Abstract
Background:
The process of development of bladder cancer features alteration of normal biological conditions caused by changes in molecular pathways. Removing control over regulation of these pathways could lead to changes in signal transduction and abnormal regulation of genes. During tumor formation and progression, genes regulate critical cellular processes, involved in cell cycling, growth and death. Here we evaluated the expression and prognostic importance of FGFR1, HRAS, CCND1, CCND3, STAT3 and FAS genes.
Methods:
Tumor tissues of 44 patients diagnosed with bladder cancer were investigated for changes in expression levels of FGFR1, HRAS, CCND1, CCND3, FAS and STAT3 genes by the RT-PCR method. Signal transduction pathways and expression of individual genes related to these pathways were analyzed using the “One Sample Test”.
Results:
There were statistically significant changes in the expression levels of HRAS, CCND1, CCND3 and STAT3, but not FGFR1 and FAS genes. Examination of associations with age, gender, smoking, chemotherapy, tumor grade and tumor growth pattern using the “Independent Samples Test”, showed importance relations between the CCND1 gene and cigarette smoking and sex.
Conclusion:
Over-expression of HRAS, CCND1, CCND3 and STAT3 genes may play roles in bladder cancer development and progression, while cigarette smoking is significantly associated with CCND1 gene expression and consequently concluded to be contributing to the development of bladder cancer.
Keywords: Signal transduction pathways, bladder cancer, gene expression, CCND, STAT3
Introduction
Bladder cancer is the fifth most frequent of all cancer types. Every year 336000 new cases are diagnosed and about 179000 patients die of bladder cancer (Cheng et al., 2011; Schulz, 2006). It is the 4th and 8th most frequent cancer in men and women, respectively. The frequency of bladder cancer in men and women diagnosed with a cancer is 8% and 3%, respectively (Jacobs et al., 2010). Patients with an invasive tumor have a worse prognosis with a 5 year survival of 50 percent (Tomlinson et al., 2009).
Various pathways are involved in signal transmission from the cell surface receptors to the nuclear transcription factors. With respect to bladder cancer, the Ras-Mitogen-Activated Protein Kinase (RAS-MAPK), Phospholipase C-Protein kinase (PLC-PKC), Janus kinase – Signal Transductor and Transcription Activator (JAK-STAT), and Phosphotidylinositol 3- kinase (PI3K) pathways are the most remarkable pathways implicated in the pathogenesis (Mitra et al., 2009). Fibroblast growh factor receptor (FGFR) gene family is known to activate the Ras-MAPK signal transmission pathway. This family consists of four cell surface receptors (FGFR1-4). It has been reported that the expression of FGFR1 is increased in bladder cancer cell lines. It has been suggested that FGFR1 induces prolonged cell life span and regulates tumor growth (Tomlinson et al., 2009).
The RAS gene family involves three functional genes: Harvey RAS (HRAS), Kristen RAS (KRAS) and Neuroblastoma RAS (NRAS) (Mundt et al., 2003). Other than point mutatins that activate RAS gene constituvely, gene amplification and other mecahanisma may also result in overexpression of the gene with continuous generation of proliferative signals (Karimianpour et al., 2008).
Cyclins are also involved in cell cycle regulation. D type cyclins (D1, 2 and 3) expression are triggered by growth factors or mitogens. The type of cyclin D to be expressed is spesific to the tissue type. The CCND1 gene is localized to chromosome 11q13 and its overexpression, due to point mutations or amplification, is frequently observed. CCND3, localized to 6p21 is deregulated in bladder tumors; however, its prognostic significance is unknown (Schulz, 2006; Lopez-Beltran et al., 2006).
The members of the Janus Kinase (JAK) family are tyrosin kinases which are activated by growth factors and cytokins. They mediate various pathways. Increased STAT activity has ben implicated in oncogenic transformation (Steelman et al., 2008). For example, increased expression of STAT3 is correlated with diminished survival and increased risk of recurrence (Mitra et al., 2009).
Expression of genes which play a role in signal transmission pathways, cell regulation and apoptosis may vary according to the stage of bladder cancer. In the present study we purposed to utilize the expression status and prognostic significance of FGFR1, HRAS, CCND1, CCND3, STAT3 and FAS genes which may be of critical importance in the development bladder cancer. We also analysed the possible relation between the expression of these genes and gender, age, pathologic staging and smoking.
Materials and Methods
Study population
Samples were collected from the Department of Urology, Istanbul Faculty of Medicine, Istanbul University and the Department of Urology, Bezmialem Vakif University Faculty of Medicine, İstanbul, Turkey. The study examined 44 bladder cancer patients. The patients who received primary other therapies such as radiotherapy or chemotherapy for bladder cancer were excluded. Tissue samples were obtained from both healthy adjacent mucosa and the tumor tissue itself during the surgery. They were urgently placed in storage at -80°C until the RNA extraction. The study protocol was approved by both the Ethical Committee of the Istanbul Faculty of Medicine (08,11,2010 No:2010/105-25) and The Scientific Research Projects Coordination Unit of Istanbul University (Project number: 9951).
The mean ages and percentage of the patients and controls are shown in Table 1. Bladder cancer patients who were diagnosed by histology or cytology. All samples recruited before treatment either surgical resection or chemoradiotherapy. All participants signed an informed consent form before enrollment, and institutional Ethics Committee approval was obtained for the study. RNA was isolated and preserved at -80°C. In the study, real time PCR was used to detect HRAS, CCND1, CCND3, STAT3, FGFR1 and FAS gene expressions in Turkish BC patients. Total RNA isolated according to the kit protocol (Ambion kit; Roche, Manheim). RNA samples were quantified using a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware, USA), and their integrity was checked electrophoretically. First strands of the cDNA samples were synthesized by using RT PCR (Cat. No. 11483188001 Roche, GmbH, D-40724 Hilden, Germany). Gene expression analysis were performed by quantitative reverse transcription (qRT)–PCR (LightCycler 1.5, Roche, Germany). The PCR reaction started with a denaturation step at 95°C for 10 second (1 cycle), followed by 45 cycles at 95°C for 10 seconds, 55°C for 5 seconds and 72°C for 20 second. Subsequently, a melting curve program was applied with continuous fluorescence measurement. Each reaction was performed duplicate. The GAPDH gene was used as reference for normalization of the gene expression levels.
Table 1.
Case and Tumor Data
| N | A | G | Localization | Growth Pattern | Histopathologic Diagnosis | Grade | Stage |
|---|---|---|---|---|---|---|---|
| 1 | 58 | M | multipl | papillary | urothelial carcinoma | high | pTa |
| 2 | 66 | M | multipl | papillary | urothelial carcinoma | low | pTa |
| 3 | 69 | M | on right wall | papillary | urothelial carcinoma | low | pTa |
| 4 | 66 | M | on left wall | squamous dif. | urothelial carcinoma | high | pTx |
| 5 | 54 | F | adverse wall | papillary | urothelial carcinoma | low | pTa |
| 6 | 70 | M | on left wall | papillary | urothelial carcinoma | low | pTa |
| 7 | 64 | M | on right wall | papillary | urothelial carcinoma | high | pT1 |
| 8 | 71 | M | on left wall | solid | urothelial carcinoma | high | pT2 |
| 9 | 56 | M | on left wall | papillary | urothelial carcinoma | high | pT1+CIS |
| 10 | 69 | M | multipl | papillary | urothelial carcinoma | low | pTa |
| 11 | 62 | M | multipl | papillary | urothelial carcinoma | high | pT1b |
| 12 | 64 | M | multipl | papillary | urothelial carcinoma | high | pT1a |
| 13 | 69 | M | on right wall | Small cell carcinoma | high | pT2 | |
| 14 | 71 | M | on left wall | squamous dif. | urothelial carcinoma | high | pT2 |
| 15 | 85 | M | on right wall | squamous dif.. | urothelial carcinoma | high | pT2 |
| 16 | 53 | F | multipl | papillary | urothelial carcinoma | high | pT1b |
| 17 | 51 | M | on right wall | papillary | urothelial carcinoma | low | pTa |
| 18 | 34 | M | on the base | papillary | urothelial carcinoma | low | pT1a |
| 19 | 66 | M | multipl | squamous dif. | urothelial carcinoma | high | pT1a |
| 20 | 83 | M | on left wall | papillary | urothelial carcinoma | high | pT1b |
| 21 | 60 | M | on the base | papillary | urothelial carcinoma | high | pT1 |
| 22 | 45 | M | multipl | papillary | urothelial carcinoma | low | pT1a |
| 23 | 70 | M | on right wall | papillary | urothelial carcinoma | high | pT1a |
| 24 | 67 | M | multipl | papillary | urothelial carcinoma | high | pT1 |
| 25 | 70 | M | on the base | papillary | urothelial carcinoma | high | pTa |
| 26 | 51 | M | on the base | papillary | urothelial carcinoma | high | pT2 |
| 27 | 63 | M | multipl | papillary | urothelial carcinoma | low | pTa |
| 28 | 68 | M | on right wall | papillary | urothelial carcinoma | low | pTa |
| 29 | 62 | M | on right wall | follicular squamous c. | urothelial carcinoma | high | pT2 |
| 30 | 54 | M | on the cap | papillary | urothelial carcinoma | low | pTa |
| 31 | 76 | M | on left wall | papillary | urothelial carcinoma | high | pTa |
| 32 | 66 | M | on left wall | papillary | urothelial carcinoma | high | pT1 |
| 33 | 74 | M | multipl | papillary | urothelial carcinoma | high | pTa |
| 34 | 58 | M | on right wall | papillary | urothelial carcinoma | high | pT1 |
| 35 | 59 | M | on right wall | papillary | urothelial carcinoma | high | pTx |
| 36 | 75 | F | on the cap | squamous dif. | urothelial carcinoma | high | pT2 |
| 37 | 67 | M | on left wall | papillary | urothelial carcinoma | high | pT1 |
| 38 | 64 | M | multipl | papillary | urothelial carcinoma | high | pT1a |
| 39 | 74 | F | on right wall | papillary | urothelial carcinoma | high | pTa |
| 40 | 75 | F | on left wall | papillary | urothelial carcinoma | high | pT1 |
| 41 | 68 | M | on the cap | papillary | urothelial carcinoma | low | pTa |
| 42 | 71 | M | adverse wall | papillary | urothelial carcinoma | high | pTa |
| 43 | 59 | M | adverse wall | papillary | urothelial carcinoma | high | pT1 |
| 44 | 68 | M | on the base | papillary | urothelial carcinoma | high | pT1 |
Statistical analysis
All statistical analyses were carried out using the SPSS version 12.0 statistical package for Windows. Numeric values were analyzed with the Student t-test. Expression ratios were transformed into fold changes and reported as relative expression. Obtained fold changes were compared between different tumor grades in addition to surrounding tissue samples. Significant fold changes were determined with t-test using SPSS 12. All tests were two-sided, and the P values less than 0.05 were considered statistically significant. The results were analyzed by the threshold cycle (Ct) numbers as fold-changes and calculated by the 2∆(∆CT) method [2geneT-N(Ct)/2 GAPDH T-N(Ct)] (N, matched surrounding tissue; T, tumor tissue). The relative associations were assessed by calculating crude Gart’s odds ratios (ORs) and 95% confidence intervals (95%CIs). A multivariate logistic regression model was used to investigate the effects of genotypes and alleles after adjustment for age. Values of P < 0.05 were considered statistically significant.
Results
FGFR1, HRAS, CCND1, CCND3, FAS and STAT3 genes are expressed in various cancer types. These genes are play an significance role in tumor differentiation, cell division and angiogenesis. Hovewer this task has not been clarified yet. In our study, were compared the expression levels of the six genes which plays a role in signal transduction pathways. We determined HRAS, CCND1, CCND3 and STAT3 genes expression levels of bladder cancer patients. We determined HRAS, CCND1, CCND3, STAT3, FGFR1 and FAS genes expression of values and statistical evaluation of cases between smoking habits. These results are shown in Table 2, Table 3 and Figure 1.
Table 2.
HRAS, CCND1, CCND3, STAT3, FGFR1 and FAS Genes Expression of Values and 95% Confidence Interval of the Difference
| Genes | Test Value = 1 | ||||
|---|---|---|---|---|---|
| t | P value | Mean Difference | 95% Confidence Interval of the Difference | ||
| Lower | Upper | ||||
| HRAS | 5.8 | <0.001* | 4.4 | 2.8 | 5.9 |
| CCND1 | 4.6 | <0.001* | 3.4 | 1.9 | 4.9 |
| CCND3 | 2.3 | 0.027* | 1.8 | 0.2 | 3.4 |
| STAT3 | 4.2 | <0.001* | 3.4 | 1.7 | 5.0 |
| FGFR1 | 0.5 | 0.633 | 0.3 | -1.1 | 1.8 |
| FAS | 1.3 | 0.213 | 1.0 | -0.6 | 2.6 |
One Sample Test; OR, (Odds Ratio)
Table 3.
HRAS. CCND1. CCND3. STAT3. FGFR1 and FAS Genes Expression of Values and Statistical Evaluation of Cases between Smoking Habits
| t-test for Equality of Means | ||||||
|---|---|---|---|---|---|---|
| t | p-value | Mean Difference | Std. Error Difference | 95% Confidence Interval of the Difference | ||
| Lower | Upper | |||||
| HRAS | 1.2 | 0.268 | 2.5 | 2.1 | -2.3 | 7.3 |
| CCND1 | 3.1 | 0.004 | 4.7 | 1.5 | 1.6 | 7.8 |
| CCND3 | 0.2 | 0.824 | 0.5 | 2.3 | -4.5 | 5.5 |
| STAT3 | 1.1 | 0.274 | 2.3 | 2.0 | -2.0 | 6.6 |
| FGFR1 | 0.5 | 0.603 | 0.9 | 1.8 | -2.8 | 4.7 |
| FAS | 1.3 | 0.230 | 2.9 | 2.2 | -2.2 | 8.0 |
Figures 1.
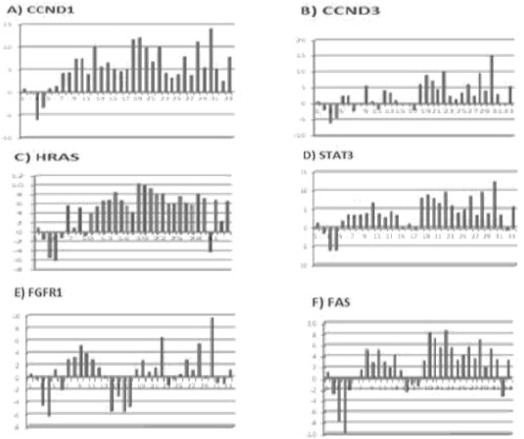
Fold Regulation of CCND1, CCND3, HRAS, STAT3, FGFR1 and FAS
Discussion
Genes which control cell cycle, growth, death and signal transduction may also be causative in tumor development and progression. The loss of control on associated pathways may alter signal transduction and abnormal gene expression (Mitra et al., 2009). It was reported that FGFR1 activation is associated with mitogenic and chemotactic response in various cell types. FGFR1 transcripts are expressed at low levels in the normal urothelium. It was shown that FGFR1 expression was increased in bladder cancer cell lines and in tumor tissue (Tomlinson et al., 2009). In the same study, the effect of increased FGFR1 expression on normal urothelium was also evaluated. It was shown that FGFR1 increases cell growth and life. On the other hand, it was found that FGFR1 regulates oncogenic transformation and cell life in bladder cancer cell lines. These findings suggest that FGFR1 plays a critical role in the malign transformation of normal bladder cells. In our study, we did not find a significance increase in FGFR1 expression. It may be suggested FGFR1 expresion may vary acoording to tumor stage or grade. In agreement with our results, Tomlinson et al., (2009) also reported that there was no association between expression level and tumor stage or grade. These findings suggest that differences in gene expression may be related to different tumor localizations.
The Ras family is another gene family which is involved in signal transduction. As well as gene amplification and point mutations, RAS dysfunction may also be related to alterations in protein level. Increased gene expression due to gene amplification or constituve activation may generate continious signals which are necessary for cell proliferation (Karimianpour et al., 2008). Boules at al., (2009) reported increased expression (50%) of KRAs and NRAS genes both. On the other hand, the HRAS expression was found to low (27%) in the same study. HRAS encodes a signal transduction protein which is located at the inner side of the cell membrane. It has been implicated in the genesis of bladder cancer. Fontana et al reported that HRAS overexpression is associated with early recurrence of superficial bladder cancer (Jung et al., 2000). HRAS was found to be overexpressed in patients with non-progressive Ta tumors when compared with those progressing to invasive disease (Mitra et al., 2009). In another study, it was suggested that HRAS is a powerful predictor of relapse and progression in non-invasive papillary tumors (Birkhahn et al., 2010). In our study we found that the expression of HRAS gene was increased in proportion to normal tissue. The product of the CCND1 gene (a candidate oncogene) contributes to the regulation of G1/S transition. It is thought to play an significance role in bladder cancer improvement. Increased CCND1 expression accelerates G1 phase and provides tumor cells with a proliferative advantage (Sgambato et al., 2002). It was reported that CCND1 overexpression is more frequent in tumors with a lower grade and stage and is associated with general survival (Sgambato et al., 2002; Niehans et al., 1999). It was observed that CCND1 alterations occur in early bladder cancer and its loss of expression is associated with recurrence (Matsushita et al., 2011; Shariat et al., 2006). In a study conducted by Lopez-Beltran et al., (2006), it was reported that tumor size and expressions of CCND1 and CCND3 are independent predictors of progression free survival. It was suggested that CCND3 expression may also influence the prognosis of high grade bladder cancer. In our study, we found that the CCND1 and CCND3 expressions were considerably increased in tumor tissue in proportion to normal tissue. However, there was no relation between expression and tumor grade or stage. These results are in agreement with previous studies. Increased STAT3 expression is a predictor of decreased survival and recurrence (Mitra et al., 2009). Also, it was reported that STAT3 expression is increased in invasive bladder cancer cell lines and it was suggested that its overexpression may be a marker of worse prognosis (Mitra et al., 2009). The increased STAT3 gene expression observed in our study is in agreement with aforementioned studies. In another study, where the effect of prolonged nicotin exposure on drug resistance was assessed, it was found that nicotin activates STAT3 and triggers cell proliferation by causing overexpression of CCND1 (Chen et al., 2010). It is of interest that we also found a significant relation between smoking and CCND1 expression.
Our findings suggest that CCND1 overexpression is significantly related with smoking and thus may play a role in the progress of bladder cancer. There was no alteration in the expression FGFR1 and FAS genes compared to normal tissue. However, we found that the expressions of HRAS, CCND1, CCNND3 and STAT3 genes were significantly increased, suggesting that these genes may be implicated in the progress and improvement of bladder cancer.
Acknowledgements
This study was supported by The Scientific Research Projects Coordination Unit of Istanbul University (Project number: 9951).
References
- Birkhahn M, Mitra AP, Williams AJ, et al. Predicting recurrence and progression of noninvasive papillary bladder cancer at initial presentation based on quantitative gene expression profiles. Eur Urol. 2010;57:12–20. doi: 10.1016/j.eururo.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulalas I, Zaravinos A, Karyotis I, Delakas D, Spandidos DA. Activation of RAS family genes in urothelial carcinoma. J Urol. 2009;181:2312–9. doi: 10.1016/j.juro.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Chen RJ, Ho YS, Guo HR, Wang YJ. Long-term nicotine exposure-induced chemoresistance is mediated by activation of Stat3 and downregulation of ERK1/2 via nAChR and beta-adrenoceptors in human bladder cancer cells. Toxicol Sci. 2010;115:118–30. doi: 10.1093/toxsci/kfq028. [DOI] [PubMed] [Google Scholar]
- Cheng L, Zhang S, MacLennan GT, et al. Bladder cancer: translating molecular genetic insights into clinical practice. Hum Pathol. 2011;42:455–81. doi: 10.1016/j.humpath.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Jacobs BL, Lee CT, Montie JE. Bladder cancer in 2010: how far have we come? CA Cancer J Clin. 2010;60:244–72. doi: 10.3322/caac.20077. [DOI] [PubMed] [Google Scholar]
- Jung I, Messing E. Molecular mechanisms and pathways in bladder cancer development and progression. Cancer Control. 2000;7:325–34. doi: 10.1177/107327480000700401. [DOI] [PubMed] [Google Scholar]
- Karimianpour N, Mousavi-Shafaei P, Ziaee AA, et al. Mutations of RAS gene family in specimens of bladder cancer. Urol J. 2008;5:237–42. [PubMed] [Google Scholar]
- Lopez-Beltran A, Requena MJ, Luque RJ, et al. Cyclin D3 expression in primary Ta/T1 bladder cancer. J Pathol. 2006;209:106–13. doi: 10.1002/path.1952. [DOI] [PubMed] [Google Scholar]
- Matsushita K, Cha EK, Matsumoto K, et al. Immunohistochemical biomarkers for bladder cancer prognosis. Int J Urol. 2011;18:616–29. doi: 10.1111/j.1442-2042.2011.02809.x. [DOI] [PubMed] [Google Scholar]
- Mitra AP, Bartsch CC, Cote RJ. Strategies for molecular expression profiling in bladder cancer. Cancer Metast Rev. 2009;28:317–26. doi: 10.1007/s10555-009-9196-5. [DOI] [PubMed] [Google Scholar]
- Mitra AP, Cote RJ. Molecular pathogenesis and diagnostics of bladder cancer. Annu Rev Pathol. 2009;4:251–85. doi: 10.1146/annurev.pathol.4.110807.092230. [DOI] [PubMed] [Google Scholar]
- Mitra AP, Pagliarulo V, Yang D, et al. Generation of a concise gene panel for outcome prediction in urinary bladder cancer. J Clin Oncol. 2009;27:3929–37. doi: 10.1200/JCO.2008.18.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt KA, Birk T, Burch MT. Critical review of the epidemiological literature on occupational exposure to perchloroethylene and cancer. Int Arch Occ Env Hea. 2003;76:473–91. doi: 10.1007/s00420-003-0457-2. [DOI] [PubMed] [Google Scholar]
- Niehans GA, Kratzke RA, Froberg MK, et al. G1 checkpoint protein and p53 abnormalities occur in most invasive transitional cell carcinomas of the urinary bladder. Brit J Cancer. 1999;80:1175–84. doi: 10.1038/sj.bjc.6990483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz WA. Understanding urothelial carcinoma through cancer pathways. Int J Cancer. 2006;119:1513–8. doi: 10.1002/ijc.21852. [DOI] [PubMed] [Google Scholar]
- Sgambato A, Migaldi M, Faraglia B, et al. Cyclin D1 expression in papillary superficial bladder cancer: its association with other cell cycle-associated proteins, cell proliferation and clinical outcome. Int J Cancer. 2002;97:671–8. doi: 10.1002/ijc.10055. [DOI] [PubMed] [Google Scholar]
- Shariat SF, Ashfaq R, Sagalowsky AI, Lotan Y. Correlation of cyclin D1 and E1 expression with bladder cancer presence, invasion, progression, and metastasis. Hum Pathol. 2006;37:1568–76. doi: 10.1016/j.humpath.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Steelman LS, Abrams SL, Whelan J, et al. Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways to leukemia. Leukemia. 2008;22:686–707. doi: 10.1038/leu.2008.26. [DOI] [PubMed] [Google Scholar]
- Tomlinson DC, Lamont FR, Shnyder SD, Knowles MA. Fibroblast growth factor receptor 1 promotes proliferation and survival via activation of the mitogenactivated protein kinase pathway in bladder cancer. Cancer Res. 2009;69:4613–20. doi: 10.1158/0008-5472.CAN-08-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]


