Abstract
Missense mutations in PIK3CA are common in breast cancers. They mostly involve exons 9 and 20 which encode kinase and helical domains of the protein and may result in its activation. PIK3CA activating mutations were previously shown to predict lower pathologic complete response (pCR) in HER2-positive breast cancer cases undergoing neoadjuvant human epidermal growth factor receptor 2-targeting therapy. Hence, the present work was conducted to estimate the mutation frequency in PIK3CA in 51 HER2-positive patients by direct sequencing. Our results showed 8 out of 51 (15.7%) to harbor PIK3CA mutations in either exon 9 or 20, or both. Three patients had mutations in both exons 9 and 20. Seven (13.7%) possess missense mutations in exon 20 which changed the amino acid sequence of the protein (H1047R, M1040I, and G1049G). Only four cases harbored mutations in exon 9, changing the codon sequences (E545K E545A, and R524K). Taking the clinicopathological data to account, the mutation frequency was greater in ductal than lobular carcinomas, in grade II rather than III and in lymph node positive lesions, with a higher HER2 score and which are ER/PR negative. However, none of the correlations proved statistically significant. In conclusion, to the best of our knowledge, the PIK3CA mutation frequency in this study is the first report regarding HER2-positive breast cancer patients in Egypt. Hereby, we highlight a moderate frequency which could be useful in the future as a predictive marker for anti-HER2 therapy.
Keywords: PIK3CA, HER2-positive, breast cancer, mutations, Egypt
Introduction
HER2-positive Breast Cancer (BC) is the most aggressive subtype of BC, where the patient has a decreased overall survival and differential responses to chemotherapeutic and hormonal agents (Berry et al., 2000). HER2-positive cancers are characterized by overexpression and/or amplification of ERBB2 gene (HER2) (Slamon et al., 1987; Slamon et al., 1989). Human epidermal growth factor receptor 2 (Her2) is a tyrosine kinase receptor. In invasive breast carcinomas, it is overexpressed or amplified in 20 to 25% of patients (Slamon et al., 1987). Her2 overexpression may lead to increased receptor homo/heterodimerization which induces phosphorylation of the intracellular domain and leading to activation of many downstream signaling molecules, including class A phosphoinositide 3-kinases (PI3K)/AKT. Activated PI3K catalyzes the phosphorylation of inositol lipids to produce phosphatidylinositol-3,4,5-trisphosphate (PIP3), which is dephosphorylated to PIP2 by phosphatase and tensin homolog (PTEN). PTEN is a lipid phosphatase which is acting directly as an inhibitory of PI3K pathway. In sporadic breast cancer, it is infrequently mutated (5%). Approximately 25% of breast cancer cases have decreased expression of PTEN (Saal et al., 2005; Saal et al., 2008). In the downstream signaling, PIP3 activates the serine/threonine kinase AKT which in turn regulates the mammalian target of rapamycin (mTOR) (Barbareschi et al., 2012).
Activating mutations in PIK3CA have been found in several cancers, including breast cancer (Slamon et al., 1987; Berry et al., 2000; Zhao and Vogt, 2008). 80% of PIK3CA mutations occurs in exons 9 (helical domain) and 20 (kinase domain) which lead to changing in the amino acids sequence and increasing the protein activity (Engelman et al., 2006; Zhao and Vogt, 2008). Mutant PIK3CA encodes the p110α catalytic subunit of PI3K enzyme leads to activation of PI3K/AKT signaling pathway which is frequently involved in several cellular processes required for breast cancer development (Isakoff et al., 2005; Engelman et al., 2006). Of note, a previous study found that patients with PIK3CA mutation and amplified HER2 are less responsive to HER2 inhibitors, reflecting prognostic and therapeutics importance of mutation testing (Berns et al., 2007; Cizkova et al., 2012; Goel and Krop, 2015).
In Egypt, so far, there is no data available about PIK3CA mutations frequency in HER2-positive breast cancer patients. Since the PIK3CA mutation status could be useful potential prognostic and chemotherapy detection marker, we analyzed PIK3CA mutations in a series of 51 patients with Her2-positive BC to investigating the mutations frequencies and their correlation to clinical and pathological parameters.
Materials and methods
Patients and samples
Formalin fixed paraffin embedded (FFPE) sections were collected from 51 female Egyptian patients, who were diagnosed with HER2 positive breast cancer between 2007 and 2013. Clinical and pathological information, including age, tumor type, tumor grade, marital and menopausal status, lymph nodes, HER2 score and ER and PR status, were collected.
The samples were recruited from the National Cancer Institute in cooperation with the Early Cancer Detection Unit of Ain Shams University Maternity Hospital. The study was approved by the ethics committee of the National Cancer Institute.
DNA extraction
Genomic DNA was extracted using QIAamp DNA extraction kit for FFPE tissues (Qiagen, Hilden, Germany) according to the manufacturer’s protocol with slight modifications. DNA yield and purity were quantitated and assessed using the Nanodrop.
PCR and Sequencing for mutation screening
Polymerase chain reaction (PCR) was performed for all DNA samples using primers designed to amplify PIK3CA (exons 9 and 20). Primers sequence were as the following:
PIK3CA exon 9 F:(ATTAGCAATGTAAAATTTATTGAAAATGTATTT GCTTTTTC)
PIK3CA exon 9 R:(TAAATTCTGCTTTATTTATTCCAATATGGT)
PIK3CA exon 20: (CTCAATGATGCTTGGCTCTG)
PIK3CA exon 20: (TGGAATCCAGAGTGAGCTTTC)
Amplification reactions were carried out using Taq polymerase (Thermoscientific, MA, USA) in accordance to the manufacturer’s protocol. Thermocycling condiotions were an initial denaturation at 95°C for 5 minutes (1 cycle), then 40 cycles of denaturation at 95°C for 1 minute, annealing at 51°C for 90 seconds, elongation at 68°C for 2 minute, followed by final elongation at 68°C for 12 minutes. Nontemplate (DNA) control represented the negative control and was included in every PCR run. The PCR product was then analyzed by gel electrophoresis performed on 2% agarose gel stained with ethidium bromide.
All sequencing reactions were accomplished using the same primers. The PCR products were sequenced using Big Dye terminator mixture. Capillary electrophoresis, sequence analysis, and data collection were done using an automated DNA sequencer. The resulted sequences were compared to human genomic DNA sequence using blast and the mutations numbers were identified using ENSEMBL and Cosmic database.
Statistical analysis
Chi-square (Fisher’s exact) test was used to examine the relation between qualitative variables as appropriate. Survival analysis was made using Kaplan-Meier method. Comparison between two survival curves used log rank test, P-value ≤0.05 was considered significant. All tests were two tailed. Overall survival (OS) was calculated from date of diagnosis till date of death or last follow up.
Results
Clinical characteristics of the study cohort
In the present study, tumor tissues from 51 breast carcinoma patients with HER-2 positive were analyzed for PIK3CA and PTEN mutations. The details of the clinicopathological data of all patients are shown in Table 1. The median age of the cohort was 48 years ranging from 28 to 71 years. According to the histopathological data, the majority of the cases were ductal carcinoma (n = 44, 86.3 %), followed by lobular (n = 2, 3.9%) and others (n = 5, 9.8 %). 84.3 % of the tumors were classified as grade II and 15.7 % as grade III. Most of the tumors were found to be node positivite (58.8 %) while about (25%) were negative. According to tumor subtyping with immunohistochemical analysis for ER and PR, 45.1% and 41.2% of the cases showed positive expression respectively.
Table 1.
Clonicopathological Data of the Study Cohort (n=51)
| Variables | n | % |
|---|---|---|
| Age (years) | ||
| Median (range) | 48 (28-71) | - |
| No. of offspring | ||
| Median (range) | 3(0-7) | - |
| Marital status | ||
| Married | 50 | 98.0 |
| Widow | 1 | 2.0 |
| Menopausal status | ||
| Premenopausal | 38 | 74.5 |
| Postmenopausal | 13 | 23.5 |
| Histology | ||
| Ductal | 44 | 86.3 |
| Lobular | 2 | 3.9 |
| Other | 5 | 9.8 |
| Tumor grade | ||
| II | 43 | 84.3 |
| III | 8 | 13.7 |
| Lymph node status | ||
| Negative | 13 | 25.5 |
| Positive | ||
| 3 | 8 | 15.7 |
| > 3 | 22 | 43.1 |
| Missed | 8 | 15.7 |
| HER2 score | ||
| 2 | 10 | 19.6 |
| 3 | 41 | 80.4 |
| ER status | ||
| Negative | 28 | 54.9 |
| Positive | 23 | 45.1 |
| PR status | ||
| Negative | 30 | 58.8 |
| Positive | 21 | 41.2 |
PIK3CA mutations type and frequency
By the mean of direct sequencing, 51 tumor samples from HER2-positive breast cancer were analyzed. In our study cohort, we found that the mutations are hitting both helical and kinase domain of the encoded protein. Eight out of 51 patients (15.7%) harbored PIK3CA mutations in either exon 9 or 20, or both. Three out of the eight patients with aberrant PIK3CA have mutations in exon 9 and 20. As shown in table 2, most of the mutations were missense mutations. Accordingly, the nucleotide substitution is predicted to change the amino acid sequence and consequently the protein function.
Table 2.
Frequency and Mutation Types of PIK3CA
| cosmic number | Nucleotide change | amino acids | type of mutation | No. of patient | % | Domain | ||
|---|---|---|---|---|---|---|---|---|
| PIK3CA | exon9 | COSM763 | c.1633G>A | p.E545K | missense | 2 | 3.9 | Helical |
| COSM297145 | c.1634A>C | p.E545A | missense | 1 | 2.0 | Helical | ||
| COSM53245 | c.1571G>A | p.R524K | missense | 1 | 2.0 | Helical | ||
| exon 20 | COSM25085 | c.3120G>A | p.M1040I | missense | 1 | 2.0 | Kinase | |
| COSM775 | c.3140A>G | p.H1047R | missense | 5 | 9.8 | Kinase | ||
| COSM87299 | c.3147T>C | p.G1049G | coding silent | 1 | 2.0 | Kinase |
Among the 51 analyzed samples, four patients (7.8%) harbored a missense mutation in exon 9. The detected mutations are changing the codon sequences as the following: (two patients possess E545K, one patients with E545A, and the 4th patient harbored R524K). The three mutations are changing the amino acid sequence of the helical domain of the protein.
Three different mutations in exon 20 were also identified in the present study that change the codon sequence in seven patients (13.7%). These mutations hit the kinase domain. Three amino acids substitution (H1047R, M1040I, and G1049G) were identified.
Mutations correlation to clinicopathological data
Table 3 displays all correlations between PIK3CA mutation status, exon9 and 20 mutation status, and every mutation found and the clinical, pathological and biological parameters of the study cohort. As noted, there is no statistical significant correlation between all PIK3CA mutations and any parameter of the clinicopathological data and also for exon 9 or 20. At the histological level, it was observed that PIK3CA mutation was frequently found in ductal carcinomas comparable to lobular carcinomas (15.9 vs. 0 %, p = 0.801). The correlation between PIK3CA mutations and tumor grade shows that patients with grade II are more frequent than grade III (16.3 vs. 1.3 %, p = 1.00). In addition, there were more cases with PIK3CA mutation in lymph node-positive tumors than negative cases (16.7 vs. 15.4 %). The opposite is for the biomarker parameters (ER and PR), The patients with ER/PR negative pattern have PIK3CA mutations more than the patients with positive ER/PR (17.9 vs. 13%, p = 1.00) and (14.3 vs. 16.7, p = 1.00) respectively. For HER-2 score we found that there is a slightly significant correlation between the higher score and exon 9 mutations (9.7 vs. 0 %, p = 0.069).
Table 3.
Correlation between the Different Types of Mutations and ClinicoPathological Data
| n | PIK3CA Exons 9 and 20 | EX9 total | c.1633 G>A | c.1634 A>C | c.1571 G>A | EX20 total | c.3140 | c.3140 | c.3147 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | mt | p value | wt | mt | p | wt | mt | p | wt | mt | p | wt | mt | p | wt | mt | p value | wt | mt | p value | wt | mt | p value | wt | mt | p value | |||
| Age (years) | |||||||||||||||||||||||||||||
| ≤50 | 29 | 25 (86.8) | 4 (13.2) | 0.713 | 27 (93.1) | 2 (6.9) | 1 | 28 (96.6) | 1 (3.4) | 1 | 29 (100) | 0 (0) | 0.431 | 28 (96.6) | 1 (3.4) | 1 | 25 (86.8) | 4 (13.2) | 1 | 25 (86.8) | 4 (13.2) | 0.375 | 29 (100) | 0 (0) | 0.431 | 29 (100) | 0 | 0.431 | |
| >50 | 22 | 18 (81.8) | 4 (18.2) | 20 (90.9) | 2 (9.1) | 21 (95.5) | 1 (4.5) | `21 (95.5) | 1 (4.5) | 22 (100) | 0 (0) | 19 (86.4) | 3 (13.6) | 21 (95.5) | 1 (4.5) | 21 (95.5) | 1 (4.5) | 21 (95.5) | 1 (4.5) | ||||||||||
| Menopausal status | |||||||||||||||||||||||||||||
| Premenopausal | 38 | 33 (86.8) | 5 (13.2) | 0.404 | 36 (94.7) | 2 (5.3) | 0.695 | 37 (97.4) | 1 (2.6) | 0.449 | 37 (97.4) | 1 (2.6) | 0.255 | 37 (97.4) | 1 (2.6) | 1 | 34 (89.5) | 4 (10.5) | 0.352 | 34 (89.5) | 4 (10.5) | 1 | 37 (97.4) | 1 (2.6) | 1 | 38 (100) | 0 | 0.255 | |
| Postmenopausal | 13 | 10 (76.9) | 3 (23.1) | 11 (84.6) | 2 (15.4) | 12 (92.3) | 1 (7.7) | 13 (100) | 0 (0) | 13 (100) | 0 (0) | 10 (76.9) | 3 (23.1) | 12 (92.3) | 1 (7.7) | 13 (100) | 0 | 12 (92.3) | 1 (7.7) | ||||||||||
| Histology | |||||||||||||||||||||||||||||
| Ductal | 44 | 37 (84.1) | 7 (15.9) | 0.801 | 40 (90.9) | 4 (9.1) | 0.708 | 42 (95.5) | 2 (4.5) | 0.847 | 43 (97.7) | 1 (2.3) | 0.922 | 43 (97.7) | 1 (2.3) | 0.922 | 38 | 6 | 0.785 | 40 (90.9) | 4 (9.1) | 0.66 | 43 (97.7) | 1 (2.3) | 0.922 | 43 (97.7) | 1 (2.3) | 0.922 | |
| lobular | 2 | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) | ||||||||||
| Other | 5 | 4 (80) | 1 (20) | 5 (100) | 0 (0) | 5 (100) | 0 (0) | 5 (100) | 0 (0) | 5 (100) | 0 (0) | 4 (80) | 1 (20) | 4 (80) | 1 (20) | 5 (100) | 0 (0) | 5 (100) | 0 (0) | ||||||||||
| Tumor grade | |||||||||||||||||||||||||||||
| II | 43 | 36 (83.7) | 7 (16.3) | 1 | 40 (93) | 3 (7) | 0.506 | 41 (95.3) | 2 (4.7) | 1 | 42 (97.7) | 1 (2.3) | 1.000 | 43 (100) | 0 (0) | 0.157 | 37 (86) | 6 (14) | 1 | 39 (90.6) | 4 (9.4) | 1 | 42 (97.7) | 1 (2.3) | 1 | 42 (97.7) | 1 (2.3) | 1 | |
| III | 8 | 7 (87.5) | 1 (12.5) | 7 (87.5) | 1 (12.5) | 8 (100) | 0 (0) | 8 (100) | 0 (0) | 7(87.5) | 1 (12.5) | 7 (87.5) | 1 (12.5) | 7 (87.5) | 1 (-12.5) | 8 (-100) | 0 | 8 (-100) | 0 | ||||||||||
| Lymph node status | |||||||||||||||||||||||||||||
| Negative | 13 | 11 (84.6) | 2 (15.4) | 0.928 | 11 (84.6) | 2 (15.4) | 0.284 | 12 (92.3) | 1 (7.7) | 0.718 | 13 (100) | 0 (0) | - | 12 (92.3) | 1 (7.7) | 0.307 | 11 (84.6) | 2 (15.4) | 0.928 | 11 (84.6) | 2 (15.4) | 0.851 | 13 (100) | 0 (0) | 0.613 | 13 (100) | 0 (0) | 0.613 | |
| Positive 1(3 | 8 | 7 (87.5) | 1 (12.5 | 8 (100) | 0 (0) | 8 (100) | 0 (0) | 8 (100) | 0 (0) | 8 (100) | 0 (0) | 7 (87.5) | 1 (12.5) | 7 (87.5) | 1 (12.5) | 8 (100) | 0 (0) | 8 (100) | 0 (0) | ||||||||||
| Positive > 3 | 22 | 18 (81.8) | 4 (18.8) | 21 (95.5) | 1 (4.5) | 21 (95.5) | 1 (4.5) | 22 (100) | 0 (0) | 22 (100) | 0 (0) | 18 (81.8) | 4 (18.8) | 20 (90.9) | 2 (9.1) | 21 (95.5) | 1 (4.5) | 21 (95.5) | 1 (4.5) | ||||||||||
| HER2 score | |||||||||||||||||||||||||||||
| 2 | 10 | 10 (100) | 0 (0) | 0.329 | 10 (100) | 0 (0) | 0.069 | 10 (100) | 0 (0) | 1 | 10 (100) | 0 (0) | 1 | 10 (100) | 0 (0) | 1 | 10 (100) | 0 (0) | 0.32 | 10 (100) | 0 | 0.569 | 10 (100) | 0 (0) | 1 | 10 (100) | 0 (0) | 1 | |
| 3 | 41 | 33 (80.5) | 8 (19.5) | 37 (90.2) | 4 (9.8) | 39 (95.1) | 2 (4.9) | 40 (97.6) | 1 (2.4) | 40 (97.6) | 1 (2.4) | 34 (82.9) | 7 (17.1) | 36 (87.8) | 5 (12.2) | 40 (-97.6) | 1 (-2.4) | 40 (-97.6) | 1 (-2.4) | ||||||||||
| ER | |||||||||||||||||||||||||||||
| positive | 23 | 20 (97) | 3 (3) | 0.715 | 22 (95.7) | 1 (4.3) | 1 | 23 (100) | 0 | 0.495 | 22 (95.7) | 1 (4.3) | 1 | 22 (95.7) | 1 (4.3) | 0.451 | 20 (97) | 3 (3) | 1 | 20 (97) | 3 (3) | 0.647 | 23 (100) | 0 | 1 | 23 (100) | 0 | 1 | |
| negative | 28 | 23 (82.1) | 5 (17.9) | 25 (89.3) | 3 (10.7) | 26 (82.9) | 2 (17.1) | 28 (100) | 0 (0) | 28 (100) | 0 (0) | 24 (85.7) | 4 (14.3) | 26 (82.9) | 2 (17.1) | 27 (94.4) | 1 (5.6) | 27 (94.4) | 1 (5.6) | ||||||||||
| PR | |||||||||||||||||||||||||||||
| positive | 21 | 18 (85.7) | 3 (14.3) | 1 | 20 (95.2) | 1 (4.8) | 1 | 21 (100) | 0 (0) | 0.506 | 21 (100) | 0 (0) | 1 | 20 (95.2) | 1 (4.8) | 0.412 | 18 (85.7) | 3 (14.3) | 1 | 18 (85.7) | 3 (14.3) | 0.637 | 21 (100) | 0 | 1 | 21 (100) | 0 | 1 | |
| negative | 30 | 25 (83.3) | 5 (16.7) | 27 (90) | 3 (10) | 28 (93.3) | 2 (6.7) | 29 (96.7) | 1 (3.3) | 30 (100) | 0 (0) | 26 (86.7) | 4 (13.3) | 28 (93.3) | 2 (6.7) | 29 (96.7) | 1 (3.3) | 29 (96.7) | 1 (3.3) | ||||||||||
Survival analysis
The median follow-up time was 35.2 months (ranging from 1.0 to 110.1months); OS of BC patients is shown in Fig (2). There is no statistically significant association between BC patients’ OS and clinicopathological data of studied cohort or between OS and PIK3 mutation as shown in Table (4).
Figure 1.
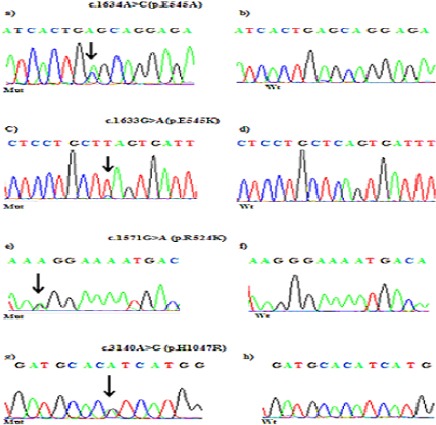
Partial Chromatograms of PIK3CA Mutations: a, c and e, PIK3CA Exon 9 Mutations (E545A, E545K and R524K); b, d, and f, Wild-Type sequences. g, PIK3CA exon 20 Mutation (H1047R); h, Wild-Type Sequence. Arrows, Position of the Missense Mutationas
Figure 2.
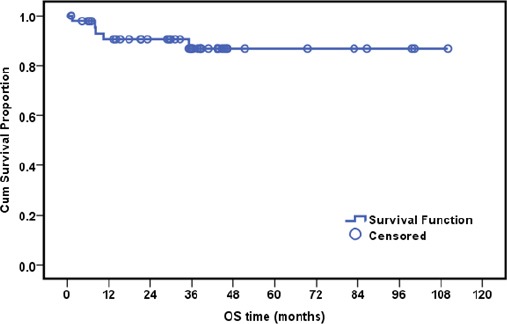
Overall Survival of the Investigated Breast Cancer Cases
Table 4.
Correlation of Overall Survival (OS) with PIK3CA Mutation Status and with Clinicopathological Data
| Variable | n | Cumulative survival at 36 months | P-value |
|---|---|---|---|
| All | 48 | 86.9% | _ |
| PIK3CA | |||
| Wild | 40 | 87.4% | 0.818 |
| Mutant | 8 | 85.7% | |
| PIK3CA exon 9 | |||
| Wild | 46 | 85.4% | * |
| Mutant | 2 | 100.0% | |
| c.1633 G>A | |||
| Wild | 46 | 100.0% | * |
| Mutant | 2 | 86.1% | |
| c.1634 A>C | |||
| Wild | 47 | 86.6% | * |
| Mutant | 1 | 100.0% | |
| c.1571 G>A | |||
| Wild | 47 | 86.5% | * |
| Mutant | 1 | 100.0% | |
| PIK3CA exon 20 | |||
| Wild | 41 | 87.6% | 0.712 |
| Mutant | 7 | 83.3% | |
| c.3147 T>C | |||
| Wild | 47 | 86.6% | * |
| Mutant | 1 | 100.0% | |
| c.3140 A>G | |||
| Wild | 43 | 88.2% | 0.423 |
| Mutant | 5 | 75.0% | |
| c.3120 G>A | |||
| Wild | 47 | 86.5% | * |
| Mutant | 1 | 100.0% | |
| Relation of overall survival (OS) and clinicopathological data | |||
| Age | |||
| ≤50 | 28 | 84.9% | 0.404 |
| >50 | 20 | 88.9% | |
| Menopausal status | |||
| Premenopausal | 36 | 87.4% | 0.827 |
| Postmenopausal | 12 | 80.0% | |
| Histology | |||
| IDC | 42 | 89.9% | 0.083 |
| Others | 6 | 66.7% | |
| Tumor grade | |||
| II | 40 | 83.3% | 0.242 |
| III | 8 | 100.0% | |
| Lymph node | |||
| Negative | 12 | 91.7% | 0.823 |
| Positive 1-3 | 8 | 85.7% | |
| Positive >3 | 22 | 79.0% | |
| HER2 score | |||
| 2 | 9 | 100.0% | 0.277 |
| 3 | 39 | 83.9% | |
| ER | |||
| Positive | 20 | 89.2% | 0.828 |
| Negative | 28 | 83.7% | |
| PR | |||
| Positive | 20 | 94.7% | 0.274 |
| Negative | 28 | 79.5 |
, no P-value because of small no of cases within subgroups; No median survival because more than half of patients were still alive till end of the study
Discussion
According to GLOBOCAN 2012, breast cancer is the second most common malignancy worldwide and the most frequent cancer in women. A slight majority of cases were in less developed countries rather than developed world. Worldwide, breast cancer is the fifth cause of malignancy death overall. In the less developed region, it is the most frequent cause of cancer death in women (Ferlay et al., 2015). Breast carcinomas are heterogeneous group of tumors which differ morphologically and at the molecular level (Samuels et al., 2005; Engelman, 2009). HER2-positive BC is the most aggressive subtype of BC, where the patient has a decreased overall survival and may have differential responses to a variety of chemotherapeutic and hormonal agents (Berry et al., 2000). In the HER2-positive patients, using therapeutic reagents that target the human epidermal growth factor receptor-2 (HER2) has improved the outcomes in combination with chemotherapy (Romond et al., 2005; Swain et al., 2015). PI3K pathway is the downstream signaling for HER2 and is frequently activated in cancer (Vivanco and Sawyers, 2002). Recently, Loibl et al., (2016) carried out a meta-analysis in which they studied whether PIK3CA activating mutations are linked to lower sensitivity to neoadjuvant therapy. They found that overall PIK3CA mutations in HER2-positive significantly decreased the pathological complete response (pCR) compared to the wild type. These findings point out that PIK3CA mutations status could potentially act as a predictive marker for HER2-positive cancer.
In the present study, we carried out a sequence analysis for most common mutated exons of PIK3CA (exons 9 and 20) in 51 patients. So, this study was executed to calculate the mutation frequency and correlation to clinicopathological data in Egyptian HER2-positive breast cancer patients. To our knowledge, our study is the first study to evaluate PIK3CA mutations in HER2-positive patients in Egypt. The only available study (on Pubmed) in Egypt was done in HER2-negative breast cancer patients and PIK3CA mutation status was tested in 24 patients. In this previous study, they found 7 out of 24 HER2-negative breast cancer patients (29.2%) in Egypt (Azim et al., 2016). In the present study, we found less frequent overall PIK3CA mutations (eight out of 51 patients) were detected in HER2-positive breast cancer (15.7%). However, it is difficult to conclude that the frequency of PIK3CA mutations is higher in HER2-negative in the Egyptian patients due to the small number of patients in both cohorts. Nevertheless, Arsenic et al. (Arsenic et al., 2014) found that the PIK3CA mutations rate was higher in HR(+)/HER2(-), whereas the lowest mutation rate was observed in HR(+)/HER2(+) in their study. In contrary, another study found that the PIK3CA somatic mutations in hormonal positive/HER2-positive are more common, though the data are not significant (Ahmad et al., 2016). The peer studies showed that PIK3CA mutations frequency is very variable across the world (Li et al., 2006; Kalinsky et al., 2009; Loibl et al., 2014; Ross et al., 2015; Ahmad et al., 2016; Sudhakar et al., 2016). The percentage of the detected mutations in our cohort is comparable to HER2-positive subtype in a recent Indian study (Ahmad et al., 2016).
As reviewed previously (Dirican et al., 2016), around 80% of PIK3CA mutations occur in exons 9 and 20. These two exons have mostly the “hot spot” of PIK3CA gene. Exons 9 and 20 encode the helical and kinase domains of the protein. the mutations of this region activate the gene with constant auto-phosphorylation. In the present data, mutations in exon 20 which encodes the kinase domain are more frequent than those in exon 9 (encodes the helical domain). These data are in alignment with several previous studies (Li et al., 2006; Loibl et al., 2014; Arsenic et al., 2015; Ahmad et al., 2016). From the detected mutations, three missense mutations were considered as hot spots which are at amino acid number 524, 545, and 1047 as described previously in the results section.
Considering the clinicopathological data, the PIK3CA mutations didn’t show any significant association with clinical and pathological data which was shown in many previous studies (Harle et al., 2013; Loibl et al., 2014; Ahmad et al., 2016). However, our results show tendency of having PIK3CA mutation in higher HER2 score (data are not significant) with higher mutations rate in exon 9 (with trend to be significant; p = 0.069). Consistently with previous studies (Dunlap et al., 2010; Kandula et al., 2013; Arsenic et al., 2015; Ahmad et al., 2016), PIK3CA mutations were more frequent in grade II tumors than patients with grade III which could be referred to involvement of the mutations at the onset of the carcinogenesis process. The survival data are not significant in our study which could be related to the limited number of patients in the investigated cohort.
In summary, the present study is the first one which investigating PIK3CA mutations in HER2-positive breast cancer patients in Egypt. The frequency reported in this study was comparable to the international frequencies. Interestingly, the frequency found in HER2-positive patients was less than HER2-negative patient reported in another study (Azim et al., 2016). However, the number of patients contributed in this study is not enough for solid conclusion. Even though, we are highlighting the frequency which could be useful in the nearby future research and clinical application of PIK3CA as a predictive marker for anti-HER2 therapy.
Compliance with ethical standard
Conflict of interest
None of the authors have any conflict of interest.
Funding
The study was funded by Ain Shams University. Also, Fatma Elwy has received a MSC funding support from BioKMT society.
Informed consent and Ethical approval
The paraffin sections were obtained from the Egyptian National Cancer Institute. A written informed consent was obtained from all patients. The study was approved by the ethics committee of the National Cancer Institute.
References
- Ahmad F, Badwe A, Verma G, et al. Molecular evaluation of PIK3CA gene mutation in breast cancer:determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol. 2016;33:74. doi: 10.1007/s12032-016-0788-y. [DOI] [PubMed] [Google Scholar]
- Arsenic R, Lehmann A, Budczies J, et al. Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol. 2014;22:50–50. doi: 10.1097/pdm.0b013e318297afea. [DOI] [PubMed] [Google Scholar]
- Arsenic R, Treue D, Lehmann A, et al. Comparison of targeted next-generation sequencing and Sanger sequencing for the detection of PIK3CA mutations in breast cancer. BMC Clin Pathol. 2015;15:20. doi: 10.1186/s12907-015-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim HA, Kassem L, Treilleux I, et al. Analysis of PI3K/mTOR pathway biomarkers and their prognostic value in women with hormone receptor-positive, HER2-negative early breast cancer. Transl Oncol. 2016;9:114–114. doi: 10.1016/j.tranon.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbareschi M, Cuorvo LV, Girlando S, et al. PI3KCA mutations and/or PTEN loss in Her2-positive breast carcinomas treated with trastuzumab are not related to resistance to anti-Her2 therapy. Virchows Arch. 2012;461:129–129. doi: 10.1007/s00428-012-1267-2. [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–395. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Berry DA, Muss HB, Thor AD, et al. HER-2/neu and p53 expression versus tamoxifen resistance in estrogen receptor-positive, node-positive breast cancer. J Clin Oncol. 2000;18:3471–3471. doi: 10.1200/JCO.2000.18.20.3471. [DOI] [PubMed] [Google Scholar]
- Cizkova M, Susini A, Vacher S, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14:R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirican E, Akkiprik M, Ozer A. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer. Tumour Biol. 2016;37:7033–7033. doi: 10.1007/s13277-016-4924-2. [DOI] [PubMed] [Google Scholar]
- Dunlap J, Le C, Shukla A, et al. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat. 2010;120:409–409. doi: 10.1007/s10549-009-0406-1. [DOI] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer:opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–550. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–606. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide:sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:359–359. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Goel S, Krop IE. Deciphering the role of phosphatidylinositol 3-kinase mutations in human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2015;33:1407–1407. doi: 10.1200/JCO.2014.60.0742. [DOI] [PubMed] [Google Scholar]
- Harle A, Lion M, Lozano N, et al. Analysis of PIK3CA exon 9 and 20 mutations in breast cancers using PCR-HRM and PCR-ARMS:correlation with clinicopathological criteria. Oncol Rep. 2013;29:1043–1043. doi: 10.3892/or.2013.2229. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Engelman JA, Irie HY, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5049. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- Kandula M, Chennaboina KK, Ys AR, et al. Phosphatidylinositol 3-kinase (PI3KCA) oncogene mutation analysis and gene expression profiling in primary breast cancer patients. Asian Pac J Cancer Prev. 2013;14:5067–5067. doi: 10.7314/apjcp.2013.14.9.5067. [DOI] [PubMed] [Google Scholar]
- Li SY, Rong M, Grieu F, et al. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–5. doi: 10.1007/s10549-005-9048-0. [DOI] [PubMed] [Google Scholar]
- Loibl S, Majewski I, Guarneri V, et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer:pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol. 2016;27:1519–1519. doi: 10.1093/annonc/mdw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loibl S, von Minckwitz G, Schneeweiss A, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32:3212–3212. doi: 10.1200/JCO.2014.55.7876. [DOI] [PubMed] [Google Scholar]
- Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1673. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- Ross JS, Ali SM, Wang K, et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat. 2015;154:155–155. doi: 10.1007/s10549-015-3592-z. [DOI] [PubMed] [Google Scholar]
- Saal LH, Gruvberger-Saal SK, Persson C, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40:102–102. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2554. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–561. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, et al. Human breast cancer:correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–177. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–707. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Sudhakar N, Priya Doss CG, Thirumal Kumar D, et al. Deciphering the impact of somatic mutations in exon 20 and exon 9 of PIK3CA gene in breast tumors among Indian women through molecular dynamics approach. J Biomol Struct Dyn. 2016;34:29–29. doi: 10.1080/07391102.2015.1007483. [DOI] [PubMed] [Google Scholar]
- Swain SM, Baselga J, Kim SB, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724–724. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–489. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5486. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]


