Abstract
Background:
Gastric cancer is the fifth most common cancer and the third top cause of cancer related death with about 1 million new cases and 700,000 deaths in 2012. The aim of this investigation was to identify important factors for outcome using a random survival forest (RSF) approach.
Materials and Methods:
Data were collected from 128 gastric cancer patients through a historical cohort study in Hamedan-Iran from 2007 to 2013. The event under consideration was death due to gastric cancer. The random survival forest model in R software was applied to determine the key factors affecting survival. Four split criteria were used to determine importance of the variables in the model including log-rank, conversation?? of events, log-rank score, and randomization. Efficiency of the model was confirmed in terms of Harrell’s concordance index.
Results:
The mean age of diagnosis was 63 ±12.57 and mean and median survival times were 15.2 (95%CI: 13.3, 17.0) and 12.3 (95%CI: 11.0, 13.4) months, respectively. The one-year, two-year, and three-year rates for survival were 51%, 13%, and 5%, respectively. Each RSF approach showed a slightly different ranking order. Very important covariates in nearly all the 4 RSF approaches were metastatic status, age at diagnosis and tumor size. The performance of each RSF approach was in the range of 0.29-0.32 and the best error rate was obtained by the log-rank splitting rule; second, third, and fourth ranks were log-rank score, conservation of events, and the random splitting rule, respectively.
Conclusion:
Low survival rate of gastric cancer patients is an indication of absence of a screening program for early diagnosis of the disease. Timely diagnosis in early phases increases survival and decreases mortality.
Keywords: Gastric cancer, random survival forest, Hamadan, Iran
Introduction
Cancer is a non-communicable disease with about 1.14 million new cases and 2.8 million deaths in 2012; it is the second cause of death after cardiovascular disease (Ferlay et al., 2013, Organization, 2015). Gastric cancer is the fifth most common cancer and the third leading cause of cancer related death with about 1 million new cases and 700,000 deaths in 2012 (Ferlay et al., 2013, Pelucchi et al., 2015). According to a global estimates, gastric cancer will be one of the main causes of death in the world by 2030; with about 2.5 million new cases and a minimum of 1.9 death by 2,050 (Torre et al., 2015). Gastric cancer is the third most common cancer after breast and skin cancers in Iran, According to national report of cancer registry in IRAN (2008) 6,886 cases of gastric cancer were recorded, which represents about 3.9% of all cancers disease (Mousavi et al., 2009). In the Middle East, Iran has highest incidence rate of Gastric cancer (Mohagheghi et al., 2009). According to studies in Iran, northern and northwestern areas of the country have the highest risk of gastric cancer, while central and southern areas of the country have moderate and low risk of stomach cancer respectively (Saidi et al., 2002, Sadjadi et al., 2003, Alireza et al., 2005). From biological point of view, symptoms of gastric cancer are unknown. The disease it is very active and progressive and incurable in most cases (Beaglehole et al., 2011). Decrease of prevalence of Helicobacter pylori infection and smoking and improved diet have caused a moderate decline in incidence rate of gastric cancer in the last three decades; however, the disease still remains a major health problem (La Vecchia and Franceschi, 2000, Boccia and La Vecchia, 2013). Survival analysis is one of the statistical methods widely used in medical studies in recent decades; it is a set of statistical procedures for data analysis in which the desired output variable is time until an event occurs (Kleinbaum and Klein, 2012). Recently, random survival forests (RSF) has been used for analyzing survival data. It is an ensemble tree method for the analysis of right censored survival data. Constructing ensembles from tree structures can significantly improve learning performance (Ishwaran and Kogalur, 2010). The results showed that the RSF model can identify complex interactions among multiple variables and outperform traditional CPH models (Omurlu et al., 2009, Kälin et al., 2011, Miao et al., 2015). Factors such as age at diagnosis, metastasis, stage of the disease, histological grade, pathological stage, metastasis, and tumor size are known as significant prognostic factors related to survival time of the patient with gastric cancer(Akhavan et al., 2013, Kakuta et al., 2014, Minami et al., 2015). Given the high prevalence of gastric cancer in the region and the lack of a reliable study to determine risk factors of the disease based on advanced statistical methods; therefore, the aim of our study is to identify important risk factors and their complex effects on Gastric cancer patients using RSF.
Materials and methods
In this historical cohort study, data from 182 patients with gastric cancer admitted in the Referral Therapy Center in Hamadan, Iran from 2007 to 2013 was analyzed. The data was extracted from the medical records. Survival status of patients was checked through telephone. Survival time was calculated from diagnosis to death or the end of the study (in months). Patients who withdrawn or lost-to-follow up for any reason during the study or patients who were still alive by the end of the study were considered as right censored. The effect of some demographic variables such as gender and age at diagnosis, as well as clinical data such as histological type (rivers - diffuse - complex), histopathology type (Adenocarcinoma - Lymphoma - Sarcoma), stage (I - II - III - IV), tumor location (Pyloric - Body - Fundus), metastatic status, number of involved lymph nodes, tumor size, type of treatment (Radiotherapy - chemotherapy), and family history of cancer on patients’ survival was evaluated. Staging was based on the tumor node metastasis system (Aronow et al., 2013).
Statistical analysis
Random survival forests
RSF is a non-parametric machine learning method for analyzing right censored survival data. The RSF-model incorporates all univariate and multivariate effects automatically. Another properties of RSF is that it can find influential covariates in highly correlated subsets of covariates, which is particularly useful in high-dimensional covariate selection problems (Ishwaran et al., 2008, Ishwaran and Kogalur, 2010).
The RSF algorithm
B bootstrap samples are randomly selected from the original dataset, while each bootstrap sample excludes 37% of the data on average and calls out-of-bag data (OOB data)(Ishwaran et al., 2008, Ishwaran and Kogalur, 2010). In this study B=1,000.
A survival tree is grown for each bootstrap sample data; q=√p candidate variables are randomly selected from all p variables for each node in survival tree to maximize the survival difference between child nodes using one of the split criteria (log-rank, conservation of events, log-rank score, and random) described in (Ishwaran et al., 2008, Ishwaran and Kogalur, 2010). In this study, 3 candidate variables were randomly selected out of all 10 variables.
The tree is grown until final node’s size reaches a minimum number of events with unique survival times (Ishwaran et al., 2008, Ishwaran and Kogalur, 2010). In this study minimum final node size was equal to 3.
For every tree the cumulative hazard function (CHF) is calculated and then the ensemble CHF is obtained by averaging CHF. The cumulative hazard function for each final node in a grown tree is estimated by Nelson-Aalen’s estimator (Ishwaran et al., 2008, Ishwaran and Kogalur, 2010).
Out-of-bag (OOB) error rate is calculated based on Harrell c-statistics for the ensemble CHF (Ishwaran et al., 2008, Ishwaran and Kogalur, 2010).
The variable importance (VIMP) for x is the prediction error for the original ensemble subtracted from the prediction error for the new ensemble obtained using randomizing x assignments (Ishwaran et al., 2008). Positive values indicate variables with predictive ability(important value), whereas zero or negative values identify non-predictive variables (not important value)(Ishwaran et al., 2008, Ishwaran and Kogalur, 2010).
In this study the four node splitting rules was used for RSF approach (log-rank splitting, conservation of events splitting, log-rank score splitting, and random).
Harrell’s concordance index
Harrell’s concordance index (C-index) is a measure of survival performance. It does not depend on choosing a fixed time for evaluation of the model and specifically takes into account censoring the individuals. The error rate is computed as 1-C, where C is the Harrell’s concordance index. Error rates are between 0 and 1, while 0.5 corresponds to a procedure doing no better than random guessing and 0 is the perfect accuracy (Ishwaran et al., 2008).
The data were analyzed using the random Survival Forest package (Ishwaran et al., 2013) by R 3.1.2. In addition, RSF drew 1000 bootstrap samples from the generated data, grew a tree for each bootstrapped data set and split a predictor using a survival splitting rule. Concordance error rates were obtained from each method for 1,000 replications and the mean of the concordance error rates were recorded.
Results
Explorative Data Analyses
The mean of age of diagnosis was 63 ±12.6 and mean and median survival time of the patients were estimated 15.1 (95%CI: 13.31, 16.99), and 12.3 (95%CI: 11, 13.4) months respectively. The one-year, two-year, and three-year survival rates of the patients were 51%, 13%, and 5% respectively (Figure 1). During the study, 146 patients died and 36 (19.8%) survived who were considered as of right censored observations. One hundred and twelve patients (61.5%) were male and 70 (38.5%) were female. The characteristics of the patients are listed in Table 1.
Figure 1.
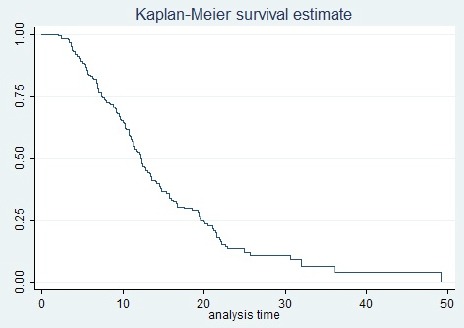
Kaplan-Meier Cumulative Survival
Table 1.
Characteristics of the Patients with Gastric Cancer and Univariate Analysis of Risk Factors
| Variables | Number | Percent | Median Survival Time(Months) | Log-Rank Test | P-value |
|---|---|---|---|---|---|
| Gender | 3.7 | 0.055 | |||
| Male | 112 | 61.5 | 11.3 | ||
| Female | 70 | 38.5 | 14.1 | ||
| Family history | 0.4 | 0.544 | |||
| Yes | 17 | 9.4 | 14.1 | ||
| No | 165 | 90.6 | 12.2 | ||
| Age at diagnosis(yr) | 8.0 | 0.018 | |||
| <60 | 73 | 40.1 | 14.1 | ||
| 61-75 | 75 | 41.2 | 10.7 | ||
| >75 | 34 | 18.7 | 12.3 | ||
| Tumor location | 5.7 | 0.057 | |||
| Pyloric | 100 | 56.8 | 12.4 | ||
| Body | 39 | 22.1 | 10.7 | ||
| Fundus | 37 | 21.1 | 14.1 | ||
| Metastatic status | 82.4 | <0.001 | |||
| No | 77 | 42.3 | 14.1 | ||
| Yes | 48 | 26.4 | 7.3 | ||
| Unknown | 57 | 31.3 | 16.2 | ||
| Number of involved lymph nodes | 15.6 | <0.001 | |||
| (1-6 number) | 102 | 75.0 | 12.3 | ||
| (7-15 number) | 34 | 25.0 | 8.3 | ||
| Histopathology type | 2.5 | 0.279 | |||
| Adenocarcinoma | 125 | 69.8 | 12.3 | ||
| Lymphoma | 29 | 16.3 | 13.6 | ||
| Sarcoma | 25 | 13.9 | 10.2 | ||
| Tumor size | 26.3 | <0.001 | |||
| T1(1 cm) | 21 | 15.2 | 22.0 | ||
| T2 (2 cm) | 48 | 34.8 | 12.2 | ||
| T3 (3 cm) | 45 | 31.9 | 11.3 | ||
| T4 (> 4cm) | 25 | 18.1 | 10.3 | ||
| Stage | 22.4 | <0.001 | |||
| I | 9 | 5.0 | 22.1 | ||
| II | 31 | 17.1 | 17.6 | ||
| III | 36 | 19.9 | 10.7 | ||
| IV | 105 | 58.0 | 11.0 | ||
| Histological type | 0.1 | 0.956 | |||
| Rivers | 91 | 53.8 | 12.1 | ||
| Diffuse | 56 | 33.1 | 11.3 | ||
| Complex | 22 | 13.1 | 11.3 | ||
| Type of treatment | 5.4 | 0.021 | |||
| Radiotherapy | 74 | 40.6 | 14.7 | ||
| Chemotherapy | 108 | 59.4 | 11.2 |
Random Survival Forrest Analyses
Informativeness of each predictor was taken into account under the log-rank splitting rule. Figure 2 shows the error rate for the RSF log-rank model as a function of the number of trees and the out-of-bag importance values for predictors. The Right part of Figure 2 depicts the importance values for all 11 predictors. From the plot, we found that the eight prognostic factors (Metastatic status, Age at diagnosis, Tumor size, Number of involved lymph nodes, Histological type, Gender, Type of treatment, Tumor location) had an effect on survival time (Positive Value). Other predictors had negative values or no effect on survival time. Concordance error rate of this RSF model was 0.2966 (Table 2).
Figure 2.
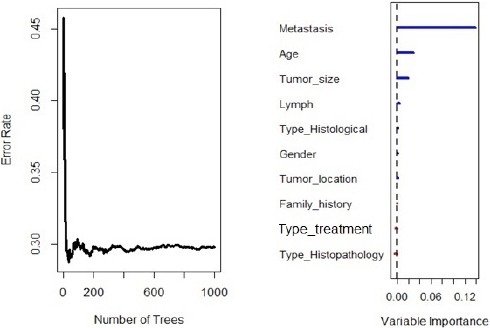
Out-of-Bag Importance Values of RSF for Log-Rank Splitting Rule
Table 2.
Harrell’s Concordance Error Rates for Methods
| Method | Error rate | |
|---|---|---|
| RSF | Log-rank | 0.297 |
| Log-rank scor | 0.301 | |
| Conservation of events | 0.304 | |
| Random | 0.325 |
Figure 3 illustrates the error rate for the RSF model as a function of the number of trees and the out-of-bag importance value for predictors. As shown the six prognostic factors (Metastatic status, Age at diagnosis, Tumor size, Gender, Type of treatment, and Family history) had an effect on survival time. Metastatic status, age at diagnosis, and tumor size were assigned with important values by RSF log-rank splitting rule. Concordance error rate of th RSF model was 0.304 (Table 2). Figure 4 illustrates the error rate for the RSF model as a function of the number of trees and the out-of-bag importance values for the predictors. This figure shows that the four prognostic factors (Metastatic status, Age at diagnosis, Tumor size, and Histological type) were positive important values and larger than all other prognostic factors. Metastatic status, age at diagnosis, and Tumor size were given important values by RSF log-rank splitting rule and RSF conservation of events splitting rule. Concordance error rate of this RSF model was 0.301 (Table 2).
Figure 3.

Out-of-Bag Importance Values of RSF for Conservation of Events Splitting Rule
Figure 4.
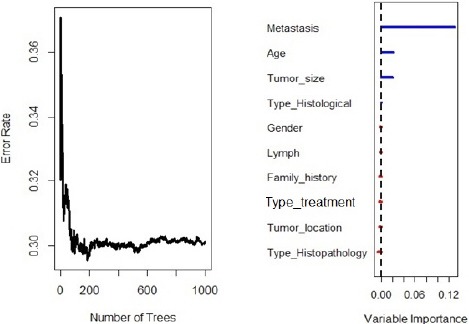
Out-of-Bag Importance Values of RSF for Log-Rank Score Splitting Rule
Figure 5 pictures the error rate for the RSF model as a function of the number of trees and the out-of-bag importance values for the predictors. As indicated in the plot, the six prognostic factors (Metastatic status, Age at diagnosis, Tumor size, and Histological type, Type of treatment, and Family history) had an effect on survival time. Metastatic status, age at diagnosis, and tumor size had positive value by RSF log-rank splitting rule, RSF conservation of events splitting rule, and log-rank score splitting rule. Concordance error rate of this RSF model was 0.31 (Table 2).
Figure 5.
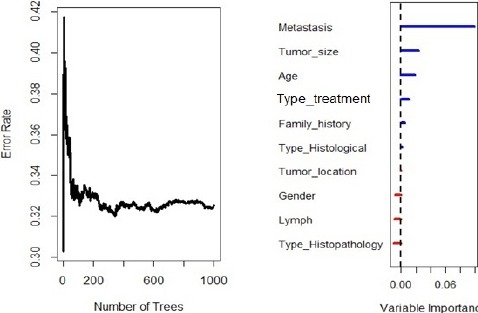
Out-of-Bag Importance Values of RSF for Random Splitting Rule
Random Survival Forrest Model Performance
The performance of each RSF approach was very similar to the best error rate (0.297) obtained by the log-rank splitting rule with 1,000 trees (Table 2). The second, third, and fourth ranks were occupied by log-rank score, conservation of events, and random splitting rule respectively.
Discussion
Each RSF approach showed a slightly different ranking order. The very important covariates in nearly all 4 RSF approaches were metastatic status, age at diagnosis, and tumor size. Unimportant covariates in nearly all 4 RSF approaches was histopathology type. The remaining covariates had positive importance values with somewhat different ranking within each RSF approach.
Age at diagnosis time had a significant effect on patients’ survival time, which is consistent with the studies carried out in Italy, China, and north of Iran (Wang et al., 2002, Bucchi et al., 2004, Yazdani-Charati et al., 2014). Metastasis was another factor that had an important value and significant effect on the survival time. This finding has been confirmed in other studies (Wang et al., 2002, Moghimi-Dehkordi et al., 2009, Maroufizadeh et al., 2012, Dixon et al., 2014). Some studies have reported that the disease stage highly influenced the patients’ survival time so that the median of survival time in stage I was more than median of survival time in stage IV (Zeraati et al., 2005, Moghimi-Dehkordi et al., 2009, Dixon et al., 2014, Yazdani-Charati et al., 2014). This is consistent with our results. Consistent with (Lin et al., 2013), we found that tumor size was an important or significant value in survival time; this means that the survival time decreases as tumor size increases. The number of involved lymph nodes was another important value in this study; by increasing the number of involved lymph nodes, the risk of death also increased; this is inconsistent with other studies including (Maroufizadeh et al., 2012). Type of the treatment was another important or significant factor in survival time. (Moghimi-Dehkordi et al., 2009) showed that the survival time of patients under chemotherapy was more than the survival time of patients who received radiotherapy (Moghimi-Dehkordi et al., 2009). Consistent with, histological type and family history were other factors with important effect. Moreover, histopathology types was not an important factor in all 4 RSF approaches without a significant effect on survival time. However, other studies have shown significant effect of this variable (Samadi et al., 2007, Moghimi-Dehkordi et al., 2009).
Low survival rate of gastric cancer patients is an indication of absence of a screening program for early diagnosis of the disease. Timely diagnosis in early phases of the disease increases survival rate and decreases mortality rate caused by the disease.
Acknowledgments
We wish to thank the staff of Referral Therapy Center in Hamadan for their helping in gathering data.
References
- Akhavan A, Binesh F, Seifaddiny A, Ghannadi F. Characteristics and survival rate of patients with gastric and gastroesophageal junction adenocarcinoma in Yazd, Iran. Middle East J Cancer. 2013;4:125–125. [Google Scholar]
- Sadjadi A, Nouraie M, Mohagheghi MA, et al. Cancer occurrence in Iran in 2002, an international perspective. Asian Pac J Cancer prev. 2005;6:359–359. [PubMed] [Google Scholar]
- Aronow ME, Portell CA, Rybicki LA, Sweetenham JW, Singh AD. Ocular adnexal lymphoma:assessment of a tumor-node-metastasis staging system. Ophthalmology. 2013;120:1915–1915. doi: 10.1016/j.ophtha.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Beaglehole R, Bonita R, Horton R, et al. Priority actions for the non-communicable disease crisis. The Lancet. 2011;377:1438–1438. doi: 10.1016/S0140-6736(11)60393-0. [DOI] [PubMed] [Google Scholar]
- Boccia S, La Vecchia C. Dissecting causal components in gastric carcinogenesis. Eur J Cancer Prev. 2013;22:489–489. doi: 10.1097/CEJ.0b013e3283603480. [DOI] [PubMed] [Google Scholar]
- Bucchi L, Nanni O, Ravaioli A, et al. Cancer mortality in a cohort of male agricultural workers from northern Italy. J Occup Environ Med. 2004;46:249–249. doi: 10.1097/01.jom.0000116804.10496.fc. [DOI] [PubMed] [Google Scholar]
- Dixon M, Mahar AL, Helyer LK, et al. Prognostic factors in metastatic gastric cancer:results of a population-based, retrospective cohort study in Ontario. Gastric Cancer. 2014;19:150–150. doi: 10.1007/s10120-014-0442-3. [DOI] [PubMed] [Google Scholar]
- Ferlay L. International Agency for Research on Cancer. Lyon: 2013. [globocan. iarc. fr (accessed 10 October 2014)]. GLOBOCAN 2012 v10, cancer incidence and mortality worldwide:IARC CancerBase, No. 11 [internet] [Google Scholar]
- Ishwaran H, Kogalur UB. Consistency of random survival forests. Stat Probab Lett. 2010;80:1056–1056. doi: 10.1016/j.spl.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H, Udaya BK, Eugene HB, Michael SL. Random survival forests. Ann Appl Stat. 2008;2:841–841. [Google Scholar]
- Kakuta T, Kosugi S, Kanda T, et al. Prognostic factors and causes of death in patients cured of esophageal cancer. Ann Surg Oncol. 2014;21:1749–1749. doi: 10.1245/s10434-014-3499-7. [DOI] [PubMed] [Google Scholar]
- Kälin M, Cima I, Schiess R, et al. Novel prognostic markers in the serum of patients with castration-resistant prostate cancer derived from quantitative analysis of the pten conditional Knockout mouse proteome. Eur Urol. 2011;60:1235–1235. doi: 10.1016/j.eururo.2011.06.038. [DOI] [PubMed] [Google Scholar]
- Kleinbaum DG, Klein M Introduction to survival analysis. ’Survival Analysis’. Springer; 2012. pp. 1–54. [Google Scholar]
- La Vecchia C, Franceschi S. Nutrition and gastric cancer. Can J Gastroenterol. 2000;14:51–51. doi: 10.1155/2000/869862. [DOI] [PubMed] [Google Scholar]
- Lin WL, Sun JL, Chang SC, et al. Factors predicting survival of patients with gastric cancer. Asian Pac J Cancer Prev. 2013;15:5835–5835. doi: 10.7314/apjcp.2014.15.14.5835. [DOI] [PubMed] [Google Scholar]
- Maroufizadeh S, Hajizadeh E, Baghestani AR, Fatemi SR. Determining the postoperative survival in patients with gastric cancer and the associated factors using Cox and Lin-Ying additive hazards models. J Arak Univ Med Sci. 2012;15:84–84. [Google Scholar]
- Miao F, Cai Y-P, Zhang Y-T, Li C-Y Is random survival forest an alternative to cox proportional model on predicting cardiovascular disease? 6th European conference of the international federation for medical and biological engineering. Springer; 2015. pp. 740–3. [Google Scholar]
- Minami Y, Kawai M, Fujiya T, et al. Family history, body mass index and survival in Japanese patients with stomach cancer:a prospective study. Int J Cancer. 2015;136:411–411. doi: 10.1002/ijc.29001. [DOI] [PubMed] [Google Scholar]
- Moghimi-Dehkordi B, Safaee A, Ghiasi S, Zali MR. Survival in gastric cancer patients:univariate and multivariate analysis. East Afr J Public Health. 2009;6:41–41. doi: 10.4314/eajph.v6i3.45773. [DOI] [PubMed] [Google Scholar]
- Mohagheghi MA, Mosavi-Jarrahi A, Malekzadeh R, Parkin M. Cancer incidence in tehran metropolis:the first report from the tehran population-based cancer registry. Arch Iran Med. 2009;12:15–15. [PubMed] [Google Scholar]
- Mousavi S, Gouya M, Ramazani R, et al. Cancer incidence and mortality in Iran. Ann Oncol. 2009;20:556–556. doi: 10.1093/annonc/mdn642. [DOI] [PubMed] [Google Scholar]
- Omurlua IK, Turea M, Tokatlib F. The comparisons of random survival forests and Cox regression analysis with simulation and an application related to breast cancer. Expert Syst Appl. 2009;36:8582–8582. [Google Scholar]
- Pelucchi C, Lunet N, Boccia S, et al. The stomach cancer pooling (StoP) project:study design and presentation. Eur J Cancer Prev. 2015;24:16–16. doi: 10.1097/CEJ.0000000000000017. [DOI] [PubMed] [Google Scholar]
- Sadjadi A, Malekzadeh R, Derakhshan MH, et al. Cancer occurrence in Ardabil:Results of a population-based Cancer Registry from Iran. Int J Cancer. 2003;107:113–113. doi: 10.1002/ijc.11359. [DOI] [PubMed] [Google Scholar]
- Saidi F, Malekzadeh R, Sotoudeh M, et al. Endoscopic esophageal cancer survey in the western part of the Caspian Littoral. Dis Esophagus. 2002;15:214–214. doi: 10.1046/j.1442-2050.2002.00250.x. [DOI] [PubMed] [Google Scholar]
- Samadi F, Babaei M, Yazdanbod A, et al. Survival rate of gastric and esophageal cancers in Ardabil province, North-West of Iran. Arch Iran Med. 2007;10:32–32. [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–87. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Wang CS1, Hsieh CC, Chao TC, et al. Resectable gastric cancer:operative mortality and survival analysis. Chang Gung Med J. 2002;25:216–216. [PubMed] [Google Scholar]
- Yazdani-Charati J, Janbabaei G, Etemadinejad S, Sadeghi S, Haghighi F. Survival of patients with stomach adenocarcinoma in North of Iran. Gastroenterol Hepatol Bed Bench. 2014;7:211–17. [PMC free article] [PubMed] [Google Scholar]
- Zeraati H, Mahmoudi M, Kazemnejad A, Mohammed K. Postoperative life expectancy in gastric cancer patients and its associated factors. Saudi Med J. 2005;26:1203–1203. [PubMed] [Google Scholar]


