for all bacterial genera, the high identity among the 16S rRNA nucleotide sequences of the Aeromonas species can impair the taxonomic classification of the Aeromonas genus [4, 5]. Therefore, housekeeping genes such as gyrB have been used for taxonomic inference of this group [6].
The pathogenesis of Aeromonas spp. is also unclear. Many virulence factors involved in different steps of infection mechanisms have been described in different Aeromonas species. These virulence factors include (1) fimbriae, flagella, and a capsule that allow attachment to the host surface; (2) toxins and enzymes such as proteases, elastases, lipases, and hemolysins that cause cell and tissue damage; (3) secretion systems that enable evasion of the host immune response; (4) iron-binding proteins called siderophores that scavenge iron from the host; (5) a capsule, S-layer, lipopolysaccharide, and porins that compromise host defenses; (6) biofilm formation that allows adherence to cell surfaces; and (7) quorum-sensing systems that can modulate bacterial virulence gene expression [1–3].
Hofer et al. [7] reported high prevalence of Aeromonas spp. as the sole pathogen in diarrheic patient feces, whereas Vibrio cholerae O1 prevalence was lower during an alleged cholera outbreak. The infections were attributed to the precariousness of the drinking water supply lacking microbiological control in the region, where a local river both receives untreated sewage and serves as a drinking water source. Despite the higher prevalence of Aeromonas isolates in patient feces, V. cholerae O1 was entailed as the etiological agent of the outbreak [8]. This issue invited further characterization of those Aeromonas isolates. Therefore, this study aimed to identify the clinical and environmental Aeromonas outbreak isolates at the species level using a combination of 16S rRNA and gyrB gene analysis to elucidate the genetic structure and virulence potential of the Aeromonas population.
Methods
Bacterial strains
This study involved 119 Aeromonas spp. isolates (103 from diarrheal patient feces and 16 from the aquatic environment) obtained from March to June 2004 during a diarrhea outbreak that occurred in São Bento do Una, a Brazilian city located in Pernambuco state [7, 8]. Clinical isolates were numbered from 251 to 374 and environmental isolates were numbered from 420 to 707. The cultures were stored at − 80 °C in Brain Heart Infusion (BHI) broth plus 25% glycerol. The reference strains of Aeromonas hydrophila subsp. hydrophila ATCC 7966T, Aeromonas veronii bv. veronii ATCC 35624T and Aeromonas caviae ATCC 15468T were used as controls.
DNA extraction
Each culture was inoculated into BHI and incubated at 37 °C for 24 h for DNA extraction, following a protocol based on Leal et al. [9]. Briefly, 1 ml of the culture was centrifuged, and the pellet was homogenized in 500 μl of TE (TrisHCl 10 mmol l−1, EDTA 1 mmol l−1) pH 8.0 and 10 μl of proteinase K (5 mg ml−1) and incubated at 60 °C for 20 min. Next, 100 μl of STE (sodium dodecyl sulfate 2.5%, TrisHCl 10 mmol l−1, pH 8.0, EDTA 0.25 mol l−1) was added, and the suspension was incubated for 15 min at 60 °C, 5 min at room temperature and 5 min on ice, followed by the addition of 130 μl of ammonium acetate (7.5 mol l−1). After 15 min on ice, the material was centrifuged, and the supernatant was transferred to a new tube and purified with phenol:chloroform:isoamyl alcohol (25:24:1). DNA in the upper phase was precipitated with an equal volume of isopropanol. The sediment was suspended in 10 μl RNAse solution (10 mg ml−1) and stored at −20 °C.
Identification of the Aeromonas spp. isolates at the species level
The DNA from the 119 Aeromonas spp. isolates was analyzed by sequencing the 16S rRNA and gyrB genes to identify isolates at the species level. Polymerase Chain Reactions (PCR) were executed as previously described [6, 8]. The amplicons (~1503 base pairs (bp) for the 16S rRNA and ~1100 bp for gyrB) were purified using ExoSAP-IT PCR Cleanup (Affymetrix, Cleveland, OH, USA) according to the vendor. The purified products were sequenced using an Applied Biosystems 3100 sequencer (Applied Biosystems, Foster City, CA, USA) using the PCR primers described in Table 1. To guarantee sequence accuracy, each nucleotide sequence was determined at least twice. Sequence assembly and editing were performed using the tools called Pregap4 and Gap4, which belong to the Staden package [10].
Table 1.
Sequences of the primers used for phylogenetic and virulence potential analysis
| Gene | Sequence (5′-3′) | Reference |
|---|---|---|
| PCR | ||
| 16S rRNAF | AGAGTTTGATCATGGCTCAG | Borrell et al. [19] |
| 16S rRNAR | GGTTACCTTGTTACGACTT | Borrell et al. [19] |
| gyrB 3F | TCCGGCGGTCTGCACGGCGT | Yáñez et al. [20] |
| gyrB 14R | TTGTCCGGGTTGTACTCGTC | Yáñez et al. [20] |
| hlyAF | GGCCGGTGGCCCGAAGATACGGG | Heuzenroeder et al. [21] |
| hlyAR | GGCGGCGCCGGACGAGACGGG | Heuzenroeder et al. [21] |
| altF | CCATCCCCAGCCTTTACGCCAT | Martínez et al. [22] |
| altR | TTTCACCGAGGTGACGCCGT | Martínez et al. [22] |
| astF | ATGCACGCACGTACCGCCAT | Martínez et al. [22] |
| astR | ATCCGGTCGTCGCTCTTGGT | Martínez et al. [22] |
| Sequencing | ||
| 16S BF | AGCAGTGGGGAATATTGCAC | This study |
| 16S BR | GGCAACAAAGGACAGGGGT | This study |
| 16S CF | ACGCAGGCGGTTGGATAAGT | This study |
| 16S CR | AAATCGACATCGTTTACGGCG | This study |
| 16S DF | AACCTTACCTGGCCTTGACA | This study |
| 16S DR | CACACACGCGGCATGGTGCATC | This study |
| gyrB 9Rs | CCTTGACCGAAATGACCGCC | Yáñez et al. [20] |
| gyrB 7F | GGGGTCTACTGCTTCACCAA | Yáñez et al. [20] |
| gyrB 9R | ACCTTGACGGAGATAACGGC | Yáñez et al. [20] |
Sequences of 1454 bp and 920 bp for 16S rRNA and gyrB genes, respectively, were used in the final analysis. These partial gene sequences were aligned using the Muscle program [11] independently and as concatenated sequences (2374 bp) with those from reference strains of published Aeromonas sequences available from the National Center for Biotechnology Information (NCBI) database at the time this study was conducted. Salmonella enterica ATCC 13311 T was included when outgroup rooting was needed for the analysis, as the two genera are closely related. The GenBank accession numbers of the utilized sequences are listed in Table 2.
Table 2.
Accession numbers of the 16S rRNA and gyrB sequences of type strains of Aeromonas spp.
| Type strains | 16S rRNA accession numbers |
gyrB
accession numbers |
|---|---|---|
| A. molluscorum LMG 22214 | AY987772.1 | AY987538.1 |
| A. tecta CECT 7082 | HQ832416.1 | HQ442662.1 |
| A. media ATCC 33907 | X74679.1 | AF417627._1 |
| A. eucrenophila ATCC 23309 | X74675.1 | AF417629.1 |
| A. encheleia CECT 4342 | HQ832414.1 | Ay101799.1 |
| A. popoffii CECT 5176 | HQ832415.1 | AY101801.1 |
| A. bestiarum LMG 13444 | AY987755.1 | AY987521.1 |
| A. piscicola CECT 7443 | HQ832417.1 | HQ442690.1 |
| A. schubertii ATCC 43700 | X60416.2 | AF417628._1 |
| A. simiae MDC 2374 | GQ860944.1 | GQ860942.1 |
| A. jandaei ATCC 49568 | X60413.2 | AF242651.3 |
| A. trota ATCC 49657 | X60415.2 | AF417633.1 |
| A. salmonicida CECT 894 | Ay987751.1 | JN711820.1 |
| A. allosaccharophila CECT 4199 | S39232.2 | HQ442733.1 |
| A. aquariorum MDC 47 | EU085557.2 | EU268444.1 |
| A. diversa CECT 4254 | GQ365710.1 | HQ442756.1 |
| A. rivuli DSM 22539 | FJ976900.1 | FJ969434.1 |
| A. bivalvium 868E | NR_043885.1 | EF465525.1 |
| A. fluvialis CECT 7401 | FJ230078.2 | FJ603455.1 |
| A. sobria JCM 2139 | AB472942.1 | AB473084.1 |
| A. sanarellii CECT 7402 | FJ230076.1 | FJ807277.1 |
| A. taiwanensis CECT 7403 | FJ230077.1 | FJ807272.1 |
| A. cavernicola MDC 2508 | HQ436040.1 | HQ442702.1 |
| Salmonella enterica subsp. enterica ATCC 13311 | NR_119108.1 | EU014643.1 |
Available in the GenBank/NCBI database: https://www.ncbi.nlm.nih.gov//
Genetic distances and clustering were determined using Kimura’s 2-parameter model [12] and phylogenetic trees were generated by the neighbor-joining method [13] with MEGA program, version 6 [14]. Neighbor-joining tree support was evaluated with 1000 bootstrap replicates.
Population genetic analyses
To assess the relationships among the 119 Aeromonas isolates and 26 reference strains, a haplotype network was constructed based on the 16S rRNA gene sequences (1454 bp) using the eBURST program version 3.0 [15, 16]. The columns containing polymorphic sites within the 16S rRNA sequence alignment were extracted, and a number N of bp was obtained for the 145 sequences analyzed.
A matrix was generated, with the numbers 1–4 replacing the adenine (A), cytosine (C), thymine (T) and guanine (G) bases, respectively, within the alignment and used as input. The eBURST analysis was performed using the default parameters. The network was defined as a group of closely related haplotypes, each one sharing a number N-1 of identical bases with at least one other member of this network. The stability of the relationships was assessed by bootstrap (1000 replicates).
Detection of virulence genes
The 119 isolates were analyzed by PCR to determine the presence of the virulence genes alt, ast and hlyA as described [8]. The sequence of primers used to amplify the target genes: alt (~338 bp), ast (260 bp) and hlyA (597 bp) are listed in Table 1. To confirm the identity of the amplified segments, PCR products of three isolates from the outbreak and the reference strain A. hydrophila subsp. hydrophila ATCC 7966 were sequenced using the same PCR primers. Sequences were compared to those available at the GenBank database using the standard nucleotide-nucleotide Basic Local Alignment Search Tool (BLAST) available at https://blast.ncbi.nlm.nih.gov/Blast.cgi.
To shed some light on the virulence panorama of the Aeromonas isolates, we analyzed the frequency pattern for seven virulence genes: alt, ast and hlyA determined in the present study and aerA, exu, lip and flaA/B, as previously determined [8].
Results
Identification of the Aeromonas isolates at the species level
Based on the phylogenetic trees generated separately by the 16S rRNA and gyrB gene sequences, a total of 31% and 11% of isolates, respectively, displayed ambiguous classification (data not shown). Therefore, the phylogenetic analysis was performed applying 16S rRNA and gyrB (2374 bp) concatenated sequences for 146 isolates. Of these isolates, 26 were reference strains and one was an outgroup. The neighbor-joining tree is shown in Fig. 1.
Fig. 1.
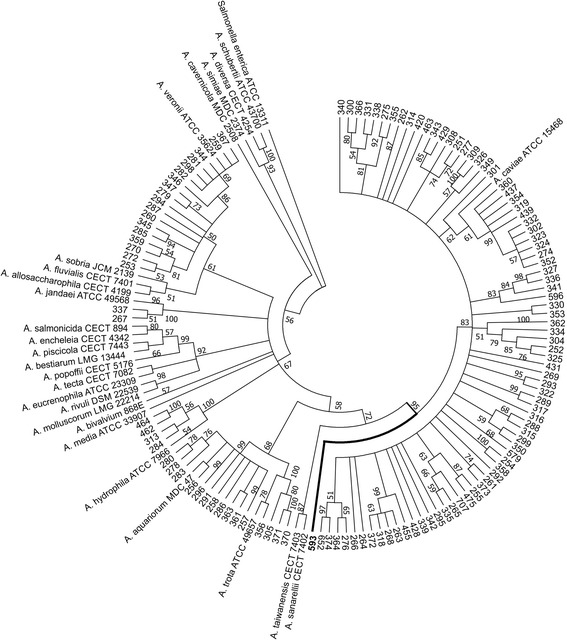
Circular phylogenetic tree (neighbor-joining) derived from concatenated 16S rRNA and gyrB (2374 bp) genes showing the relationship among 119 Aeromonas isolates, 26 known species of this genus and a Salmonella strain representing the outgroup. Numbers near the nodes indicate the bootstrap values ≥50 (percentage of 1000 replicates). The isolate 593 branch is highlighted
In total, 118 isolates clustered with the reference strains of six Aeromonas species: A. caviae (79 isolates = 66.9%), A. veronii (18 isolates = 15.3%), A. aquariorum (11 isolates = 9.3%), A. hydrophila (4 isolates = 3.4%), A. trota (4 isolates = 3.4%) and A. jandaei (2 isolates = 1.7%). All strains clustered with a reference strain in the tree with bootstrap values ≥50% (percentage of 1000 replicates). Just one environmental isolate, number 593, formed an independent branch distinct from the assessed species.
The clinical isolates fit into six species: A. caviae (66/103; 64.1%), A. veronii (18/103; 17.5%), A. aquariorum (11/103; 10.7%), A. trota (4/103; 3.9%), A. hydrophila (2/103; 1.9%) and A. jandaei (2/103; 1.9%). The environmental isolates fit into only two species: A. caviae (13/16; 81.3%) and A. hydrophila (2/16; 12.5%), plus one non-identified isolate.
Population genetic analysis
The relationship among the isolates was also assessed through the construction of a haplotype network based on the 16S rRNA gene sequences (1454 bp) of the 119 Aeromonas isolates and 26 reference strains using eBURST. This analysis clustered the 145 Aeromonas spp. sequences into 59 haplotypes or groups (G1-G59) as shown in Table 3. Out of these 59 haplotypes, 17 haplotypes distributed among 84 isolates and 3 reference strains clustered into a network represented by a radial diagram, in which each edge represents a nucleotide substitution (Fig. 2). 42 haplotypes distributed among 35 isolates (seven A. caviae, three A. trota, and all A. veronii, A. hydrophila and A. jandaei isolates) and 23 reference strains did not fit within the network and were categorized as satellite haplotypes.
Table 3.
Distribution of 59 haplotypes among 119 isolates and 26 type strains of Aeromonas species by eBURST
| Genotype | Isolates and type strains of Aeromonas spp. |
|---|---|
| 1 | 260, 282, A. veronii biov. Veronii ATCC 35624 |
| 2 | A. schubertii ATCC 43700 |
| 3 | 279 |
| 4 | A. salmonicida subsp. salmonicida CECT 894 |
| 5 | 299, 350 |
| 6 | 277 |
| 7 | 255, 475 |
| 8 | A. sobria JCM 2139 |
| 9 | 284, 313, A. hydrophila subsp. hydrophila ATCC 7966 |
| 10 | 278 |
| 11 | A. bestiarum LMG 13444, A. piscicola CECT 7443 |
| 12 | 253 |
| 13 | A. rivuli DSM 22539 |
| 14 | 335 |
| 15 | A. fluvialis CECT 7401 |
| 16 | A. simiae MDC 2374 |
| 17 | A. bivalvium 868E |
| 18 | 349 |
| 19 | 305 |
| 20 | 280 |
| 21 | 289 |
| 22 | 340 |
| 23 | 316 |
| 24 | 431 |
| 25 | 370, 371 |
| 26 | 294 |
| 27 | 267, A. jandaei ATCC 49568 |
| 28 | 455 |
| 29 | A. eucrenophila ATCC 23309 |
| 30 | 270, 272, 287 |
| 31 | 579 |
| 32 | 346 |
| 33 | 317 |
| 34 | 341 |
| 35 | 373 |
| 36 | 359 |
| 37 | A. allosaccharophila CECT 4199 |
| 38 | A. molluscorum LMG 22214 |
| 39 | 262, 264, 266, 276, 292, 293, 308, 314, 327, 331, 334, 336, 338, 339, 364 |
| 40 | 285 |
| 41 | 322 |
| 42 | 356, A. trota ATCC 49657 |
| 43 | 291 |
| 44 | A. media ATCC 33907 |
| 45 | 462, 464 |
| 46 | 259, 281, 298, 344, 345, 347, 367 |
| 47 | 337 |
| 48 | A. encheleia CECT 4342 |
| 49 | A. diversa CECT 4254 |
| 50 | 257, 286, 296 |
| 51 | 251, 254, 261, 263,265, 268, 269, 274, 275, 288, 295, 300, 301, 302, 304, 309, 315, 318, 319, 323, 324, 326, 330, 332,342, 352, 353, 354, 355, 358, 360, 362, 366, 372, 374, 420, 428, 429, 437, 439, 463, 596, 652,707, A. caviae ATCC 15468 |
| 52 | A. tecta CECT 7082 |
| 53 | 256, 258, 283, 361, 363, A. aquariorum MDC 47 |
| 54 | A. popoffii CECT 5176 |
| 55 | 252,325,343 |
| 56 | A. sanarellii CECT 7402 |
| 57 | 593 |
| 58 | A. taiwanensis CECT 7403 |
| 59 | A. cavernicola MDC 2508 |
Fig. 2.
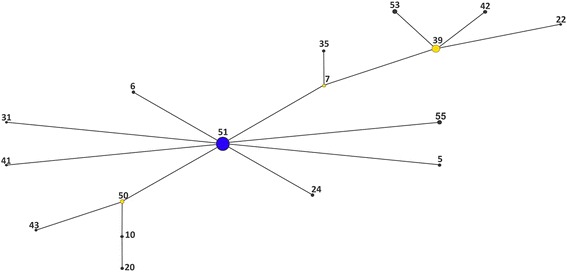
eBURST diagram based on 16S rRNA sequences from Aeromonas isolates and reference strains. Blue circles represent the population founder haplotype, yellow circles represent subgroup founder haplotypes, and black circles represent descending haplotypes
G51 (represented as a blue node in Fig. 2) clustered a higher number of strains (44/119; 37% of A. caviae isolates and the A. caviae ATCC 15468T) and is directly linked to eight derived haplotypes represented in yellow nodes in Fig. 2. G39 is the second most frequent haplotype and harbored two derived haplotypes closely related to A. caviae. G35 and G22 related to G7 and G39 respectively and are composed of A. caviae isolates. G50 and its related haplotypes (G10, G20 and G43) grouped only A. aquariorum isolates.
Some clinical and environmental A. caviae isolates were both closely related in the phylogenetic tree based on 16S rRNA and gyrB concatenated sequences and clustered into the same haplotypes in the network: 319, 354, 360, 437, 439 (G51); 374, 652 (G51); 255, 475 (G7); 265, 707 (G51).
Detection of virulence genes
The gene alt was amplified in 81% (82 clinical and 15 environmental) of isolates (97/119), the gene ast was amplified in 11% (8 clinical and 5 environmental) of isolates (13/119), and the gene hlyA was amplified in 12% (11 clinical and 3 environmental) of isolates (14/119).
The identity of the three gene segments (alt, ast and hlyA) amplified from representative isolates and one reference strain was confirmed through sequencing. The alignment with sequences deposited in the GenBank database resulted in 98 to 100% identity with the corresponding genes of Aeromonas.
The analysis of the gene profiles based on the distribution of the seven virulence-associated genes (aerA, exu, lip, flaA/B, alt, ast and hlyA) among the clinical and environmental Aeromonas species revealed 29 virulence patterns and is summarized in Table 4. All the isolates were positive for at least one of the genes, and three isolates (one environmental and one clinical A. hydrophila and one environmental A. caviae) amplified all seven genes. The pattern exu + lip + flaA/B+ alt + was the most frequent and particular of A. caviae occurring among 28 isolates. The second most frequent pattern (aerA+ exu + lip + flaA/B+ alt+) was found among 13 A. caviae, six A. veronii and two A. aquariorum isolates (Table 4).
Table 4.
Distribution of virulence-associated genes among 119 clinical and environmental isolates of Aeromonas species
| Species | aerA | exu | lip | fla A/B | alt | ast | hlyA | No. of isolates | |
|---|---|---|---|---|---|---|---|---|---|
| Clinical | A. caviae (n = 66) | − | − | − | − | + | − | − | 1 |
| − | − | + | − | − | − | − | 1 | ||
| − | + | − | − | + | − | − | 2 | ||
| − | − | + | − | + | − | − | 2 | ||
| − | + | − | + | − | − | − | 1 | ||
| − | − | + | + | + | − | − | 3 | ||
| − | + | + | − | + | − | − | 7 | ||
| − | + | + | + | − | − | − | 3 | ||
| − | + | − | + | + | − | − | 3 | ||
| − | + | + | + | + | − | − | 21 | ||
| − | + | + | + | − | + | − | 3 | ||
| − | − | + | + | + | + | − | 1 | ||
| + | − | + | + | + | − | − | 3 | ||
| + | + | + | − | + | − | − | 1 | ||
| + | + | + | + | + | − | − | 11 | ||
| + | + | + | − | + | + | − | 1 | ||
| − | + | + | + | + | + | − | 1 | ||
| + | + | + | + | + | + | − | 1 | ||
| A. hydrophila (n = 2) | + | + | − | + | + | − | + | 1 | |
| + | + | + | + | + | + | + | 1 | ||
| A. aquariorum (n = 11) | − | + | − | + | + | − | + | 1 | |
| − | + | − | + | − | + | + | 1 | ||
| + | − | + | + | + | − | + | 2 | ||
| + | + | + | + | + | − | − | 2 | ||
| + | + | + | + | + | − | + | 4 | ||
| − | + | + | + | + | + | + | 1 | ||
| A. trota (n = 4) | − | + | − | + | + | − | − | 1 | |
| + | + | − | + | − | − | − | 2 | ||
| + | + | − | + | + | − | − | 1 | ||
| A. jandaei (n = 2) | + | + | − | + | − | − | − | 1 | |
| + | + | + | + | − | − | − | 1 | ||
| A. veronii (n = 18) | − | − | + | + | + | − | − | 2 | |
| + | + | + | + | − | − | − | 9 | ||
| + | + | + | − | + | − | − | 1 | ||
| + | + | + | + | + | − | − | 6 | ||
| Environmental | A. caviae (n = 13) | − | + | + | + | + | − | − | 7 |
| − | + | + | − | + | + | − | 1 | ||
| + | + | + | + | + | − | − | 2 | ||
| − | + | + | + | + | + | − | 2 | ||
| + | + | + | + | + | + | + | 1 | ||
| A. hydrophila (n = 2) | − | + | + | + | + | + | + | 1 | |
| + | + | + | + | + | + | + | 1 | ||
| + | + | + | − | + | − | − | 1 |
Discussion
The taxonomy, virulence potential and pathogenic mechanisms of the genus Aeromonas are still unclear. Therefore, in the present study, we attempted to accurately identify a set of Aeromonas isolates from a diarrheal outbreak in a county in Northeast of Brazil at the species level and to assess their virulence potential.
In previous work [7], all Aeromonas spp. isolates had been submitted to unreliable biochemical identification. Some isolates, but not all (n = 77), had also been classified through Restriction Fragment Length Polymorphism (RFLP) [8], but many showed atypical banding patterns. Considering this, and since the RFLP results were different from the biochemical classification for some isolates (n = 23), in the present work we proposed to identify the Aeromonas spp. isolates at the species level through a phylogenetic analysis performed on concatenated 16S rRNA and gyrB gene sequences.
First, we conducted the analysis with the two genes sequences individually which provided ambiguous classification of the isolates. Although 16S gene sequences can be routinely used to distinguish and establish relationships between genera and well-resolved bacterial species, this gene alone is not sufficient to classify bacteria at the species level once it did not differentiate very recently diverged species [17].
On the other hand, a phylogenetic analysis based on concatenated sequences of the 16S rRNA and gyrB genes identified distinct clusters of species with high bootstrap confidence values. A. caviae was the most prevalent species, followed by A. veronii and A. aquariorum among the clinical isolates. Among the environmental samples A. caviae and A. hydrophila were most frequently identified.
The species A. hydrophila, A. veronii, and A. caviae had been formerly categorized as major human pathogens of the Aeromonas genus. More recently, A. aquariorum has been associated with gastroenteritis [18]. Despite the pathogenic importance of A. hydrophila, only two clinical isolates of this species were identified in our study. On the other hand, A. trota and A. jandaei rarely reported from diarrhea isolates occurred among the study clinical isolates.
The haplotype network built by the eBURST algorithm revealed a prevalent and probably well-adapted A. caviae clone (G51) connected to several less prevalent variant haplotypes. Specific point mutations allowed distinguishing the Aeromonas species based on 16S rRNA sequences. Actually, except for the grouping of A. bestiarum ATCC 13444T and A. piscicola CECT 7443T, no haplotype included different species, even the satellite genotypes.
The environmental isolate (number 593) was grouped in a phylogenetic branch different from the reference strains, and therefore, it is probably an unknown Aeromonas species. However, further studies are needed to confirm this assumption since at the time this study was conducted, not all currently described species had 16S rRNA and gyrB sequences deposited in the NCBI database. This analysis was also performed with gyrB sequences but was unsuccessful due to wide nucleotide variation.
Although their roles in human infections are not conclusively established, a number of putative virulence-associated genes were reported in several Aeromonas isolates [1–3]. In this work, we performed an analysis on 119 Aeromonas isolates for the presence and distribution of seven virulence-associated genes widely used to determine the pathogenicity potential of Aeromonas spp.: the genes coding for a cytotonic heat-labile (alt) and cytotonic heat-stable (ast) enterotoxins, a cytotoxic enterotoxin (hlyA), the cytolysin aerolysin (aerA), a DNAse (exu), a lipase (lip) and a flagellin (flaA/B) were assessed.
The patterns generated by the combined presence of the alt, ast and hlyA genes determined in this study and the genes aerA, exu, lip and flaA/B reported by Mendes-Marques et al. [8] produced 29 profiles according to the presence of the seven genes assessed. Taking into account the number of virulence genes present in the majority of the isolates, the species A. hydrophila and A. aquariorum exhibit higher virulence potential among the species studied, followed by A. caviae and A. veronii species. In spite of its higher prevalence among the clinical isolates compared to other species from the outbreak, A. caviae harbored a lower content of virulence-associated genes. However, different species could use different pathogenicity mechanisms, which may justify different gene contents. On the other hand, although some strains may harbor a certain gene, it may only be expressed in certain growth conditions [3].
Conclusion
Taking into account their potential as a public health threat, an accurate characterization of Aeromonas species is crucially important. The diversity of haplotypes and their close relationship in the phylogenetic network, even between clinical and environmental isolates, suggest that Aeromonas spp. are well established in the study region. Furthermore, the virulence gene content highlights the potential role of these bacteria in some diarrheic processes. These results emphasize the importance of monitoring Aeromonas spp. in the environment to effectively treat and control any future public health event.
Acknowledgments
The authors thank the Núcleo de Plataformas Tecnológicas (NPT) from Instituto Aggeu Magalhães (IAM/Fiocruz PE) for use of the sequencing platform.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author by request.
Abbreviations
- BHI
Brain Heart Infusion
- BLAST
Basic Local Alignment Search Tool
- NCBI
National Center for Biotechnology Information
- PCR
Polymerase Chain Reaction
- RFLP
Restriction Fragment Length Polymorphism
Authors’ contributions
NCL and LCAS conceived the study. LCAS cultured bacterial strains, did the PCR reactions, prepared samples for sequencing, and did sequence data analysis. AMR did the haplotype network. BSTM did the phylogenetic tree and figures. NCL and AMPA drafted the manuscript. CLMM and TCLB helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Concerning Ethics and Consent, no need for formal approval for the study was required according to IAM, Fiocruz PE policy. However, the work was conducted following the recommended FIOCRUZ guidelines for work with pathogenic organisms (BRASIL. Ministério da Saúde. Fundação Oswaldo Cruz. Procedimentos para a manipulação de microrganismos patogênicos e/ou recombinantes na FIOCRUZ. Rio de Janeiro, Comissão Técnica de Biossegurança da Fiocruz, 2005).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lívia Christina Alves da Silva, Email: liviacalves@yahoo.com.br.
Tereza Cristina Leal-Balbino, Email: cristina@cpqam.fiocruz.br.
Beatriz Souza Toscano de Melo, Email: biatoscano@gmail.com.
Carina Lucena Mendes-Marques, Email: clmendes@gmail.com.
Antonio Mauro Rezende, Email: antoniomauro@gmail.com.
Alzira Maria Paiva de Almeida, Email: aalmeida@cpqam.fiocruz.br.
Nilma Cintra Leal, Email: nilmacleal@gmail.com.
References
- 1.Batra P, Mathur P, Misra MC. Aeromonas spp.: an emerging nosocomial pathogen. J Lab Physicians. 2016;8:14. doi: 10.4103/0974-2727.176234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23:3573. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tomas JM. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012; Article ID 256261. doi: 10.5402/2012/256261 [DOI] [PMC free article] [PubMed]
- 4.Alperi A, Figueras MJ, Inza I, Martínez-Murcia AJ. Analysis of 16S rRNA gene mutations in a subset of Aeromonas strains and their impact in species delineation. Int Microbiol. 2008;11:185–194. [PubMed] [Google Scholar]
- 5.Morandi A, Zhaxybayeva O, Gogarten JP, Graf J. Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J Bacteriol. 2005;187:6561–6564. doi: 10.1128/JB.187.18.6561-6564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yáñez MA, Catalán V, Apráiz D, Figueras MJ, Martínez-Murcia AJ. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol. 2003;53:875–883. doi: 10.1099/ijs.0.02443-0. [DOI] [PubMed] [Google Scholar]
- 7.Hofer E, Reis CMF, Theophilo GND, Cavalcanti VO, Lima NV, Henriques MFCM. Envolvimento de Aeromonas em surto de doença diarréica aguda em São Bento do Una, Pernambuco. Rev Soc Bras Med Trop. 2006;39:217–220. doi: 10.1590/S0037-86822006000200016. [DOI] [PubMed] [Google Scholar]
- 8.Mendes-Marques CL, Nascimento LM, Theophilo GN, Hofer E, Melo Neto OP, Leal NC. Molecular characterization of Aeromonas spp. and Vibrio cholerae O1 isolated during a diarrhea outbreak. Rev Inst Med Trop São Paulo. 2012;54:299–304. doi: 10.1590/S0036-46652012000600001. [DOI] [PubMed] [Google Scholar]
- 9.Leal NC, da Silva SC, Cavalcanti VO, Figueiroa ACTA, Nunes VVF, Miralles IS, Hofer E. Vibrio parahaemolyticus serovar O3:K6 gastroenteritis in northeast Brazil. J Appl Microbiol. 2008;105:691697. doi: 10.1111/j.1365-2672.2008.03782.x. [DOI] [PubMed] [Google Scholar]
- 10.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:17921797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M. A simple method for estimating evolutionary rates of base substitutions t+hrough comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 13.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:15181530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ. Displaying the relatedness among isolates of bacterial species the eBURST approach. FEMS Microbiol Lett. 2004;241:129–134. doi: 10.1016/j.femsle.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puthucheary SD, Puah SM, Chua KH. Molecular characterization of clinical isolates of Aeromonas species from Malaysia. PLoS One. 2012;7:e30205. doi: 10.1371/journal.pone.0030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrell N, Acinas SG, Figueras MJ, Martínez-Murcia AJ. Identification of Aeromonas clinical isolates by Restriction Fragment Length Polymorphism of PCR-Amplified 16S rRNA genes. J Clin Microbiol. 1997;35:1671-1674. [DOI] [PMC free article] [PubMed]
- 20.Yáñez MA, Catalán V, Apráiz D, Figueras MJ, Martínez-Murcia AJ. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol. 2003;53:875-883. [DOI] [PubMed]
- 21.Heuzenroeder MW, Wong CYF, Flower RLP. Distribution of two hemolytic toxin genes in clinical and environmental isolates of spp.: correlation with virulence in a suckling mouse model . FEMS Microbiol Lett. 1999;174(1):131-136. [DOI] [PubMed]
- 22.Martínez O, Rodríguez-Calleja JM, Santos JA, Otero A, García-López ML. Foodborne and Indicator Bacteria in Farmed Molluscan Shellfish before and after Depuration. J Food Prot. 2009;72(7):1443-1449. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author by request.


