Abstract
Cell‐penetrating peptides (CPPs) are small peptides capable of crossing cellular membranes while carrying molecular cargo. Although they have been widely studied for their ability to translocate nucleic acids, small molecules, and proteins into mammalian cells, studies of their interaction with fungal cells are limited. In this work, we evaluated the translocation of eleven fluorescently labeled peptides into the important human fungal pathogens Candida albicans and C. glabrata and explored the mechanisms of translocation. Seven of these peptides (cecropin B, penetratin, pVEC, MAP, SynB, (KFF)3K, and MPG) exhibited substantial translocation (>80% of cells) into both species in a concentration‐dependent manner, and an additional peptide (TP‐10) exhibiting strong translocation into only C. glabrata. Vacuoles were involved in translocation and intracellular trafficking of the peptides in the fungal cells and, for some peptides, escape from the vacuoles and localization in the cytosol were correlated to toxicity toward the fungal cells. Endocytosis was involved in the translocation of cecropin B, MAP, SynB, MPG, (KFF)3K, and TP‐10, and cecropin B, penetratin, pVEC, and MAP caused membrane permeabilization during translocation. These results indicate the involvement of multiple translocation mechanisms for some CPPs. Although high levels of translocation were typically associated with toxicity of the peptides toward the fungal cells, SynB was translocated efficiently into Candida cells at concentrations that led to minimal toxicity. Our work highlights the potential of CPPs in delivering antifungal molecules and other bioactive cargo to Candida pathogens.
Keywords: Cell‐penetrating peptides, Candida, translocation, drug delivery
Introduction
Candidiasis is a human fungal infection caused by Candida species, often C. albicans or C. glabrata. Infections include relatively minor mucosal infections, such as skin infections and oral thrush, as well as severe systemic infections that can occur in people with compromised immune systems.1 Available antifungal drugs to treat candidiasis act on a limited number of cellular targets, and the incidence of drug resistance is rising,2 spurring the demand for novel therapeutic approaches.
One potential approach to improving antifungal therapeutics is to use a drug delivery vehicle to improve cellular recognition and intracellular delivery of new or existing antifungal agents. Cell‐penetrating peptides (CPPs) are promising candidates for delivering therapeutic agents and have been widely studied for intracellular delivery of bioactive molecules into mammalian cells. CPPs are typically short, cationic peptides with 30 amino acids or fewer3 and have the ability to cross cell membranes. A variety of bioactive molecules have been successfully attached to CPPs and delivered intracellularly, including nucleic acids (both DNA and RNA),4, 5, 6 fluorescent proteins,7, 8 and antibodies.9, 10
Although CPPs show great potential in drug delivery, the structural and functional diversity of CPPs complicates studies of their interactions with cells. Previous mechanistic research on CPP translocation has focused on translocation into mammalian cells and suggests some peptides penetrate cells via an energy‐dependent endocytosis process. These CPPs include MAP (synthetic, highly amphiphilic model peptide11), TP‐10 (fragment of transportan12), hCT (derivative of calcitonin13), SynB (derivative of the antimicrobial peptide protegrin 114), and PAF26 (hexapeptide with antimicrobial activity15). In contrast, other CPPs may enter cells via macropinocytosis,16 including pVEC (derivative of murine vascular endothelium cadherin17) and penetratin (fragment of antennaedia homeodomain18). The mechanism of translocation may also include transient pore formation,19 which is suspected for MPG (derivative of two viruses20) and Pep‐1 (synthetic peptide21). MPG and Pep‐1 also contain nuclear localization sequences, which promote the translocation efficacy and solubility of the peptides.19 For other CPPs, such as (KFF)3K (synthetic peptide22), additional work is required to elucidate the mechanism of translocation. These examples highlight the diversity of CPPs as delivery vehicles and the challenges in understanding their interaction with cells.
Most studies on the translocation mechanisms of CPPs have focused on translocation in mammalian cells, and studies of the interactions of CPPs with fungal cells, including Candida cells, are very limited.7, 8, 23, 24, 25, 26 To expand the application of CPPs to delivering molecules to Candida species, an improved understanding of the interaction of CPPs with Candida cells is required. One key structural difference between mammalian cells and fungal cells is the presence of a cell wall in fungal cells. The cell wall is composed of chitin, glucans, mannans, and glycoproteins and provides an additional barrier for CPP transport into fungal cells compared to mammalian cells.27 Another key difference is that fungal cells have vacuoles, which are involved in a number of biological processes in fungal cells, including endocytosis, pH and salt balance maintenance, and phosphate degradation.28 The effect of these structures on CPP translocation and trafficking has not been described previously, and an understanding of their role will facilitate the use and design of CPPs for delivering molecular cargo to fungal cells.
To improve the understanding of how CPPs translocate into fungal cells, we studied the translocation of known CPPs into two Candida pathogens, C. albicans and C. glabrata. We evaluated the translocation and toxicity of the CPPs and explored their mechanisms of translocation. Some peptides previously shown to enter mammalian cells were also translocated into Candida species, while others exhibited little to no translocation. Our analysis of subcellular localization of CPPs provides insight into intracellular trafficking of the peptides, as well as translocation mechanisms. Further experiments to explore the translocation mechanisms indicate the translocation of some CPPs in fungal cells may differ from the mechanisms proposed for mammalian cells. Our data suggest translocation of CPPs into fungal cells often correlates with toxicity toward the cells, but some peptides are taken up by Candida cells with little effect on viability.
Results and Discussion
Translocation of CPPs into Candida species
To study the interaction of CPPs with Candida species, we selected peptides representing a variety of structures and native origins to study CPP translocation into Candida cells (Table 1). All peptides were synthesized commercially with an N‐terminal 5‐carboxyfluorescein label (FAM), which served as the cargo for the CPP and as the reporter to detect translocation. Our set of peptides includes peptides shown previously to translocate into mammalian cells (all CPPs) or microbial cells, including bacteria and fungi (pVEC, (KFF)3K, penetratin, MAP, PAF26, and TP‐10).7, 8, 23, 24, 25, 26 The CPPs pVEC, (KFF)3K, penetratin, and TP‐10 were shown to enter C. albicans previously.7, 23 Some peptides are thought to be transported by energy‐dependent endocytic mechanisms (MAP, hCT, TP‐10, SynB, and PAF26) or macropinocytosis (pVEC and penetratin), while others undergo pore formation (penetratin, MPG and Pep‐1) or unknown mechanisms ((KFF)3K).11, 12, 13, 14, 15, 16, 17, 18, 19 We also included cecropin B, a well‐known antimicrobial peptide with antifungal activity, to compare its translocation with peptides previously identified as CPPs. C. albicans and C. glabrata were selected as target cells, because they are frequently isolated from patients with candidiasis.29
Table 1.
CPPs tested in this study
| Peptide | Sequence | MW (Da) | Net chargea |
|---|---|---|---|
| pVEC | LLIILRRRIRKQAHAHSK | 2209.7 | +8 |
| (KFF)3K | KFFKFFKFFK | 1413.8 | +4 |
| Penetratin | RQIKIWFQNRRMKWKK | 2246.8 | +7 |
| MAP | KLALKLALKALKAALKLA | 1876.0 | +5 |
| Pep‐1 | KETWWETWWTEWSQPKKKRKV | 2848.3 | +2 |
| hCT | LGTYTQDFNKTFPQTAIGVGAP | 2326.6 | 0 |
| SynB | RGGRLSYSRRRFSTSTGR | 2100.3 | +6 |
| MPG | GALFLGFLGAAGSTMGAWSQPKKKRKV | 2807.4 | +5 |
| PAF26 | RKKWFW | 950.2 | +3 |
| TP‐10 | AGYLLGKINLKALAALAKKIL | 2182.8 | +4 |
| Cecropin B (antimicrobial peptide) | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKAL | 3835.7 | +9 |
Includes only charges due to amino acid side chains (pH 7) and not N‐terminal FAM (‐3).
To screen the CPPs for translocation into Candida species, we incubated C. albicans and C. glabrata cells with each of the CPPs. We identified the CPPs that cross the barriers of these fungal cells using fluorescence microscopy to visualize the location of the fluorescein‐labeled peptides [Fig. 1(A,B)]. Cells were treated with trypsin prior to imaging to remove peptide associated with the cell surface.7, 30 A high level of translocation efficacy was observed in both types of fungal cells for several peptides with a relatively high net charge (≥ +4), including penetratin, pVEC, MAP, SynB, (KFF)3K, and MPG. PAF26 and Pep‐1, which have a lower net charge (< +4), showed limited levels of translocation, while hCT (no net charge) could not be detected entering the Candida cells. The antimicrobial peptide cecropin B (+ 9) exhibited a high level of translocation, suggesting cecropin B can function as a CPP in addition to an antimicrobial peptide.
Figure 1.
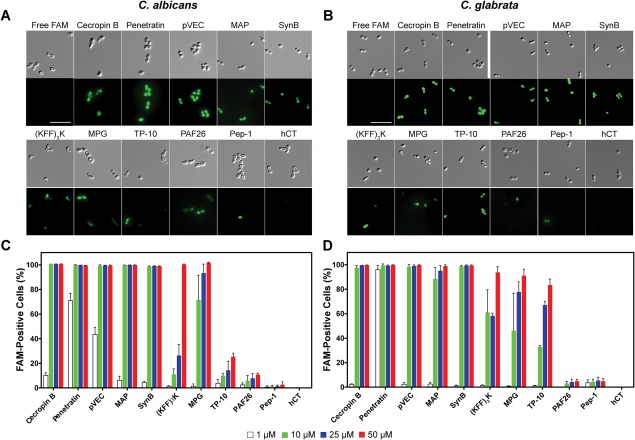
Translocation of FAM‐labeled CPPs into Candida cells. DIC and FAM fluorescence images for translocation into (A) C. albicans and (B) C. glabrata. Flow cytometry data for translocation into (C) C. albicans and (D) C. glabrata. Cells were incubated for 1 h at 30°C with 25 µM peptides for imaging or serial dilutions of peptides (1–50 µM) for flow cytometry. Surface‐bound peptides were removed by trypsin prior to imaging or flow cytometry, and controls with free FAM were included. For (A) and (B), scale bar = 10 µm. For (C) and (D), error bars represent the standard error of the mean for three separate experiments (N = 3).
We quantified translocation and the effect of peptide concentration on translocation in C. albicans and C. glabrata using flow cytometry. Single cells were identified, and the percentage of these cells positive for FAM fluorescence was determined for each species [Fig. 1(C,D)]. We detected a dose‐dependent fluorescence signal for each of the peptides that showed substantial translocation in the fluorescence microcopy assay. The flow cytometry data confirmed the limited translocation for PAF26 and Pep‐1 and the lack of translocation for hCT that we observed by fluorescence microscopy.
For most of the peptides, the behavior in C. albicans and C. glabrata was very similar. The exception to this was TP‐10, which showed significantly enhanced translocation in C. glabrata compared to C. albicans. Although these two species are closely related, they do have unique characteristics that could affect the interaction of the cells with CPPs. For example, they express different membrane‐anchored proteases and other proteins,31, 32 which may alter degradation of CPPs and their interaction with the cell membrane.
Role of net charge in translocation
For Candida cells, our data indicate that net charge of the peptides plays a role in translocation, with higher levels of positive charge generally leading to higher levels of translocation. As the phosphate heads of the membrane lipids are negatively charged, positively charged residues in peptides will lead to electrostatic interactions that bring peptide in close contact with the cell membrane. CPPs with higher net charges (≥+4) including pVEC, penetratin, MAP, SynB, (KFF)3K, MPG, TP‐10 and Cecropin B would have stronger electrostatic interactions, leading to a stronger interaction with the cell membrane and explaining their higher translocation efficacy. In our experiments, we used a FAM label as the cargo for our peptides. This FAM label has a net negative charge of −3 at the pH of our experiments, which would reduce the overall net charge of the peptide‐cargo constructs. Additional studies evaluating other cargo molecules will be important in understanding the impact of the peptide charge alone and in determining whether properties of cargo molecules strongly influence the translocation effects of the peptides.
Comparison to translocation into mammalian cells
Although each of the CPPs was previously shown to translocate into mammalian cells,30, 33 only pVEC, (KFF)3K, penetratin, and TP‐10 were previously shown to translocate into Candida cells.7, 23 Our results indicate that many of the CPPs that function in mammalian cells do function in C. albicans and C. glabrata, but translocation into mammalian cells does not guarantee translocation in fungal cells. Three of the peptides tested showed little to no translocation in the two Candida species. In the case of Pep‐1, association of the peptide with the cell wall contributed to the low level of translocation: when cells incubated with Pep‐1 were imaged prior to trypsin treatment, the peptide was observed on the cell surface (Supporting Information Fig. S1). However, for hCT and PAF26, the origin of the difference is less clear, since these peptides did not localize at the cell surface.
Subcellular localization of peptides
To gain insight into the intracellular trafficking of the CPPs, we examined our microscopy images (Fig. 1). In some images, peptides appeared to localize in the vacuole [e.g. see FAM image for PAF26 in Fig. 1(A)]. In other images, the vacuoles that typically are easily visible were no longer present [e.g. see differential interference contrast (DIC) images for MAP and penetratin in Fig. 1(B)]. Yeast vacuoles are very important for maintaining homeostasis, and they are highly involved in transmembrane transport.28 To better understand the relationship between CPP trafficking, vacuoles, and vacuole loss, we used CellTracker Blue, a yeast vacuole stain, to track the vacuoles during incubation of C. albicans cells with the peptide pVEC [Fig. 2(A)]. When cells were incubated with a low concentration of pVEC, they retained their vacuoles and exhibited colocalization of a low level of FAM fluorescence with vacuole stain fluorescence. At higher concentrations of peptide, we observed total loss of vacuole stain fluorescence, an enhancement of FAM fluorescence intensity, and a shift to cytosolic fluorescence. For cells treated with 10 μM of pVEC, the differential FAM fluorescence was most apparent. Cells retaining their vacuoles showed a brighter vacuole stain signal but a weaker FAM fluorescence, whereas cells that lost vacuoles had a stronger cytosolic FAM signal and no vacuolar fluorescence. The differential fluorescence intensity arises from the sensitivity of the 5‐FAM fluorescein derivative we used. 5‐FAM is very sensitive to pH and exhibits higher fluorescence intensity at pH 7.5 and lower intensity at pH 6.6.34 Because the cytosolic pH (∼7.4) is relatively higher than the one inside vacuoles (∼6.2),35 the fluorescence intensity of the peptides is lower in vacuoles than in the cytosol.36 Vacuole loss can also be identified by using flow cytometry to observe the FAM signal shift as the peptides move from the vacuoles to the cytosol at higher concentrations [Fig. 2(B)]. We quantified the cellular localization by gating two distinct populations in the flow cytometry data: one with a stronger FAM intensity but no vacuole stain fluorescence and the other with both vacuole stain fluorescence and lower intensity FAM fluorescence. These data show that vacuole loss is associated with high levels of translocation of pVEC.
Figure 2.
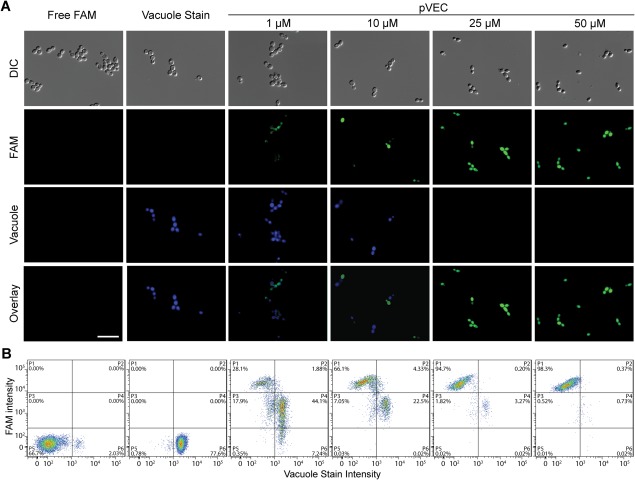
Intracellular distribution of pVEC in C. albicans at different peptide concentrations. (A) DIC and fluorescence microscopy images showing location of FAM‐labeled pVEC and location of vacuoles in cells. (B) Flow cytometry data illustrating shift of FAM and vacuolar fluorescence. Cells were incubated with serial dilutions of pVEC (1–50 µM) at 30°C for 1 h and treated with trypsin to remove surface‐bound peptide. CellTracker Blue vacuolar stain was added, and the samples were incubated at room temperature for 10 min prior to analysis. For (A), scale bar = 10 µm.
Based on the results for pVEC, we used flow cytometry to evaluate the subcellular localization in C. albicans and C. glabrata for the peptides with significant translocation (Fig. 3). Subcellular vacuole localization was observed for each of the CPPs, suggesting vacuoles are generally involved in the translocation mechanism of CPPs for fungal cells. However, the proportion of FAM‐positive cells that exhibited vacuole localization was lower for some peptides (e.g. pVEC, penetratin) compared with other peptides (e.g. SynB, MPG, (KFF)3K) (Supporting Information Fig. S2), indicating differences in translocation mechanisms and/or trafficking amongst the peptides.
Figure 3.
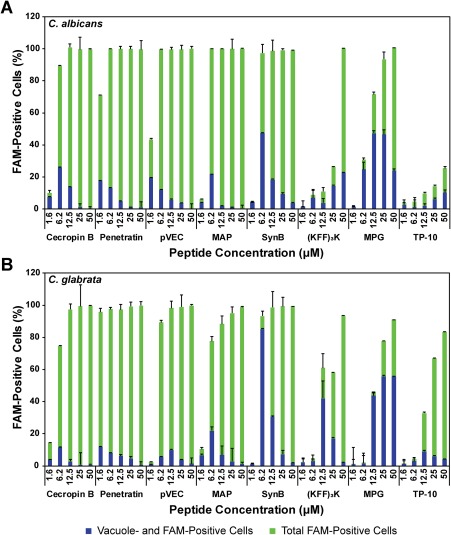
Quantification of cellular location of CPPs in (A) C. albicans and (B) C. glabrata. Cells were incubated with serial dilutions of peptide (1–50 µM) at 30°C for 1 h, washed with trypsin, and incubated with CellTracker Blue vacuolar stain at room temperature for 10 min. Flow cytometry data were collected for FAM (peptide) fluorescence and vacuolar stain fluorescence. The percentage of cells with FAM fluorescence and with both FAM and vacuolar fluorescence were quantified. Error bars represent the standard error of the mean for three separate experiments (N = 3).
Mechanisms for translocation into fungal cells
Our localization experiments revealed the important relationship between vacuoles and translocation. Previous work with yeast vacuoles suggested vacuoles are involved in endocytosis,28, 37 so we next studied endocytosis of the CPPs. To explore whether the mechanisms of translocation in fungal cells involve endotytosis, we first studied the translocation process in C. albicans under conditions that inhibit ATP synthesis. Most ATP‐dependent processes, including endocytosis, are limited at 4°C.38 Sodium azide (NaN3) also inhibits energy‐dependent endocytosis by inhibiting the function of cytochrome‐c‐oxidase for ATP synthesis.39, 40 We observed a significant reduction in translocation for six of the eight peptides (cecropin B, MAP, SynB, MPG, (KFF)3K, and TP‐10) in the presence of 25 mM NaN3 and at low temperature (Fig. 4), suggesting these peptides utilize energy‐dependent endocytosis as their translocation mechanisms. pVEC and penetratin were not highly affected by the addition of NaN3 or the lower temperature, suggesting an energy‐independent translocation process.
Figure 4.
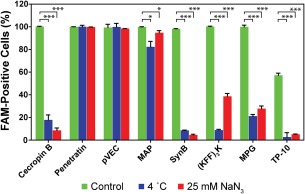
CPP translocation into C. albicans under conditions that inhibit energy‐dependent endocytosis. Cells were incubated with peptides (10 µM) for 1 h. Control samples were incubated at 30°C. Endocytosis was inhibited by adding 25 mM NaN3 or by changing the incubation temperature to 4°C. The percentage of cells exhibiting FAM fluorescence was quantified by flow cytometry. Error bars represent the standard error of the mean for three separate experiments (N = 3). Statistical significance was analyzed with a one‐way ANOVA test (α = 0.1), and the number of asterisks indicates the level of significance (* for P ≤ 0.01 and *** for P ≤ 0.0001).
Membrane destabilization is also involved in the translocation mechanism for some CPPs.14, 15, 41 To evaluate whether membrane destabilization plays a role in translocation of the peptides in our study, we used propidium iodide (PI) to identify pore formation in the membranes during CPP translocation into C. albicans and C. glabrata. Cells are normally impermeable to PI, but PI fluorescence can be detected in cells with destabilized membranes, which typically represents cells losing viability.42 Using fluorescence microscopy, we observed that pVEC, penetratin, MAP, and cecropin B contributed to membrane permeabilization of C. albicans at a moderate concentration (10 µM) [Fig. 5(A)]. We confirmed these results by flow cytometry [Fig. 5(B,C)] and found almost all cells incubated with these peptides were both PI‐ and FAM‐positive, indicating translocation of these peptides is correlated to cell permeabilization. In contrast, (KFF)3K, SynB, and MPG showed FAM fluorescence with little to no PI fluorescence, suggesting these CPPs did not lead to general defects in membrane integrity at this concentration.
Figure 5.
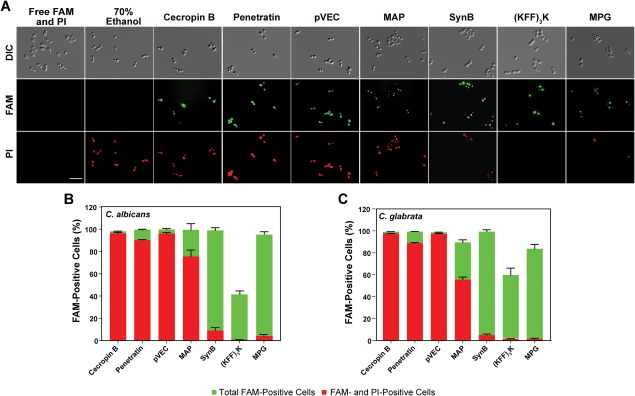
Effect of CPPs on integrity of cell membrane. (A) DIC and fluorescence microscopy images showing CPP translocation and PI uptake in C. albicans. (B) Flow cytometry data indicating the percentage of cells with CPP translocation (FAM fluorescence) and with PI uptake in C. albicans. (C) Flow cytometry data indicating the percentage of cells with CPP translocation and with PI uptake in C. glabrata. Following incubation with FAM‐labeled CPPs (10 µM), cells were treated with trypsin and then incubated with PI for 1 min immediately prior to imaging or flow cytometry. For (A), scale bar = 10 µm. For (B) and (C), error bars represent the standard error of the mean for three separate experiments (N = 3).
Relationship between subcelluar localization and translocation
Our results for subcellular localization, membrane stability, and endocytic inhibition together provide insight into the translocation mechanisms for the CPPs in fungal cells (Table 2). The combination of significant vacuolar localization, lack of strong membane destabilization, and inhibition of translocation by NaN3 and low temperature suggests endocytosis is involved in the translocation of a peptide; we observed these characteristics for the translocation of SynB and (KFF)3K. Stronger cytosolic localization along with high PI permeability, as we observed at high concentrations for cecropin B, pVEC, penetratin, and MAP, indicates the possibility of direct translocation into the cytosol, though endocytosis might still be possible at concentrations resulting in vacular localization.
Table 2.
Summary of subcellular localization and potential translocation mechanism of CPPs
| Cecropin B | Penetratin | pVEC | MAP | SynB | (KFF)3K | MPG | TP‐10 | |
|---|---|---|---|---|---|---|---|---|
| Vacuolar localization | Low | Low | Low | Low | High | High | High | High |
| PI permeability a | ++ | ++ | ++ | ++ | + | – | – | – |
| NaN3/4°C endocytosis inhibition | Yes | No | No | Yes | Yes | Yes | Yes | Yes |
| Potential mechanism b | E/D | M/D | E/M | E/D | E | E | E | E |
| Previously reported mechanism b | E/D43 | E/D14 | M17 | D44 | E14 | n/a | D41 | D44 |
Strong PI permeability is indicated by ++, moderate permeability by +, and no significant permeability by –.
Translocation mechanisms are as follows: endocytosis (E), macropinocytosis (M), direct translocation/pore formation (D), or no data available (n/a).
Interestingly, the data for several of the peptides indicate multiple mechanisms may be involved in their translocation into fungal cells. For example, the NaN3 and low temperature data for pVEC suggest an energy‐independent mechanism, such as macropinocytosis or direct pore formation, which is consistent with the PI data. However, the microscopy images and flow cytometry data (Fig. 2) also indicate vacuolar trafficking, which is more consistent with energy‐dependent endocytosis. The possibility of multiple mechanisms is consistent with prior work with pVEC in mammalian cells. Elmquist et al.45 found that a clathrin‐dependent endocytic pathway is involved in the translocation of pVEC, yet they still observed modest translocation at low temperatures to also implicate non‐endocytic pathways. Likewise, MAP and cecropin B have high PI permeability consistent with direct translocation, yet they exhibit the energy‐dependent translocation and vacuolar trafficking consistent with endocytosis, which could indicate multiple mechanisms are also involved in translocation of these peptides.
For some peptides, our results implicate a translocation mechanism in Candida cells that differs from previous studies with mammalian cells. MPG and TP‐10 were previously suggested to destabilize and make pores in the membrane of HeLa and melanoma cells, respectively.41, 44 However, our PI data, NaN3 assay, and localization images indicate an endocytic process is involved for Candida cells. The difference in structure of the cell types may help explain this discrepancy. Hallbrink et al.44 and Simeoni et al.41 showed the translocation and pore formation of MPG, Pep‐1, and TP‐10 were associated with the close interaction between peptides and cell membranes. However, the additional barrier of the cell wall in fungal cells could interfere with this close interaction. This idea is consistent with our observation that Pep‐1 associated with the surface and did not translocate into fungal cells (Supporting Information Fig. S1). Additionally, differences in cell‐surface properties and proteins could change the overall structure and behavior of CPPs. For example, Candida cells could have a receptor that recognizes MPG and TP‐10, causing a shift in the translocation mechanism in fungal cells to endocytosis, even though membrane destabilization occurs in mammalian cells.
Toxicity of CPPs toward Candida cells
Yeast vacuoles are necessary for cells to maintain homeostasis, so the loss of vacuoles due to CPPs may lead to toxicity toward fungal cells. To examine the toxicity of CPPs toward C. albicans and C. glabrata, we incubated the cells with serial dilutions of the peptides. At high concentrations of peptide, we observed significant toxicity toward both types of Candida cells for each peptide that exhibited substantial translocation (Fig. 6), with minimum inhibitory concentrations (MIC50s) ranging from 1 µM to 50 µM (Table 3). Little to no antifungal activity could be detected for CPPs that exhibited very low levels of translocation (i.e., Pep‐1, PAF26, and hCT in both species and TP‐10 in C. albicans) (Fig. 6), and MIC50s for these peptides were above 50 µM (Table 3). Incubation with free FAM did not lead to any viability loss of Candida cells compared to the cells incubated with only Na2HPO4 buffer (data not shown). Interestingly, the CPPs pVEC, penetratin, and MAP exhibited even stronger antifungal activity than the well‐known antimicrobial peptide cecropin B. For many of the peptides, the loss of viability could be due to the vacuole loss we observed in our microscopy and flow cytometry data, since vacuoles are essential for fungal cells in controlling the cellular osmotic pressure, salt balance, and pH.28, 46 For the peptides that exhibited substantial PI permeability (pVEC, penetratin, and MAP), the loss of viability could also be due to membrane permeabilization, though it is not clear from our data whether the permeabilization is a cause or an effect of cell death. Between the two Candida species, C. glabrata tended to be less sensitive to toxicity of the peptides compared to C. albicans, which is consistent with previous research of antimicrobial peptides and other antifungal agents.47, 48, 49 This difference in toxicity of CPPs and antimicrobial peptides between Candida species is not yet fully understood, though work has suggested that it may be due to the different compositions of the cell membranes and the presence of different membrane proteins.47, 49
Figure 6.
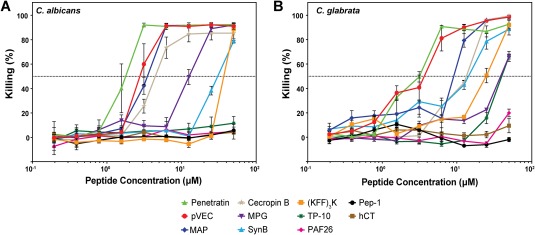
Toxicity of CPPs toward (A) C. albicans and (B) C. glabrata. Cells were incubated with serial dilutions of peptides (0.2–50 µM) for 1 h at 30°C. Samples were diluted, mixed with YPD medium, and incubated at 30°C for 16 h. Optical density (OD600) of the cultures was measured and converted to killing percentage. Error bars represent the standard error of the mean for three separate experiments (N = 3). Dotted line represents 50% killing, which was used to determine the MIC50 (Table III).
Table 3.
Antimicrobial activity of peptides
| Minimum Inhibitory Concentration (MIC50, µM)a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cecropin B | Penetratin | pVEC | MAP | SynB | (KFF)3K | MPG | TP‐10 | PAF26 | Pep‐1 | hCT | |
| C. albicans | 4 | 1 | 2 | 4 | 50 | 50 | 10 | > 50 | > 50 | > 50 | > 50 |
| C. glabrata | 25 | 2 | 4 | 8 | 25 | 50 | 50 | 32 | > 50 | > 50 | > 50 |
MIC50 values are defined as the minimum concentration of peptide required to reduce growth of cells by 50%. The highest concentration tested was 50 µM.
The toxicity of CPPs toward Candida pathogens could prove to be a positive feature of peptides targeted to fungal pathogens, as one motivation for studying CPPs in fungal cells is to use them to deliver antifungal molecules, and the cytotoxicity of the CPP vehicles would potentially increase the therapeutic effects of the cargo being delivered. To function as a delivery vehicle for antifungal therapeutics, toxicity of the CPPs to host cells would need to be minimal. Previous studies suggest TP‐10, penetratin, MAP, and cecropin B negatively affect mammalian cell viability,33, 50, 51, 52, 53 which would complicate their use in delivery of antifungal therapeutics. In contrast, pVEC and MPG did not significantly reduce viability of mammalian cells,33, 54 and we observed both strong translocation and strong toxicity toward fungal cells, suggesting these peptides would be promising candidates for delivering antifungal agents or could potentially serve as antifungal agents themselves.
Although toxicity of CPPs may be desirable in the case of delivering antifungal molecules, CPPs also have potential in delivering non‐toxic bioactive cargo to study the effect of the cargo on cellular function.55, 56 In this case, toxicity from CPP vehicles would need to be as low as possible. While most of the peptides we studied would not be suitable for this type of application, SynB is the exception. At a concentration of 10 μM, essentially 100% of Candida cells were positive for peptide translocation [Fig. 1(C)], while the same concentration led to a loss of viability for only 10% of C. albicans and 5% of C. glabrata cells (Fig. 6). SynB also has a low level of toxicity toward mammalian cells,57 further increasing its potential applications in studying the biology of Candida pathogens.
Our data for the toxicity of the CPPs highlight the importance of considering the goal of cargo delivery in selecting a CPP. The level of CPP translocation, along with the toxicity toward the target cells and any other cells that may be present, must be evaluated to identify a CPP with the desired delivery and toxicity profiles.
Conclusion
We have identified a number of CPPs that are able to translocate into the fungal pathogens C. albicans and C. glabrata. We also explored the intracellular distribution of the CPPs following translocation into fungal cells and found that vacuoles play a significant role in CPP trafficking. Our results suggest that translocation of CPPs into fungal cells may involve multiple mechanisms and that these mechanisms may lead to the toxicity observed for some peptides. Further work to explore the translocation mechanisms of CPPs could improve their efficacy and toxicity profiles to make CPPs viable therapeutic delivery agents. By increasing the translocation of bioactive cargo using CPPs, effectiveness of antifungal agents could be improved and new classes of molecules that currently lack the ability to cross cell membranes could be explored as antifungal agents.
Materials and Methods
Peptides
The peptides listed in Table 1 were commercially synthesized at >95% purity with an N‐terminal 5‐carboxyfluorescein (FAM) (Genscript, Piscataway, NJ). The lyophilized peptides were reconstituted in sterile, ultrapure H2O and diluted to a final concentration of 10 mM Na2HPO4 buffer.
Strains and culture conditions
C. albicans strain SC5314 and C. glabrata strain ATCC2001 were purchased from American Type Culture Collection (ATCC, Manassas, VA). Candida cells were inoculated from yeast‐peptone‐dextrose (YPD) agar plates (1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose, and 2% w/v agar) into 5 mL of liquid YPD medium (1% w/v yeast extract, 2% w/v peptone, and 2% w/v glucose) and grown overnight at 30°C while shaking at 230 rpm. The cells in the overnight culture were subcultured into 5 mL of fresh YPD medium at OD600=0.1 (equivalent to ∼2 × 106 CFU/mL). The culture was then grown at 30°C to OD600 = 0.5 (equivalent to ∼1 × 107 CFU/mL) while shaking. Cells were harvested by centrifugation at 4,000 × g for 10 min and washed twice with 10 mM Na2HPO4 buffer before use in downstream assays.
Fluorescence imaging
For each peptide, 100 µL of peptide solution (2–100 µM, depending on the experiment) was prepared in 10 mM Na2HPO4 buffer, mixed with 100 µL of cell suspension containing 5 × 105 cells in 10 mM Na2HPO4 and incubated at 30°C for 60 min. Cells were collected by centrifugation at 5000 × g for 10 min at 4°C and washed once with 10 mM Na2HPO4. The cell pellet was then incubated with 200 µL of 0.025% trypsin (Invitrogen, Waltham, MA) at 37°C for 10 min to remove surface‐bound peptide.30 Cells were collected and washed again with 10 mM Na2HPO4. For vacuole staining, 1 µM of CellTracker Blue CMAC (Invitrogen Molecular Probes, Waltham, MA) was added into the washed cell suspension and incubated at ambient temperature for 10 min. To prepare the cells for imaging, cells were collected and resuspended in 5 µL of 10 mM Na2HPO4. The suspension was transferred to a glass slide and imaged using an Olympus IX83 fluorescence microscopy system (Olympus, Center Valley, PA). PI (1 mg·mL−1; Invitrogen, Waltham, MA) was added immediately before imaging as needed to examine the membrane integrity. DIC, GFP fluorescence, vacuolar stain fluorescence, and/or PI fluorescence images were taken using the automatic process manager of the CellSens Dimension software (Olympus), and images were analyzed using NIH ImageJ software.58
Quantification of translocation
To prepare the Candida cells for quantification by flow cytometry, procedures analogous to those for microscopy were followed. Fresh cells were incubated with dilutions of each peptide (1–50 µM) and then treated with trypsin. After washing the cells with 10 mM Na2HPO4, the cells were resuspended in 150 µL of 10 mM Na2HPO4. Cell suspensions were analyzed for FAM and PI fluorescence using a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA). Only single cells were selected for analysis, and the analysis was performed using FlowJo software (FLOWJO Inc., Ashland, OR).
Antifungal activity assay
In order to assess the antimicrobial activities of the peptides, a microdilution assay was performed. After subculturing cells and growing the culture to OD600 = 0.5, a 5 × 105 cells/mL cell suspension was prepared in 10 mM Na2HPO4. Serial dilutions (20 µL) of the peptides were prepared at 0.4–100 µM in 96‐well plates. A control containing 100 µM of free FAM in 10 mM Na2HPO4 buffer was also added to the plate. The cell suspension (20 µL) was then added into each well, and the plate was incubated at 30°C with vigorous shaking for 60 min. Treated cells were diluted 20‐fold in 10 mM Na2HPO4 buffer, and 100 µL of diluted cell suspension was added into 100 µL fresh YPD medium in a new 96‐well plate. Plates were incubated at 30°C with vigorous shaking for 16 h, and the OD600 of the wells was measured using a 96‐well plate reader (BioTek, Winooski, VT). The percentage of killing was calculated from
| (1) |
Minimal inhibitory concentrations were determined as the minimum concentration resulting in a 50% reduction in cell viability (MIC50).
Supporting information
Supporting Information.
Acknowledgments
The authors thank Dr. Xiangbin Zeng and Prof. Christopher Jewell for assistance with flow cytometry training and data processing. This research was supported by the National Science Foundation (CBET 1511718).
References
- 1. Richardson JP, Moyes DL (2015) Adaptive immune responses to Candida albicans infection. Virulence 6:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention (2013) Antibiotic resistance threats in the United States, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/. Accessed April 2017.
- 3. Mae M, Langel U (2006) Cell‐penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr Opin Pharmacol 6:509–514. [DOI] [PubMed] [Google Scholar]
- 4. Stetsenko DA, Gait MJ (2000) Efficient conjugation of peptides to oligonucleotides by “native ligation”. J Org Chem 65:4900–4908. [DOI] [PubMed] [Google Scholar]
- 5. El‐Andaloussi S, Johansson H, Magnusdottir A, Jarver P, Lundberg P, Langel U (2005) TP10, a delivery vector for decoy oligonucleotides targeting the Myc protein. J Control Release 110:189–201. [DOI] [PubMed] [Google Scholar]
- 6. Muratovska A, Eccles MR (2004) Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett 558:63–68. [DOI] [PubMed] [Google Scholar]
- 7. Gong Z, Walls MT, Karley AN, Karlsson AJ (2016) Effect of a flexible linker on recombinant expression of cell‐penetrating peptide fusion proteins and their translocation into fungal cells. Mol Biotechnol 58:838–849. [DOI] [PubMed] [Google Scholar]
- 8. Rajarao GK, Nekhotiaeva N, Good L (2002) Peptide‐mediated delivery of green fluorescent protein into yeasts and bacteria. FEMS Microbiol Lett 215:267–272. [DOI] [PubMed] [Google Scholar]
- 9. Theisen DM, Pongratz C, Wiegmann K, Rivero F, Krut O, Kronke M (2006) Targeting of HIV‐1 Tat traffic and function by transduction‐competent single chain antibodies. Vaccine 24:3127–3136. [DOI] [PubMed] [Google Scholar]
- 10. Kameyama S, Horie M, Kikuchi T, Omura T, Takeuchi T, Nakase I, Sugiura Y, Futaki S (2006) Effects of cell‐permeating peptide binding on the distribution of 125I‐labeled Fab fragment in rats. Bioconjug Chem 17:597–602. [DOI] [PubMed] [Google Scholar]
- 11. Oehlke J, Scheller A, Wiesner B, Krause E, Beyermann M, Klauschenz E, Melzig M, Bienert M (1998) Cellular uptake of an alpha‐helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior non‐endocytically. Biochim Biophysic Acta 1414:127–139. [DOI] [PubMed] [Google Scholar]
- 12. Soomets U, Lindgren M, Gallet X, Hallbrink M, Elmquist A, Balaspiri L, Zorko M, Pooga M, Brasseur R, Langel U (2000) Deletion analogues of transportan. Biochim Biophys Acta 1467:165–176. [DOI] [PubMed] [Google Scholar]
- 13. Lopez‐Garcia B, Perez‐Paya E, Marcos JF (2002) Identification of novel hexapeptides bioactive against phytopathogenic fungi through screening of a synthetic peptide combinatorial library. Appl Environ Microbiol 68:2453–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Drin G, Cottin S, Blanc E, Rees AR, Temsamani J (2003) Studies on the internalization mechanism of cationic cell‐penetrating peptides. J Biol Chem 278:31192–31201. [DOI] [PubMed] [Google Scholar]
- 15. Munoz A, Marcos JF, Read ND (2012) Concentration‐dependent mechanisms of cell penetration and killing by the de novo designed antifungal hexapeptide PAF26. Mol Microbiol 85:89–106. [DOI] [PubMed] [Google Scholar]
- 16. Mager I, Eiriksdottir E, Langel K, El Andaloussi S, Langel U (2010) Assessing the uptake kinetics and internalization mechanisms of cell‐penetrating peptides using a quenched fluorescence assay. Biochim Biophys Acta 1798:338–343. [DOI] [PubMed] [Google Scholar]
- 17. Elmquist A, Lindgren M, Bartfai T, Langel U (2001) VE‐cadherin‐derived cell‐penetrating peptide, pVEC, with carrier functions. Exp Cell Res 269:237–244. [DOI] [PubMed] [Google Scholar]
- 18. Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A (1996) Cell internalization of the third helix of the antennapedia homeodomain is receptor‐independent. J Biol Chem 271:18188–18193. [DOI] [PubMed] [Google Scholar]
- 19. Fischer R, Fotin‐Mleczek M, Hufnagel H, Brock R (2005) Break on through to the other side‐biophysics and cell biology shed light on cell‐penetrating peptides. ChemBioChem 6:2126–2142. [DOI] [PubMed] [Google Scholar]
- 20. Morris MC, Vidal P, Chaloin L, Heitz F, Divita G (1997) A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res 25:2730–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morris MC, Depollier J, Mery J, Heitz F, Divita G (2001) A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol 19:1173–1176. [DOI] [PubMed] [Google Scholar]
- 22. Vaara M, Porro M (1997) Group of peptides that act synergistically with hydrophobic antibiotics against gram‐negative enteric bacteria. Antimicrob Agents Chemother 41:496–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holm T, Netzereab S, Hansen M, Langel U, Hallbrink M (2005) Uptake of cell‐penetrating peptides in yeasts. FEBS Lett 579:5217–5222. [DOI] [PubMed] [Google Scholar]
- 24. Parenteau J, Klinck R, Good L, Langel U, Wellinger RJ, Elela SA (2005) Free uptake of cell‐penetrating peptides by fission yeast. FEBS Lett 579:4873–4878. [DOI] [PubMed] [Google Scholar]
- 25. Palm C, Netzereab S, Hallbrink M (2006) Quantitatively determined uptake of cell‐penetrating peptides in non‐mammalian cells with an evaluation of degradation and antimicrobial effects. Peptides 27:1710–1716. [DOI] [PubMed] [Google Scholar]
- 26. Munoz A, Harries E, Contreras‐Valenzuela A, Carmona L, Read ND, Marcos JF (2013) Two functional motifs define the interaction, internalization and toxicity of the cell‐penetrating antifungal peptide PAF26 on fungal cells. PLoS One 8:e54813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bowman SM, Free SJ (2006) The structure and synthesis of the fungal cell wall. BioEssays 28:799–808. [DOI] [PubMed] [Google Scholar]
- 28. Klionsky DJ, Herman PK, Emr SD (1990) The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 54:266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paluchowska P, Tokarczyk M, Bogusz B, Skiba I, Budak A (2014) Molecular epidemiology of Candida albicans and Candida glabrata strains isolated from intensive care unit patients in Poland. Mem Inst Oswaldo Cruz 109:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B (2003) Cell‐penetrating peptides. A reevaluation of the mechanism of cellular uptake. J Biol Chem 278:585–590. [DOI] [PubMed] [Google Scholar]
- 31. Meiller TF, Hube B, Schild L, Shirtliff ME, Scheper MA, Winkler R, Ton A, Jabra‐Rizk MA (2009) A novel immune evasion strategy of Candida albicans: proteolytic cleavage of a salivary antimicrobial peptide. PLoS One 4:e5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sikora M, Dabkowska M, Swoboda‐Kopec E, Jarzynka S, Netsvyetayeva I, Jaworska‐Zaremba M, Pertkiewicz M, Mlynarczyk G (2011) Differences in proteolytic activity and gene profiles of fungal strains isolated from the total parenteral nutrition patients. Folia Microbiol 56:143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saar K, Lindgren M, Hansen M, Eiriksdottir E, Jiang Y, Rosenthal‐Aizman K, Sassian M, Langel U (2005) Cell‐penetrating peptides: a comparative membrane toxicity study. Anal Biochem 345:55–65. [DOI] [PubMed] [Google Scholar]
- 34. Lorenz JN, Gruenstein E (1999) A simple, nonradioactive method for evaluating single‐nephron filtration rate using FITC‐inulin. Am J Physiol 276:F172–F177. [DOI] [PubMed] [Google Scholar]
- 35. Preston RA, Murphy RF, Jones EW (1989) Assay of vacuolar pH in yeast and identification of acidification‐defective mutants. Proc Natl Acad Sci U S A 86:7027–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mochon AB, Liu H (2008) The antimicrobial peptide histatin‐5 causes a spatially restricted disruption on the Candida albicans surface, allowing rapid entry of the peptide into the cytoplasm. PLoS Pathog 4:e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Kaplan J (2004) A mitochondrial‐vacuolar signaling pathway in yeast that affects iron and copper metabolism. J Biol Chem 279:33653–33661. [DOI] [PubMed] [Google Scholar]
- 38. Jiao CY, Delaroche D, Burlina F, Alves ID, Chassaing G, Sagan S (2009) Translocation and endocytosis for cell‐penetrating peptide internalization. J Biol Chem 284:33957–33965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lichstein HC, Soule MH (1944) Studies of the effect of sodium azide on microbic growth and respiration: I. The action of sodium azide on microbic growth. J Bacteriol 47:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Noumi T, Maeda M, Futai M (1987) Mode of inhibition of sodium azide on H+‐ATPase of Escherichia coli . FEBS Lett 213:381–384. [DOI] [PubMed] [Google Scholar]
- 41. Simeoni F, Morris MC, Heitz F, Divita G (2003) Insight into the mechanism of the peptide‐based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res 31:2717–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Halder S, Yadav KK, Sarkar R, Mukherjee S, Saha P, Haldar S, Karmakar S, Sen T (2015) Alteration of Zeta potential and membrane permeability in bacteria: a study with cationic agents. Springerplus 4:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Epand RM, Vogel HJ (1999) Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophysic Acta 1462:11–28. [DOI] [PubMed] [Google Scholar]
- 44. Hallbrink M, Floren A, Elmquist A, Pooga M, Bartfai T, Langel U (2001) Cargo delivery kinetics of cell‐penetrating peptides. Biochim Biophys Acta 1515:101–109. [DOI] [PubMed] [Google Scholar]
- 45. Elmquist A, Hansen M, Langel U (2006) Structure‐activity relationship study of the cell‐penetrating peptide pVEC. Biochim Biophys Acta 1758:721–729. [DOI] [PubMed] [Google Scholar]
- 46. Shirahama K, Yazaki Y, Sakano K, Wada Y, Ohsumi Y (1996) Vacuolar function in the phosphate homeostasis of the yeast Saccharomyces cerevisiae . Plant Cell Physiol 37:1090–1093. [DOI] [PubMed] [Google Scholar]
- 47. Helmerhorst EJ, Venuleo C, Beri A, Oppenheim FG (2005) Candida glabrata is unusual with respect to its resistance to cationic antifungal proteins. Yeast 22:705–714. [DOI] [PubMed] [Google Scholar]
- 48. Pfaller MA, Messer SA, Hollis RJ, Jones RN, Diekema DJ (2002) In vitro activities of ravuconazole and voriconazole compared with those of four approved systemic antifungal agents against 6,970 clinical isolates of Candida spp. Antimicrob Agents Chemother 46:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raman N, Lee MR, Lynn DM, Palecek SP (2015) Antifungal activity of 14‐helical beta‐peptides against planktonic cells and biofilms of Candida species. Pharmaceuticals (Basel) 8:483–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. El‐Andaloussi S, Jarver P, Johansson HJ, Langel U (2007) Cargo‐dependent cytotoxicity and delivery efficacy of cell‐penetrating peptides: a comparative study. Biochem J 407:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dinca A, Chien WM, Chin MT (2016) Intracellular delivery of proteins with cell‐penetrating peptides for therapeutic uses in human disease. Int J Mol Sci 17:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Suttmann H, Retz M, Paulsen F, Harder J, Zwergel U, Kamradt J, Wullich B, Unteregger G, Stockle M, Lehmann J (2008) Antimicrobial peptides of the cecropin‐family show potent antitumor activity against bladder cancer cells. BMC Urol 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bacalum M, Radu M (2015) Cationic antimicrobial peptides cytotoxicity on mammalian cells: an analysis using therapeutic index integrative concept. Int. J Peptide Res Therapeut 21:47–55. [Google Scholar]
- 54. Mussbach F, Pietrucha R, Schaefer B, Reissmann S (2011) Internalization of nucleoside phosphates into live cells by complex formation with different CPPs and JBS‐nucleoducin. Methods Mol Biol 683:375–389. [DOI] [PubMed] [Google Scholar]
- 55. Alhakamy NA, Nigatu AS, Berkland CJ, Ramsey JD (2013) Noncovalently associated cell‐penetrating peptides for gene delivery applications. Therap Deliv 4:741–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang QY, Lai LH, Shen J, Wang QQ, Xu FJ, Tang GP (2011) Gene delivery to tumor cells by cationic polymeric nanovectors coupled to folic acid and the cell‐penetrating peptide octaarginine. Biomaterials 32:7253–7262. [DOI] [PubMed] [Google Scholar]
- 57. Tian XH, Wei F, Wang TX, Wang P, Lin XN, Wang J, Wang D, Ren L (2012) In vitro and in vivo studies on gelatin‐siloxane nanoparticles conjugated with SynB peptide to increase drug delivery to the brain. Int J Nanomedicine 7:1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Meth 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.


