Abstract
The expression of phase-II detoxification and antioxidant enzymes is governed by a cis-acting regulatory element named the antioxidant response element (ARE). ARE-containing genes are regulated by the nuclear factor erythroid-2-related factor 2 (Nrf2), a member of the Cap’n’Collar basic-leucine-zipper family of transcription factors. ARE-regulated genes are preferentially activated in astrocytes, which consequently have more efficient detoxification and antioxidant defences than neurons. Astrocytes closely interact with neurons to provide structural, metabolic and trophic support, as well as actively participating in the modulation of neuronal excitability and neurotransmission. Therefore, functional alterations in astrocytes can shape the interaction with surrounding cells, such as neurons and microglia. Activation of Nrf2 in astrocytes protects neurons from a wide array of insults in different in vitro and in vivo paradigms, stressing the role of astrocytes in determining the vulnerability of neurons to noxious stimuli. Here, we review the current data supporting Nrf2 activation in astrocytes as a viable therapeutic approach, not only in acute neuronal damage, but also in chronic neurodegeneration related to oxidative stress.
In mammalian cells, reactive oxygen species (ROS) and reactive nitrogen species (RNS) are continuously generated as a consequence of normal cell metabolism. The superoxide radical is formed by processes mediated by enzymes such as NAD(P)H oxidases and xanthine oxidase or nonenzymatically, by redox-reactive compounds such as the semiubiquinone compound of the mitochondrial electron transport chain. Superoxide dismutases (SODs) convert superoxide into hydrogen peroxide and oxygen. In the presence of reduced transition metals (e.g. ferrous or cuprous ions), hydrogen peroxide can be converted into the highly reactive hydroxyl radical, which is capable of causing several oxidation-mediated modifications in biomolecules (Ref. 1). The nitric oxide radical is produced in higher eukaryotes by the oxidation of L-arginine in a reaction catalysed by nitric oxide synthases (Ref. 2). Nitric oxide itself is not particularly toxic in vivo, but it can react with superoxide to form the powerful oxidant peroxynitrite (Ref. 3). Oxidative stress arises when the rate of ROS and RNS production exceeds their rate of clearance by antioxidant compounds and enzymes (Ref. 4). Owing to their high oxygen consumption and high lipid content, neural tissues are particularly sensitive to oxidative stress. It is therefore perhaps unsurprising that oxidative stress has been implicated in the pathogenesis of several neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS), Alzheimer disease (AD), Parkinson disease (PD) and Huntington disease (HD) (Refs 5, 6, 7, 8).
Phase II detoxifying and antioxidant enzymes are the primary means by which neural cells protect themselves from toxic ROS and RNS, and electrophiles (compounds with an electron-deficient center). The induction of such enzymes depends almost exclusively on de novo gene transcription, which is governed by the transcription factor nuclear factor erythroid-2-related factor 2 (Nrf2, encoded by NFE2L2) (Refs 9, 10), which acts on the electrophile-response element or antioxidant-response element (ARE; Ref. 11). Nrf2 activation protects neurons and astrocytes from oxidative damage in many different acute paradigms, and it is a promising therapeutic target in chronic neurodegeneration.
Nrf2 activation and regulation
Nrf2 is a redox-sensitive transcription factor and a member of a protein family characterised by the presence of a conserved basic region leucine zipper (bZip) dimerisation domain, which was first identified in the Drosophila transcription factor Cap’n’Collar [CNC (Ref. 12)]. The CNC–bZip protein family includes the p45 subunit of the nuclear factor erythroid 2 (NF-E2), which was originally identified as an erythroid-restricted DNA-binding protein that recognises regulatory elements in the globin gene (Refs 13,14). Other CNC–bZip family members are Nrf1 (NFE2L1), Nrf3 (NFE2L3), BACH1 and BACH2. CNC–bZip transcription factors must heterodimerise with members of the Maf proto-oncogene family in order to bind to regulatory elements in the DNA (Refs 15, 16).
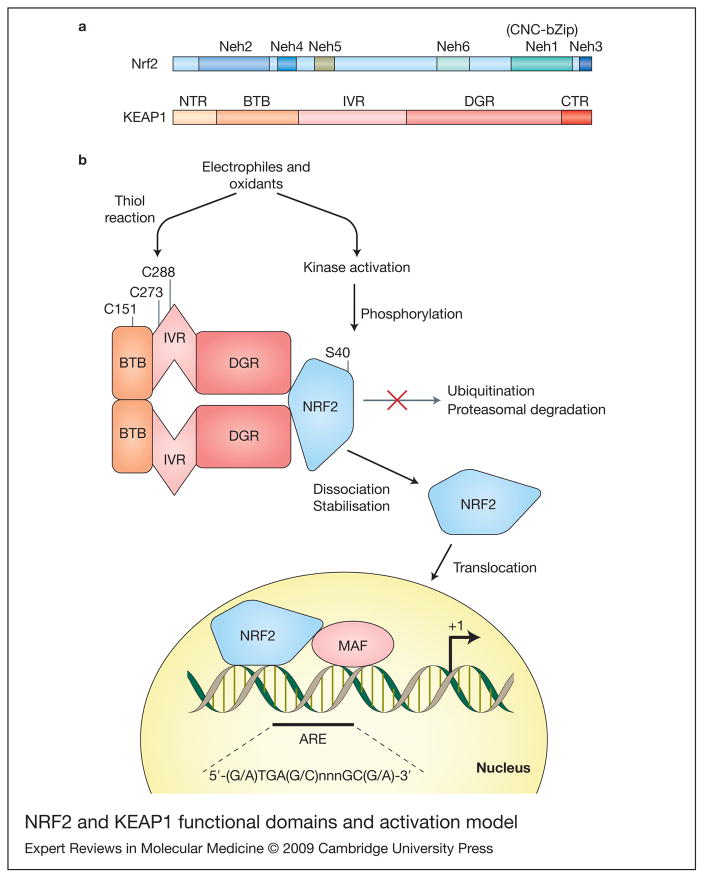
Comparison of the human Nrf2 (Ref. 9) and the chicken homologue ECH (erythroid-derived CNC homology protein) (Ref. 10), led to the identification of Nrf2–ECH homology domains (Neh; Fig. 1). One of these domains (Neh2) was found to mediate the association of Nrf2 with the cytoplasmic protein KEAP1 [kelch-like ECH-associated protein 1 (Ref. 17)]. When the murine Keap1 gene was disrupted, constitutive activation of Nrf2 and its target genes caused juvenile lethality as a result of hyperkeratotic lesions in the esophagus and forestomach. In vivo evidence of an interaction between Nrf2 and KEAP1 was obtained when the lethality of the Keap1-null mutation was reversed by the simultaneous knockout of Nrf2 (Ref. 18). The mechanism of this interaction and that of Nrf2 transactivation remains a topic of active research. It has been postulated that under basal conditions Nrf2 remains in the cytoplasm associated with the actin cytoskeleton through Keap1 (Ref. 17). In unstressed conditions, Nrf2 is rapidly degraded and displays a half-life of between 10 and 40 min (Refs 17, 19, 20, 21). KEAP1 has been shown to function as an adaptor for the Cullin-3-based E3 ligase in the cytoplasm, where it targets Nrf2 for ubiquitin-mediated proteasomal degradation (Refs 22–24). In the presence of ARE inducers, the Nrf2–KEAP1 interaction is suppressed, and Nrf2 translocates to the nucleus, where it dimerises with small Maf proteins to increase the transcription rate of ARE-driven genes (Fig. 1).
Figure 1. Nrf2 and KEAP1 functional domains and activation model.
(a) Diagram representing the functional domains of Nrf2 and Keap1. Nrf2 has six highly conserved regions named Neh1–Neh6 (Nrf2-ECH homology). The Neh1 contains the CNC homology region and basic-leucine zipper domain (CNC-bZip). The N-terminus (Neh2) and C-terminus (Neh3) of the proteins are also highly conserved. Keap1 binds Nrf2 at Neh2 and also the serine40 (S40) it is located in this domain. Additionally, there are two conserved acidic domains (Neh4 and Neh5) as well as a serine-rich conserved region (Neh6). KEAP1 presents two characteristic domains, the bricabrac, tramtrack and broad complex (BTB) domain and the double glycine repeat (DGR) domain. KEAP1 bridges the Cullin-3-based E3 ligase and Nrf2 using its BTB and the central intervening region (IVR) to bind Cul3 and its DGR to bind the Neh2 domain of Nrf2. Two additional regions are present in Nrf2: the N-terminal region (NTR) and the C-terminal region (CTR). (b) A model for KEAP1–Nrf2 interaction and activation. The BTB domain participates in KEAP1 dimerisation. Under basal conditions Nrf2 is continuously degraded. Electrophiles and oxidants directly modify reactive cysteines residues in KEAP1, disrupting its dimerisation or KEAP1–Cul3 interaction, and ubiquitination of Nrf2 is interrupted. Alternative activated kinases can phosphorylate Nrf2 at S40 and disrupt KEAP1–Nrf2 interaction. Nrf2 then translocates to the nucleus and increases ARE-driven transcription.
Since several electrophilic ARE inducers are able to modify sulfhydryl groups by alkylation or oxidation, and because KEAP1 contains 25 cysteines that are conserved in human, mouse and rat, it was proposed that disruption of the Nrf2–KEAP1 complex might result from the direct modification of cysteine thiols in KEAP1 (Ref. 25). These cysteine residues are highly reactive to electrophiles and, at least in cell culture, Cys151, Cys273 and Cys288 are essential for proper KEAP1 function. Upon modification, an increase in Nrf2 activity is observed (Refs 26, 27, 28, 29). In addition to direct modification of thiol groups in KEAP1, activation of several kinases in response to electrophilic or oxidative stress causes translocation of Nrf2 to the nucleus. Phosphorylation of Nrf2 at serine and threonine residues by protein kinase C, phosphatidylinositol 3-kinase, PKR-like endoplasmic reticulum kinase, JNK, ERK and p38 MAPK (MK14) has been reported to facilitate the release of Nrf2 from KEAP1 (Ref. 30).
Interestingly, it has also been proposed that KEAP1 shuttles between the nucleus and the cytoplasm, thus limiting Nrf2 activity by escorting the transcription factor out of the nucleus (Refs 31, 32, 33). However, the subcellular distribution of KEAP1 remains under investigation (Ref. 34). In general, it has become clear that the major regulatory steps in Nrf2 activation are increased protein stability (by dissociating from KEAP1) and its translocation to the nucleus prompted by post-translational modifications of Nrf2 or KEAP1. Hence, most chemical inducers increase Nrf2 protein without affecting Nrf2 mRNA levels. An exception to the above mechanism is observed with some members of the fibroblast growth factor family (FGF-1, FGF-7, FGF-10), which increased Nrf2 protein levels, at least in part, by increasing gene transcription (Refs 35, 36, and therefore might contribute to the activation of ARE-driven genes without any additional post-translational modification of Nrf2 or KEAP1.
Genes regulated by the Nrf2-ARE pathway
Nrf2 activation can prevent or reduce cellular damage associated with several types of injury in many different tissues and organs (Ref. 37). Nrf2-mediated protection depends on the expression of a battery of genes with detoxification, antioxidant and anti-inflammatory capacity. Although it is clear that the coordinated expression of this group of genes has an important cytoprotective role, two components are particularly important for the prevention of oxidative-mediated damage in nervous cells: the haem oxygenase (HO) system and a group of enzymes involved in glutathione synthesis and utilisation.
Haem oxygenase
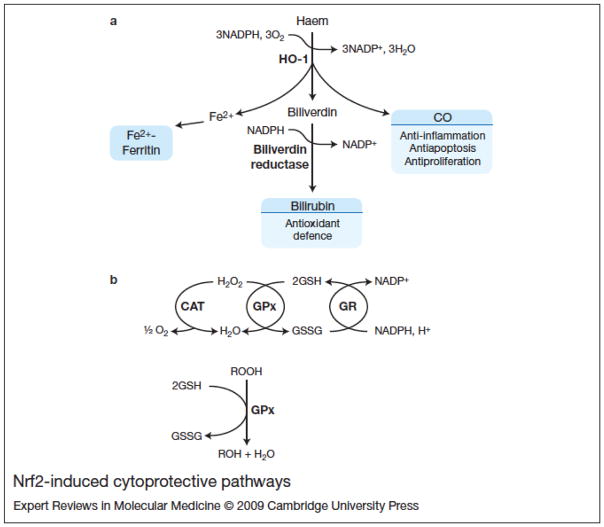
Haem oxygenase is a microsomal enzyme that, in concert with the NADPH cytochrome P450 reductase, catalyses the degradation of the haem group to yield biliverdin, iron and carbon monoxide (Refs 38, 39). Biliverdin is subsequently converted to bilirubin by biliverdin reductase (Fig. 2A). Two isoforms of haem oxygenase have been characterised: a constitutive isoform, HO-2 (HMOX2) and an inducible enzyme, HO-1 (HMOX1; Refs 40, 41). HO-2 protein is widely expressed throughout the rodent nervous system, whereas basal HO-1 expression in the normal brain is confined to small groups of scattered neurons and glial cells (Ref. 42) The induction of HO-1 is considered to counteract oxidative damage and confer cytoprotection, as suggested by studies with either deficient or increased HO-1 expression (Refs 43–45). In the nervous system, HO-1 can be highly induced in glia by its substrate haem, Nrf2 activation, and by a variety of pro-oxidant and inflammatory stimuli (Refs 46–52).
Figure 2. Nrf2-induced cytoprotective pathways.
(a) Haem degradation pathway. Haem oxygenase-1 (HO-1) catalyses the degradation of the haem group to iron [Fe(II)], biliverdin and carbon monoxide (CO). Iron is sequestered by ferritin and biliverdin is subsequently converted to bilirubin by biliverdin reductase. Biliverdin and bilirubin are potent antioxidants. (b) Disposal of hydrogen peroxide (H202) and organic peroxides (ROOH) by glutathione (GSH)-utilising enzymes. In the reaction catalysed by the GSH peroxidase (GPx), GSH serves as an electron donor in the reduction of hydrogen peroxide to water, and in the reduction of organic hydroperoxides to the corresponding alcohol. The oxidised glutathione disulfide (GSSG) is then recycled by the glutathione reductase (GR) in a NADPH-consuming reaction. The peroxisomal catalase (CAT) also participates in the disposal of H202 to water and oxygen.
The products of haem degradation – biliverdin, iron and carbon monoxide – are all biologically active molecules. Induction of HO-1 might protect cells against oxidative damage by augmenting the breakdown of the pro-oxidant haem group to the radical-scavenging bile pigments, biliverdin and bilirubin (Refs 53, 54). Furthermore, both these pigments, as well as carbon monoxide, have been shown to have potent immunomodulatory properties. Experimental observations indicate that the extent of HO-1 induction may be crucial because excessive haem degradation could result in toxic levels of carbon monoxide, bilirubin and, more importantly, iron. However, cytoprotection by HO-1 is attributable to its augmentation of iron efflux from the cell (Ref. 55). Moreover, in many tissues co-induction of ferritin provides a ‘sink’ for the excess of intracellular iron. In some situations, haem-derived iron and carbon monoxide might exacerbate intracellular oxidative stress and cellular injury by promoting free radical generation within the mitochondrial compartment (Refs 56, 57). Increased HO-1 expression has been reported in neurodegenerative diseases involving glial activation, such as AD, PD, multiple sclerosis (Refs 58–60) and ALS (Refs 36, 61). Although the cytoprotective role of HO-1 is not yet completely understood, it is likely that the elevated HO-1 levels represent an attempt to restore the redox homeostasis and attenuate inflammation (Ref. 62).
Glutathione
SOD activity is a major defence mechanism that combats oxygen toxicity by converting superoxide to hydrogen peroxide. The efficacy of SOD as an antioxidant relies on the decomposition of hydrogen peroxide by catalase (CAT) and glutathione peroxidase (GPx) to prevent its conversion to the hydroxyl radical. The peroxisomal CAT quickly decomposes hydrogen peroxide to water and molecular oxygen, but CAT specific activity in the brain is much lower than in peripheral tissues (Refs 63, 64). GPx (a selenium-dependent enzyme) probably has a major role in disposing of hydrogen peroxide and organic hydroperoxides in neural tissue. Since there is no CAT in the mitochondria, glutathione is particularly important for peroxide detoxification in this organelle during normal or pathological conditions. Although mitochondrial glutathione comprises only about 10% of the total glutathione in the cell, it represents a distinctly regulated pool with major implications for antioxidant protection (Ref. 65).
In the reaction catalysed by GPx, the tripeptide glutathione [γ-l-glutamyl-l-cysteinylglycine (GSH)] serves as an electron donor in the reduction of hydrogen peroxide to water and organic hydroperoxides to the corresponding alcohol (Ref. 66). The oxidised GSH – glutathione disulfide, (GSSG) – is then recycled by glutathione reductase in an NADPH-consuming reaction (Fig. 2B). In addition to its role in peroxide detoxification, GSH functions as the main cysteine storage in the cell, maintains the cellular redox homeostasis (sustaining the thiol status of proteins) and is a key component in the detoxification of xenobiotics and their metabolites through GSH conjugation. The glutathione S-transferases (GSTs) form a group of multigene isoenzymes involved in the cellular detoxification of both xenobiotic and endobiotic compounds (Ref. 67). Catalysis occurs by conjugation with GSH and the less toxic and more hydrophilic conjugated products are then eliminated from the cell by the trans-membrane multidrug resistance-associated proteins (MRPs). In particular, MRP1 and MRP2 can export GSH conjugates to the extracellular compartment (Ref. 68).
GSH is synthesised by the consecutive action of the enzymes glutamate cysteine ligase (GCL) and glutathione synthase. GCL is the rate-limiting enzyme in GSH synthesis and it is feedback inhibited by GSH (Ref. 69). Nrf2 regulates the basal and inducible levels of GCL-modifier subunit (GCLM), GCL-catalytic subunit (GCLC), glutathione synthase and glutathione reductase (Refs 70, 71, 72, 73); hence Nrf2 activation might regulate GSH homeostasis by affecting de novo synthesis and/or GSH redox state. Nrf2 also regulates the expression of several GSTs (Ref. 74), MRP1 (Ref. 75) and the cysteine–glutamate exchange transporter that maintains intracellular GSH levels by regulating cysteine influx (Refs 76, 77). Following Nrf2 activation, the cell will experience an increase in the enzymes involved in GSH synthesis and utilisation, which will have a major role in resistance to oxidative stress. Remarkably, even in the absence of a net increase in the amount of GSH, in a cell with enough redox potential (a supply of NADPH), the increase in glutathione reductase and GSH-utilising enzymes induced by Nrf2 activation should still provide increased antioxidant defences.
Nrf2 in cell survival
The cytoprotective effect of Nrf2 expression has been studied in many different cell models. Nrf2-deficient cells are more sensitive to peroxides, nitric oxide, mitochondrial toxins, endoplasmic reticulum stress and glucose deprivation (Ref. 37). Although Nrf2-knockout mice develop normally, aged females develop a systemic autoimmune disease (Refs 78, 79), and it is possible that Nrf2 has an important role in modulating the innate immunological response (Ref. 80). In vivo, electron paramagnetic resonance analyses of the disappearance of the spin probe Carbamoyl-PROXYL in Nrf2-knockout mice demonstrated a primary decrease of tissue-reducing activity and thus an impaired capacity to deal with electrophiles and ROS detoxification (Ref. 81). Consequently, Nrf2-deficient mice are more sensitive to pulmonary inflammatory diseases, chemical hepatotoxicity and carcinogenesis, and chemopreventive agents have no or reduced efficacy (Ref. 82). It has also been proposed that the decline in the cellular defences against oxidative, toxicological and pathological insults associated with ageing is due to a decrease in Nrf2-mediated GCL expression (Ref. 83). However, this observation was restricted to changes in hepatic GSH levels and it remains to be determined whether this phenomenon is relevant to other tissues.
Nrf2-dependent GSH levels regulate the sensitivity of the cells to Fas and TNF-α-induced apoptosis (Refs 84, 85). There is an inverse correlation between the intracellular levels of glutathione and the ability of the receptor to initiate apoptotic downstream signaling pathways, suggesting that GSH levels regulate the sensitivity of the cell to death receptor signals. In motor neurons, apoptosis induced by p75NTR (another member of the death receptor family, also known as TNR16) or mediated by Fas involves increased production of ROS and RNS (Refs 86, 87). Accordingly, increased GSH levels mediated by Nrf2 activation completely prevents p75NTR- and Fas-mediated motor neuron apoptosis in motor neurons (Ref. 88).
Role of the astrocyte–neuron interaction in neuroprotection
In the presence of astrocytes, neurons are more resistant to the oxidative stress induced by several compounds (Refs 89, 90, 91). GSH levels are key in this observation, because GSH depletion in co-cultures caused the loss of astrocyte-mediated neuroprotection (Refs 92, 93). Astrocytes have more efficient GSH synthesis systems and higher GSH content than neurons (Refs 94, 95, 96, 97). One of the reasons for this difference resides in the fact that Nrf2-driven genes are preferentially activated in astrocytes over neurons (Refs 98, 99, 100). Consequently, astrocytes have more enzymes involved in GSH synthesis (GCL and GS), and can better increase its production and release to the extracellular medium upon stimulation. The coordinated increase in GSH biosynthesis and release is a fundamental component in the neuronal protection conferred by Nrf2 activation in astrocytes (Refs 99, 100, 101).
Neurons cannot take up GSH, so how can an Nrf2-mediated increase in GSH release from astrocytes protect neighbouring neurons? GSH will react nonenzymatically with a variety of radicals, and therefore might function as a first line of defence against ROS and RNS in the extracellular space (Ref. 102). Because GSH possesses unique and specific binding sites in membranes of both neurons and astrocytes, it has also been proposed that GSH might be a glia-derived neuromodulator and/or neurotransmitter (Refs 103–105). However, the best understood protective mechanism is that the increased secretion of GSH by astrocytes boosts the content of GSH in the co-cultured neurons (Refs 106, 107).
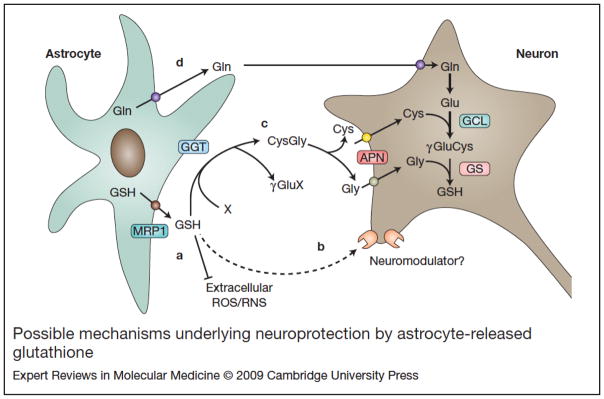
Cysteine is extremely unstable extracellularly and rapidly auto-oxidises to cystine (Ref. 108). Because neurons cannot use cystine, they must rely on the presence of cysteine as a GSH precursor (Ref. 107). Extracellular GSH is the most important source of cysteine for neurons. Most cells readily take up cysteine, and the availability of cysteine determines the level of neuronal GSH (Ref. 107). GSH and GSH-conjugates are exported from astrocytes by MRP1 (Refs 109, 110, 111) and the release of GSH from astrocytes is probably the process that consumes most of the intracellular GSH. Glutamate and cysteine in GSH are linked through the γ-carboxyl group of glutamate rather than the α-carboxyl group. This unconventional link is subject to hydrolysis by γ-glutamyltranspeptidase (GGT), which is present on the external surfaces of astrocytes (Refs 107, 112). GGT produces the dipeptide cysteinyl-glycine, which can then be hydrolysed by aminopeptidase N (Ref. 113) to release cysteine and glycine. Astrocytes also release glutamine, and hence make available all the necessary precursors for neuronal GSH synthesis (Fig. 3). Although all the available data supporting this working model come from cell culture systems, we have recently provided in vivo evidence that modification of the astrocytic glutathione system might help surrounding neurons to cope with noxious stimuli (Ref. 114). Transgenic overexpression of Nrf2 in astrocytes resulted in ~25% increase in glutathione content in the spinal cord and protected motor neurons from the toxicity of ALS-linked mutant SOD1 (see below).
Figure 3. Possible mechanisms underlying neuroprotection by astrocyte-released glutathione.
At least three different mechanisms could account for the neuroprotective effect of increased glutathione (GSH) release from astrocytes. (a) First, GSH reacts non-enzymatically with a variety of radicals and might function as a first line of defence against ROS/RNS in the extracellular space. (b) Second, it has been proposed that GSH might be a glia-derived neuromodulator and/or neurotransmitter. (c) Third, released GSH is hydrolysed by the γ-glutamyltranspeptidase (GGT) present on the external surfaces of astrocytes. The GGT produces the dipeptide cysteinyl-glycine, which then can be hydrolysed by the aminopeptidase N (APN) to release cysteine and glycine. (d) Astrocytes also release glutamine and hence make available all the necessary precursors for neuronal GSH synthesis. X, any acceptor of the γ-glutamyl moiety; MRP1, multidrug-resistance-associated protein 1; GCL, glutamate cysteine ligase; GS, glutathione synthase.
Nrf2-mediated neuroprotection
Although the role of oxidative stress in the etiology of neurodegenerative diseases remains controversial, it is clear that oxidative stress markers (for example, protein oxidation and nitration, lipid peroxidation and nucleic acid oxidation) are found in many neurodegenerating tissues (Refs 8, 115, 116). Oxidative stress might occur as the primary pathological mechanism or as a secondary event, originating from insults such as mitochondrial dysfunction and excitoxicity. In addition, antioxidant defences decrease with ageing, which is the greatest risk factor for neurodegenerative diseases. In consequence, much attention has been given to Nrf2 as a promising therapeutic target to combat oxidative stress in neurodegeneration (Refs 117, 118, 119, 120).
Amyotrophic lateral sclerosis
ALS is the most common adult-onset motor neuron disease, caused by the progressive degeneration of motor neurons in the spinal cord, brainstem and motor cortex (Ref. 121). The aetiology of most ALS cases remains unknown (sporadic ALS); however, ~10% are inherited in a dominant manner (familial ALS). Both forms of ALS share the same pathological features, including progressive muscle weakness, atrophy and spasticity. ALS is usually fatal within 3–5 years of onset because of denervation of the respiratory muscles and diaphragm. Approximately 10–20% of familial ALS is caused by a toxic gain-of-function induced by mutations in the Cu/Zn-superoxide dismutase (SOD1) (Ref. 122). Rodents overexpressing mutated forms of human SOD1 generally develop an ALS-like phenotype (Refs 123, 124). Several hypotheses, including oxidative stress, glutamate excitotoxicity, formation of high molecular weight aggregates, defective axonal transport, decreased trophic support and mitochondrial dysfunction have all been proposed to explain the toxic effect of mutated human SOD1 (Refs 125, 126, 127, 128).
Although the molecular mechanism underlying the selective death of motor neurons remains unknown, toxicity to motor neurons requires the expression of mutant human SOD1 in non-neuronal cells, as well as in motor neurons. The proposed non-cell-autonomous mechanism of the disease suggests that, although the expression of the mutant enzyme in motor neurons affects disease onset (Ref. 129), expression within the glial compartment influences disease progression (Refs 129, 130, 131). Whether it is a primary etiological component or a secondary event, it is clear that oxidative stress occurs in ALS, and probably affects the course of the disease (Refs 3, 5, 115). Both nitric oxide and superoxide are produced by motor neurons in response to apoptotic stimuli, such as trophic factor deprivation and the activation of Fas and p75NTR pathways (Refs 86, 87, 132). Additionally, in ALS patients and rodent models, a strong glial reaction typically surrounds degenerating motor neurons and oxidative markers are present in these cells, as well as in motor neurons (Ref. 133).
Recently Sarlette and co-workers (Ref. 134) reported a reduction of expression of Nrf2 mRNA and protein in neurons from primary motor cortex and spinal cord from ALS postmortem tissue samples. A similar decrease in mRNA encoding Nrf2 was observed in embryonic motor neurons isolated from ALS model rats (Ref. 88). However, no altered expression of Nrf2 or ARE-driven genes was found in laser-capture microdissected motor neurons from ALS model mice (Ref. 135). Despite these apparent discrepancies, it is clear that there is an increase in ARE-driven genes in the spinal cord of animal models for ALS upon disease onset. A strong increase in HO-1 was found at onset in model rats (Ref. 36), whereas a direct assessment of ARE activation was obtained by mating the model mice with mice transgenic for ARE-driven human placental alkaline phosphatase activity (Ref. 136). This protective response might prevent neuronal degeneration in early stages of the disease, but it is overwhelmed by other mechanisms that drive apoptosis.
Transgenic mice expressing mutant human SOD1 exclusively in astrocytes displayed astrocytosis but failed to develop the disease (Ref. 137). However, astrocytes isolated from rats (Ref. 101) or mice (Refs 114, 138) expressing mutated human SOD1G93A are toxic to cocultured wild-type motor neurons. Overexpression of Nrf2 completely reverses the toxicity of ALS astrocytes to motor neurons in vitro. Moreover, overexpression of Nrf2 under the control of the astrocyte-specific promoter for the glial fibrillary acidic protein delays onset and increases the lifespan in ALS mouse models (Ref. 114). These results suggest that the astrocytic Nrf2-mediated increase in GSH content and secretion does not directly modify the toxic component of astrocytes but instead improves the ability of motor neurons to cope with them. Increased glutathione secretion from astrocytes might also alter the way in which they interact with other cells and contribute to the decreased microglial response observed (Ref. 114). This was the first in vivo evidence that Nrf2 activation in astrocytes can be beneficial to protect neurons in chronic neurodegeneration and a proof of principle demonstrating that Nrf2 activation should be a suitable therapeutic target in ALS. In addition, since Nrf2 activation in motor neurons critically modulate apoptotic pathways (Ref. 88), the direct activation of Nrf2 in neurons deserves further investigation in the context of ALS.
Parkinson disease
PD is associated with progressive loss of dopaminergic neurons in the substantia nigra pars compacta, which project to the striatum, as well as with more-widespread neuronal changes that cause complex and variable motor and nonmotor symptoms (Ref. 139). Decreased activity of mitochondrial complex I, as well as increased lipid, protein and DNA oxidation, are found in individuals affected with PD (Refs 140, 141, 142). Studies of familial PD have revealed a prominent role for mitochondrial dysfunction in the pathogenesis of the disease, and impaired mitochondrial function increases oxidative stress (Refs 143, 144). In many cases, Nrf2 status has been inferred from ARE-driven gene expression and distribution. The enzyme NAD(P)H:quinone oxidodreductase (NQO1) catalyses the two-electron reduction of quinones, preventing their participation in redox cycling and subsequent generation of ROS. NQO1 is a prototypical ARE-driven gene, which, along with HO-1, is strongly upregulated in glial cells in postmortem PD brain (Refs 145, 146). Nrf2 distribution is mainly nuclear in parkinsonian nigral neurons – a pattern that is consistent with a neuronal response to increased oxidative stress (Ref. 147).
Nrf2 activation protects against the neurotoxic dompamine analogue 6-hydroxydopamine and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) in vitro (Refs 148, 149, 150). Moreover, MPTP administration resulted in a greater loss of dopamine transporter levels in the striatum of Nrf2-knockout mice (Ref. 151), whereas intrastriatal transplantation of Nrf2-overexpressing astrocytes protects against 6-hydroxydopamine in mice (Ref. 149). Nrf2 deficiency also increases MPTP sensitivity. Moreover, Nrf2 overexpression restricted to astrocytes in a Nrf2-knockout background is sufficient to completely protect against MPTP toxicity (Ref. 152), indicating that modulation of the Nrf2 ARE pathway in astrocytes represents a suitable therapeutic approach to reduce or prevent neuronal degeneration in PD.
Alzheimer disease
AD is a complex and genetically heterogeneous disease, characterised by progressive memory deficit, cognitive impairment and personality changes, which are accompanied by specific structural abnormalities in the brain. Loss of neurons and synapses in the neocortex, hippocampus and other subcortical regions of the brain are common features of AD (Ref. 153). The main histological features of AD are extracellular protein deposits termed β-amyloid (or senile) plaques, β-amyloid deposits in blood vessels and intraneuronal neurofibrillary tangles. The β-amyloid peptide is organised in fibrils intermixed with non-fibrillar forms of the peptide and forms the major component of the amyloid deposits (Ref. 153). Altered metal biology, mitochondrial dysfunction and increased ROS and RNS production by reactive glial cells are believed to contribute to the oxidative damage in lipids, proteins and DNA observed in AD (Refs 154, 155, 156, 157, 158). In postmortem temporal cortex and hippocampus of patients with AD, the percentage of astrocytes expressing HO-1 is significantly greater than in non-demented individuals (Ref. 156). Moreover, NQO1 activity and expression is increased in neurons and astrocytes in AD (Refs 159, 160, 161). However, Nrf2 immunostaining was found to be predominantly cytoplasmic in hippocampal neurons from individuals with AD (Ref. 147), unlike the nuclear distribution in dopaminergic neurons in PD. This differential distribution of Nrf2 in AD and PD, might reflect a difference in the way that affected neurons in these diseases respond to oxidative stress.
In a mouse model of AD (APP/PS1 mice) the decrease in the expression of mRNA encoding Nrf2, NQO1, GCLC and GCLM correlates with an increased accumulation of β-amyloid deposits (Ref. 162). Recently, the GSH:GSSG ratio was found to be reduced in brain homogenates in a different mouse model of AD (3×Tg-AD; Ref. 163). In addition, Nrf2 activation protects neural cells against β-peptide-induced neurotoxicity in vitro (Refs 162, 164), suggesting that activation of the Nrf2 ARE pathway might be beneficial in the context of AD pathology.
Huntington’s disease
HD is an autosomal dominant neurodegenerative disorder that results from a polyglutamine repeat expansion in the first exon of the huntingtin gene (HTT). HD is characterised by the progressive development of involuntary choreiform movements, cognitive impairment, neuropsychiatric symptoms and premature death (Ref. 8). Neuronal degeneration is mainly observed in the striatum and cerebral cortex, and it is believed to be due to the effect of mutant huntingtin on mitochondrial function and energy metabolism, but the exact mechanism remains unknown. Severe deficits in mitochondrial complex II, III and IV activity were observed in the striatum of postmortem HD brains (Refs 165, 166) and reduced complex IV activity, together with elevated nitric oxide and superoxide production, has been observed in one genetic mouse model of HD (Refs 167, 168). Although, there is no available information on the status of the Nrf2 ARE pathway in HD, the protective effect of Nrf2 activation has been demonstrated in models of mitochondrial complex II toxicity, where ROS production increases upon disruption of the electron transport chain. Mitochondrial complex II inhibition with 3-nitropropionic acid (3NP) or malonate produces characteristic striatal degeneration, which is similar to that observed in HD (Refs 169, 170). Nrf2-knockout mice are more sensitive to the striatal lesions caused by administration of these inhibitors (Refs 171, 172). Chemical activation or viral overexpression of Nrf2 significantly reduces lesion size caused by 3NP (Ref. 171) and intrastriatal transplantation of Nrf2-overexpressing astrocytes has a remarkable neuroprotective effect against malonate toxicity in vivo (Ref. 172).
In addition to its role in neurodegenerative diseases, Nrf2 activation protects neurons from ischaemia and intracerebral haemorrhage (Refs 173, 174, 175, 176). Interestingly, in the case of ischaemia or reperfusion injury, activation of Nrf2 and increased expression of HO-1, mainly in neurons and not astrocytes, seems to be central to the observed protection (Refs 177, 178). Moreover, Nrf2 activation attenuates the disruption in the blood–brain barrier observed in traumatic brain injury, by preventing the loss of tight junction proteins and endothelial cell death (Refs 179, 180). Since neuronal function is intimately linked to vascular function, it might be necessary to activate Nrf2 in endothelial, glial and neuronal cells, to effectively protect the neurovascular unit and achieve significant protection against trauma and neurodegenerative diseases.
Nrf2 activation through an increase in GSH also helps cells to cope with the oxidative stress associated with viral infection of the central nervous system (CNS) (Refs 181, 182). Although microglial production of intracellular and extracellular ROS is intrinsic to the role of microglia in the CNS, persistent ROS and cytokine release from microglia will result in chronic neuroinflammation and neuronal damage. Neuroinflammation is a hallmark of viral infection, ischaemia, and trauma, and the role of Nrf2 – in particular the role of the astrocytic Nrf2 ARE pathway – in modulating the magnitude and duration of the microglial response is a very interesting possibility (Refs 183, 184).
Conclusions and outstanding questions
It is clear that Nrf2 activation represents an exceptional defence mechanism for many cell types. Particularly in the CNS, astrocytic Nrf2 activation has a major role in protecting neurons from noxious stimuli. Many laboratories, including ours, have generated a considerable amount of data firmly indicating the potential therapeutic value of Nrf2 activation in acute lesions of the CNS. It is likely that more evidence of the beneficial role of Nrf2 activation in chronic neurodegeneration will soon be available. Although potent activators of Nrf2 are available, the therapeutic challenge resides in identifying chemicals that, in addition to their role in activation of Nrf2, can efficiently penetrate the blood–brain barrier to reach astrocytes. In cell-replacement therapies, the intrinsic complications of replacing highly specialised cells such as neurons, make astrocytes a much more appealing target. Glial-restricted precursors (GRPs) are an exciting candidate for this purpose. However, in the context of a chronic neurodegenerative disease, transplantation of GRPs that go on to produce astrocytes might not be enough; Nrf2-overexpressing GRPs might offer a much better solution. Because of the extraordinary capacity of astrocytes to activate the Nrf2 ARE pathway, most of the research so far has concentrated in this particular aspect of Nrf2 protection. Although the exact mechanism governing astrocytic Nrf2-mediated neuroprotection is still under investigation, enough data exists to support the proof-of-principle for this concept and now the challenge ahead is to determine the role of neuronal Nrf2 activation in neurodegeneration.
Acknowledgments
We thank the reviewers for their comments and suggestions. This work was supported by grants from the ALS Association and the Robert Packard Center for ALS Research at Johns Hopkins, ES08089 and ES10042 from the National Institute of Environmental Health Sciences (NIEHS) to J.AJ. M.R.V. is a recipient of the Milton Safenowitz postdoctoral fellowship for ALS research.
References
- 1.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiological Reviews. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 2.Palmer RMJ, Rees DD, Ashton DS, Moncada S. L-Arginine is the physiological precursor for the formation of nitric oxide in endothelium dependent relaxation. Biochemical and Biophysical Research Communications. 1988;153:1251–1256. doi: 10.1016/s0006-291x(88)81362-7. [DOI] [PubMed] [Google Scholar]
- 3.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiological Reviews. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutteridge JM, Halliwell B. Free radicals and antioxidants in the year 2000. A historical look to the future. Annals of the New York Academy of Science. 2000;899:136–147. doi: 10.1111/j.1749-6632.2000.tb06182.x. [DOI] [PubMed] [Google Scholar]
- 5.Goodall EF, Morrison KE. Amyotrophic lateral sclerosis (motor neuron disease): proposed mechanisms and pathways to treatment. Expert Reviews in Molecular Medicine. 2006;8:1–22. doi: 10.1017/S1462399406010854. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell B. Oxidative stress and neurodegeneration: where are we now? Journal of Neurochemistry. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- 7.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 8.Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxidants and Redox Signalling. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 9.Moi P, Chan K, Asunis I, et al. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh K, Igarashi K, Hayashi N, et al. Cloning and characterization of a novel erythroid cell-derived CNC familytranscription factor heterodimerizing with the small Maf family proteins. Molecular and Cellular Biology. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. Journal of Biological Chemistry. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 12.Andrews NC, Erdjument-Bromage H, Davidson MB, et al. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 13.Ney PA, Sorrentino BP, Lowrey CH, Nienhuis AW. Inducibility of the HS II enhancer depends on binding of an erythroid specific nuclear protein. Nucleic Acids Research. 1990;18:6011–6017. doi: 10.1093/nar/18.20.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strauss EC, Orkin SH. In vivo protein-DNA interactions at hypersensitive site 3 of the human beta-globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:5809–5813. doi: 10.1073/pnas.89.13.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and Biophysical Research Communications. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 16.Motohashi H, O’Connor T, Katsuoka F, et al. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 17.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes and Development. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakabayashi N, Itoh K, Wakabayashi J, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nature Genetics. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 19.Stewart D, Killeen E, Naquin R, et al. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. Journal of Biological Chemistry. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 20.Alam J, Killeen E, Gong P, et al. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. American Journal of Physiology Renal Physiology. 2003;284:F743–F752. doi: 10.1152/ajprenal.00376.2002. [DOI] [PubMed] [Google Scholar]
- 21.McMahon M, Thomas N, Itoh K, et al. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. Journal of Biological Chemistry. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 22.Cullinan SB, Gordan JD, Jin J, et al. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Molecular and Cellular Biology. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi A, Kang MI, Okawa H, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and Cellular Biology. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon M, Thomas N, Itoh K, et al. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. Journal of Biological Chemistry. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 25.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Molecular and Cellular Biology. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levonen AL, Landar A, Ramachandran A, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. The Biochemical Journal. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto T, Suzuki T, Kobayashi A, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Molecular and Cellular Biology. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Advances in Enzyme Regulation. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Molecular and Cellular Biology. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen T, Sherratt PJ, Nioi P, et al. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. Journal of Biological Chemistry. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 33.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Molecular and Cellular Biology. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watai Y, Kobayashi A, Nagase H, et al. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes to Cells. 2007;12:1163–1178. doi: 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- 35.Braun S, Hanselmann C, Gassmann MG, et al. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Molecular and Cellular Biology. 2002;22:5492–5505. doi: 10.1128/MCB.22.15.5492-5505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vargas MR, Pehar M, Cassina P, et al. Fibroblast growth factor-1 induces heme oxygenase-1 via nuclear factor erythroid 2-related factor 2 (Nrf2) in spinal cord astrocytes: consequences for motor neuron survival. Journal of Biological Chemistry. 2005;280:25571–25579. doi: 10.1074/jbc.M501920200. [DOI] [PubMed] [Google Scholar]
- 37.Lee JM, Li J, Johnson DA, et al. Nrf2, a multi-organ protector? The FASEB Journal. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 38.Tenhunen R, Marver HS, Schmid R. The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proceedings of the National Academy of Sciences of the United States of America. 1968;61:748–755. doi: 10.1073/pnas.61.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maines MD. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. The FASEB Journal. 1988;2:2557–2568. [PubMed] [Google Scholar]
- 40.Shibahara S, Müller R, Taguchi H, Yoshida T. Cloning and expression of cDNA for rat heme oxygenase. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:7865–7869. doi: 10.1073/pnas.82.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maines MD, Trakshel GM, Kutty RK. Characterization of two constitutive forms of rat liver microsomal heme oxygenase. Only one molecular species of the enzyme is inducible. Journal of Biological Chemistry. 1986;261:411–419. [PubMed] [Google Scholar]
- 42.Barañano DE, Snyder SH. Neural roles for heme oxygenase: contrasts to nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10996–11002. doi: 10.1073/pnas.191351298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poss KD, Tonegawa S. Reduced stress defense in heme oxygenase 1-deficient cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10925–10930. doi: 10.1073/pnas.94.20.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yachie A, Niida Y, Wada T, et al. Oxidative stress causes enhanced endothelial cell injury in human heme oxygenase-1 deficiency. Journal of Clinical Investigation. 1999;103:129–135. doi: 10.1172/JCI4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, Gunter K, Maines MD. Neurons overexpressing heme oxygenase-1 resist oxidative stress-mediated cell death. Journal of Neurochemistry. 2000;75:304–313. doi: 10.1046/j.1471-4159.2000.0750304.x. [DOI] [PubMed] [Google Scholar]
- 46.Dwyer BE, Nishimura RN, De Vellis J, Yoshida T. Heme oxygenase is a heat shock protein and PEST protein in rat astroglial cells. Glia. 1992;5:300–305. doi: 10.1002/glia.440050407. [DOI] [PubMed] [Google Scholar]
- 47.Dwyer BE, Nishimura RN, Lu SY. Differential expression of heme oxygenase-1 in cultured cortical neurons and astrocytes determined by the aid of a new heme oxygenase antibody. Response to oxidative stress. Brain Research Molecular Brain Research. 1995;30:37–47. doi: 10.1016/0169-328x(94)00273-h. [DOI] [PubMed] [Google Scholar]
- 48.Kitamura Y, Furukawa M, Matsuoka Y, et al. In vitro and in vivo induction of heme oxygenase-1 in rat glial cells: possible involvement of nitric oxide production from inducible nitric oxide synthase. Glia. 1998;22:138–148. [PubMed] [Google Scholar]
- 49.Schipper HM, Bernier L, Mehindate K, Frankel D. Mitochondrial iron sequestration in dopamine-challenged astroglia: role of heme oxygenase-1 and the permeability transition pore. Journal of Neurochemistry. 1999;72:1802–1811. doi: 10.1046/j.1471-4159.1999.0721802.x. [DOI] [PubMed] [Google Scholar]
- 50.Regan RF, Guo Y, Kumar N. Heme oxygenase-1 induction protects murine cortical astrocytes from hemoglobin toxicity. Neuroscience Letters. 2000;282:1–4. doi: 10.1016/s0304-3940(00)00817-x. [DOI] [PubMed] [Google Scholar]
- 51.Mehindate K, Sahlas DJ, Frankel D, et al. Proinflammatory cytokines promote glial heme oxygenase-1 expression and mitochondrial iron deposition: implications for multiple sclerosis. Journal of Neurochemistry. 2001;77:1386–1395. doi: 10.1046/j.1471-4159.2001.00354.x. [DOI] [PubMed] [Google Scholar]
- 52.Fauconneau B, Petegnief V, Sanfeliu C, et al. Induction of heat shock proteins (HSPs) by sodium arsenite in cultured astrocytes and reduction of hydrogen peroxide-induced cell death. Journal of Neurochemistry. 2002;83:1338–1348. doi: 10.1046/j.1471-4159.2002.01230.x. [DOI] [PubMed] [Google Scholar]
- 53.Stocker R, Yamamoto Y, McDonagh AF, et al. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 54.Doré S, Snyder SH. Neuroprotective action of bilirubin against oxidative stress in primary hippocampal cultures. Annals of the New York Academy of Science. 1999;890:167–172. doi: 10.1111/j.1749-6632.1999.tb07991.x. [DOI] [PubMed] [Google Scholar]
- 55.Ferris CD, Jaffrey SR, Sawa A, et al. Haem oxygenase-1 prevents cell death by regulating cellular iron. Nature Cell Biology. 1999;1:152–157. doi: 10.1038/11072. [DOI] [PubMed] [Google Scholar]
- 56.Song W, Su H, Song S, Paudel HK, Schipper HM. Over-expression of heme oxygenase-1 promotes oxidative mitochondrial damage in rat astroglia. Journal of Cellular Physiology. 2006;206:655–663. doi: 10.1002/jcp.20509. [DOI] [PubMed] [Google Scholar]
- 57.Schipper HM. Brain iron deposition and the free radical-mitochondrial theory of ageing. Ageing Research Reviews. 2004;3:265–301. doi: 10.1016/j.arr.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Takeda A, Itoyama Y, Kimpara T, et al. Heme catabolism and heme oxygenase in neurodegenerative disease. Antioxidants and Redox Signalling. 2004;6:888–894. doi: 10.1089/ars.2004.6.888. [DOI] [PubMed] [Google Scholar]
- 59.Schipper HM. Heme oxygenase expression in human central nervous system disorders. Free Radical Biology and Medicine. 2004;37:1995–2011. doi: 10.1016/j.freeradbiomed.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 60.van Horssen J, Schreibelt G, Drexhage J, et al. Severe oxidative damage in multiple sclerosis lesions coincides with enhanced antioxidant enzyme expression. Free Radical Biology and Medicine. 2008;45:1729–1737. doi: 10.1016/j.freeradbiomed.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 61.Ferrante RJ, Shinobu LA, Schulz JB, et al. Increased 3-nitrotyrosine and oxidative damage in mice with a human copper/zinc superoxide dismutase mutation. Annals of Neurology. 1997;42:326–334. doi: 10.1002/ana.410420309. [DOI] [PubMed] [Google Scholar]
- 62.Cuadrado A, Rojo AI. Heme oxygenase-1 as a therapeutic target in neurodegenerative diseases and brain infections. Current Pharmaceutical Design. 2008;14:429–442. doi: 10.2174/138161208783597407. [DOI] [PubMed] [Google Scholar]
- 63.Ho YS, Magnenat JL, Bronson RT, et al. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. Journal of Biological Chemistry. 1997;272:16644–16651. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 64.Ho YS, Xiong Y, Ma W, Spector A, Ho DS. Mice lacking catalase develop normally but show differential sensitivity to oxidant tissue injury. Journal of Biological Chemistry. 2004;279:32804–32812. doi: 10.1074/jbc.M404800200. [DOI] [PubMed] [Google Scholar]
- 65.Lash LH. Mitochondrial glutathione transport: physiological, pathological and toxicological implications. Chemico-Biological Interactions. 2006;163:54–67. doi: 10.1016/j.cbi.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dringen R, Pawlowski PG, Hirrlinger J. Peroxide detoxification by brain cells. Journal of Neuroscience Research. 2005;79:157–165. doi: 10.1002/jnr.20280. [DOI] [PubMed] [Google Scholar]
- 67.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annual Review of Pharmacology and Toxicology. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 68.Cole SP, Deeley RG. Transport of glutathione and glutathione conjugates by MRP1. Trends in Pharmacological Sciences. 2006;27:438–446. doi: 10.1016/j.tips.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 69.Meister A. Glutathione metabolism and its selective modification. Journal of Biological Chemistry. 1988;263:17205–17208. [PubMed] [Google Scholar]
- 70.Moinova HR, Mulcahy RT. Up-regulation of the human gamma-glutamylcysteine synthetase regulatory subunit gene involves binding of Nrf-2 to an electrophile responsive element. Biochemical and Biophysical Research Communications. 1999;261:661–668. doi: 10.1006/bbrc.1999.1109. [DOI] [PubMed] [Google Scholar]
- 71.Wild AC, Moinova HR, Mulcahy RT. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. Journal of Biological Chemistry. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 72.Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochimica et Biophysica Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- 73.Harvey CJ, Thimmulappa RK, Singh A, et al. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radical Biology and Medicine. 2009;46:443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chanas SA, Jiang Q, McMahon M, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. The Biochemical Journal. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochemical and Biophysical Research Communications. 2003;310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 76.Sasaki H, Sato H, Kuriyama-Matsumura K, et al. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. Journal of Biological Chemistry. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 77.Shih AY, Erb H, Sun X, et al. Cystine/glutamate exchange modulates glutathione supply for neuroprotection from oxidative stress and cell proliferation. Journal of Neuroscience. 2006;26:10514–23. doi: 10.1523/JNEUROSCI.3178-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoh K, Itoh K, Enomoto A, et al. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney International. 2001;60:1343–1353. doi: 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 79.Li J, Stein TD, Johnson JA. Genetic dissection of systemic autoimmune disease in Nrf2-deficient mice. Physiological Genomics. 2004;18:261–272. doi: 10.1152/physiolgenomics.00209.2003. [DOI] [PubMed] [Google Scholar]
- 80.Thimmulappa RK, Lee H, Rangasamy T, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. Journal of Clinical Investigation. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirayama A, Yoh K, Nagase S, et al. EPR imaging of reducing activity in Nrf2 transcriptional factor-deficient mice. Free Radical Biology and Medicine. 2003;34:1236–1242. doi: 10.1016/s0891-5849(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 82.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annual Review of Pharmacology and Toxicology. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 83.Suh JH, Shenvi SV, Dixon BM, et al. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kotlo KU, Yehiely F, Efimova E, et al. Nrf2 is an inhibitor of the Fas pathway as identified by Achilles’ Heel Method, a new function-based approach to gene identification in human cells. Oncogene. 2003;22:797–806. doi: 10.1038/sj.onc.1206077. [DOI] [PubMed] [Google Scholar]
- 85.Morito N, Yoh K, Itoh K, et al. Nrf2 regulates the sensitivity of death receptor signals by affecting intracellular glutathione levels. Oncogene. 2003;22:9275–9281. doi: 10.1038/sj.onc.1207024. [DOI] [PubMed] [Google Scholar]
- 86.Raoul C, Estévez AG, Nishimune H, et al. Motoneuron death triggered by a specific pathway downstream of Fas. potentiation by ALS-linked SOD1 mutations. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 87.Pehar M, Cassina P, Vargas MR, et al. Astrocytic production of nerve growth factor in motor neuron apoptosis: implications for amyotrophic lateral sclerosis. Journal of Neurochemistry. 2004;89:464–473. doi: 10.1111/j.1471-4159.2004.02357.x. [DOI] [PubMed] [Google Scholar]
- 88.Pehar M, Vargas MR, Robinson KM, et al. Mitochondrial superoxide production and nuclear factor erythroid 2-related factor 2 activation in p75 neurotrophin receptor-induced motor neuron apoptosis. Journal of Neuroscience. 2007;27:7777–7785. doi: 10.1523/JNEUROSCI.0823-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bronstein DM, Perez-Otano I, Sun V, et al. Glia-dependent neurotoxicity and neuroprotection in mesencephalic cultures. Brain Research. 1995;704:112–116. doi: 10.1016/0006-8993(95)01189-7. [DOI] [PubMed] [Google Scholar]
- 90.Desagher S, Glowinski J, Premont J. Astrocytes protect neurons from hydrogen peroxide toxicity. Journal of Neuroscience. 1996;16:2553–2562. doi: 10.1523/JNEUROSCI.16-08-02553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanaka J, Toku K, Zhang B, et al. Astrocytes prevent neuronal death induced by reactive oxygen and nitrogen species. Glia. 1999;28:85–96. doi: 10.1002/(sici)1098-1136(199911)28:2<85::aid-glia1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 92.Drukarch B, Schepens E, Jongenelen CA, et al. Astrocyte-mediated enhancement of neuronal survival is abolished by glutathione deficiency. Brain Research. 1997;770:123–130. doi: 10.1016/s0006-8993(97)00790-7. [DOI] [PubMed] [Google Scholar]
- 93.Gegg ME, Clark JB, Heales SJ. Co-culture of neurones with glutathione deficient astrocytes leads to increased neuronal susceptibility to nitric oxide and increased glutamate-cysteine ligase activity. Brain Research. 2005;1036:1–6. doi: 10.1016/j.brainres.2004.11.064. [DOI] [PubMed] [Google Scholar]
- 94.Sagara J, Miura K, Bannai S. Cystine uptake and glutathione level in fetal brain cells in primary culture and in suspension. Journal of Neurochemistry. 1993;61:1667–1671. doi: 10.1111/j.1471-4159.1993.tb09801.x. [DOI] [PubMed] [Google Scholar]
- 95.Sagara J, Makino N, Bannai S. Glutathione efflux from cultured astrocytes. Journal of Neurochemistry. 1996;66:1876–1881. doi: 10.1046/j.1471-4159.1996.66051876.x. [DOI] [PubMed] [Google Scholar]
- 96.Dringen R. Metabolism and functions of glutathione in brain. Progress in Neurobiology. 2000;62:649–671. doi: 10.1016/s0301-0082(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 97.Sun X, Shih AY, Johannssen HC, et al. Two-photon imaging of glutathione levels in intact brain indicates enhanced redox buffering in developing neurons and cells at the cerebrospinal fluid and blood-brain interface. Journal of Biological Chemistry. 2006;281:17420–17431. doi: 10.1074/jbc.M601567200. [DOI] [PubMed] [Google Scholar]
- 98.Lee JM, Calkins MJ, Chan K, et al. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. Journal of Biological Chemistry. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 99.Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. Journal of Neuroscience. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. Journal of Neuroscience. 2004;24:1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vargas MR, Pehar M, Cassina P, et al. Increased glutathione biosynthesis by Nrf2 activation in astrocytes prevents p75NTR-dependent motor neuron apoptosis. Journal of Neurochemistry. 2006;97:687–696. doi: 10.1111/j.1471-4159.2006.03742.x. [DOI] [PubMed] [Google Scholar]
- 102.Drukarch B, Schepens E, Stoof JC, et al. Astrocyte-enhanced neuronal survival is mediated by scavenging of extracellular reactive oxygen species. Free Radical Biology and Medicine. 1998;25:217–220. doi: 10.1016/s0891-5849(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 103.Guo N, Shaw C. Characterization and localization of glutathione binding sites on cultured astrocytes. Brain Research Molecular Brain Research. 1992;15:207–215. doi: 10.1016/0169-328x(92)90110-w. [DOI] [PubMed] [Google Scholar]
- 104.Janáky R, Ogita K, Pasqualotto BA, et al. Glutathione and signal transduction in the mammalian CNS. Journal of Neurochemistry. 1999;73:889–902. doi: 10.1046/j.1471-4159.1999.0730889.x. [DOI] [PubMed] [Google Scholar]
- 105.Hermann A, Varga V, Oja SS, et al. Involvement of amino-acid side chains of membrane proteins in the binding of glutathione to pig cerebral cortical membranes. Neurochemical Research. 2002;27:389–394. doi: 10.1023/a:1015599830320. [DOI] [PubMed] [Google Scholar]
- 106.Bolaños JP, Heales SJ, Peuchen S, et al. Nitric oxide-mediated mitochondrial damage: a potential neuroprotective role for glutathione. Free Radical Biology and Medicine. 1996;21:995–1001. doi: 10.1016/s0891-5849(96)00240-7. [DOI] [PubMed] [Google Scholar]
- 107.Dringen R, Pfeiffer B, Hamprecht B. Synthesis of the antioxidant glutathione in neurons: supply by astrocytes of CysGly as precursor for neuronal glutathione. Journal of Neuroscience. 1999;19:562–569. doi: 10.1523/JNEUROSCI.19-02-00562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meister A. Glutathione metabolism. Methods in Enzymology. 1995;251:3–7. doi: 10.1016/0076-6879(95)51106-7. [DOI] [PubMed] [Google Scholar]
- 109.Hirrlinger J, König J, Keppler D, et al. The multidrug resistance protein MRP1 mediates the release of glutathione disulfide from rat astrocytes during oxidative stress. Journal of Neurochemistry. 2001;76:627–636. doi: 10.1046/j.1471-4159.2001.00101.x. [DOI] [PubMed] [Google Scholar]
- 110.Hirrlinger J, Dringen R. Multidrug resistance protein 1-mediated export of glutathione and glutathione disulfide from brain astrocytes. Methods in Enzymology. 2005;400:395–409. doi: 10.1016/S0076-6879(05)00023-6. [DOI] [PubMed] [Google Scholar]
- 111.Minich T, Riemer J, Schulz JB, et al. The multidrug resistance protein 1 (Mrp1), but not Mrp5, mediates export of glutathione and glutathione disulfide from brain astrocytes. Journal of Neurochemistry. 2006;97:373–384. doi: 10.1111/j.1471-4159.2006.03737.x. [DOI] [PubMed] [Google Scholar]
- 112.Dringen R, Kranich O, Hamprecht B. The gamma-glutamyl transpeptidase inhibitor acivicin preserves glutathione released by astroglial cells in culture. Neurochemical Research. 1997;22:727–733. doi: 10.1023/a:1027310328310. [DOI] [PubMed] [Google Scholar]
- 113.Dringen R, Gutterer JM, Gros C, Hirrlinger J. Aminopeptidase N mediates the utilization of the GSH precursor CysGly by cultured neurons. Journal of Neuroscience Research. 2001;66:1003–1008. doi: 10.1002/jnr.10042. [DOI] [PubMed] [Google Scholar]
- 114.Vargas MR, Johnson DA, Sirkis DW, et al. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. Journal of Neuroscience. 2008;28:13574–13581. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ischiropoulos H, Beckman JS. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? Journal of Clinical Investigation. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nunomura A, Moreira PI, Lee HG, et al. Neuronal death and survival under oxidative stress in Alzheimer and Parkinson diseases. CNS and Neurological Disorders Drug Targets. 2007;6:411–423. doi: 10.2174/187152707783399201. [DOI] [PubMed] [Google Scholar]
- 117.van Muiswinkel FL, Kuiperij HB. The Nrf2-ARE Signalling pathway: promising drug target to combat oxidative stress in neurodegenerative disorders. Current Drug Targets CNS Neurol Disord. 2005;4:267–281. doi: 10.2174/1568007054038238. [DOI] [PubMed] [Google Scholar]
- 118.Calkins MJ, Johnson DA, Townsend JA, et al. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxidants and Redox Signalling. 2008 doi: 10.1089/ars.2008.2242. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.de Vries HE, Witte M, Hondius D, et al. Nrf2-induced antioxidant protection: a promising target to counteract ROS-mediated damage in neurodegenerative disease? Free Radical Biology and Medicine. 2008;45:1375–1383. doi: 10.1016/j.freeradbiomed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 120.Johnson JA, Johnson DA, Kraft AD, et al. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Annals of the New York Academy of Science. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. New England Journal of Medicine. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 122.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 123.Gurney ME, Pu H, Chiu AY, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 124.Howland DS, Liu J, She Y, et al. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS) Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beckman JS, Estévez AG, Crow JP, Barbeito L. Superoxide dismutase and the death of motoneurons in ALS. Trends in Neurosciences. 2001;24:S15–S20. doi: 10.1016/s0166-2236(00)01981-0. [DOI] [PubMed] [Google Scholar]
- 126.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nature Reviews Neuroscience. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 127.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annual Review of Neuroscience. 2004;27:723–479. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 128.Manfredi G, Xu Z. Mitochondrial dysfunction and its role in motor neuron degeneration in ALS. Mitochondrion. 2005;5:77–87. doi: 10.1016/j.mito.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 129.Boillée S, Yamanaka K, Lobsiger CS, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 130.Beers DR, Henkel JS, Xiao Q, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2003;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yamanaka K, Chun SJ, Boillee S, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nature Neuroscience. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Estévez AG, Spear N, Manuel SM, et al. Nitric oxide and superoxide contribute to motor neuron apoptosis induced by trophic factor deprivation. Journal of Neuroscience. 1998;18:923–931. doi: 10.1523/JNEUROSCI.18-03-00923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Barbeito LH, Pehar M, Cassina P, et al. A role for astrocytes in motor neuron loss in amyotrophic lateral sclerosis. Brain Research Brain Research Rev. 2004;47:263–274. doi: 10.1016/j.brainresrev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 134.Sarlette A, Krampfl K, Grothe C, et al. Nuclear erythroid 2-related factor 2-antioxidative response element signaling pathway in motor cortex and spinal cord in amyotrophic lateral sclerosis. Journal of Neuropathology and Experimental Neurology. 2008;67:1055–1062. doi: 10.1097/NEN.0b013e31818b4906. [DOI] [PubMed] [Google Scholar]
- 135.Ferraiuolo L, Heath PR, Holden H, et al. Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. Journal of Neuroscience. 2007;27:9201–9219. doi: 10.1523/JNEUROSCI.1470-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kraft AD, Resch JM, Johnson DA, Johnson JA. Activation of the Nrf2-ARE pathway in muscle and spinal cord during ALS-like pathology in mice expressing mutant SOD1. Experimental Neurology. 2007;207:107–117. doi: 10.1016/j.expneurol.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gong YH, Parsadanian AS, Andreeva A, et al. Restricted expression of G86R Cu/Zn superoxide dismutase in astrocytes results in astrocytosis but does not cause motoneuron degeneration. Journal of Neuroscience. 2000;20:660–665. doi: 10.1523/JNEUROSCI.20-02-00660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nagai M, Re DB, Nagata T, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nature Neuroscience. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Thomas B, Beal MF. Parkinson’s disease. Human Molecular Genetics. 2007;16:R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 140.Alam ZI, Daniel SE, Lees AJ, et al. A generalised increase in protein carbonyls in the brain in Parkinson’s but not incidental Lewy body disease. Journal of Neurochemistry. 1997;69:1326–1329. doi: 10.1046/j.1471-4159.1997.69031326.x. [DOI] [PubMed] [Google Scholar]
- 141.Alam ZI, Jenner A, Daniel SE, et al. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. Journal of Neurochemistry. 1997;69:1196–1203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- 142.Dexter DT, Holley AE, Flitter WD, et al. Increased levels of lipid hydroperoxides in the parkinsonian substantia nigra: an HPLC and ESR study. Movement Disorders. 1994;9:92–97. doi: 10.1002/mds.870090115. [DOI] [PubMed] [Google Scholar]
- 143.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 144.Clements CM, McNally RS, Conti BJ, et al. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.van Muiswinkel FL, de Vos RA, Bol JG, et al. Expression of NAD(P)H:quinone oxidoreductase in the normal and Parkinsonian substantia nigra. Neurobiology of Aging. 2004;25:1253–1262. doi: 10.1016/j.neurobiolaging.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 146.Schipper HM. Heme oxygenase expression in human central nervous system disorders. Free Radical Biology and Medicine. 2004;37:1995–2011. doi: 10.1016/j.freeradbiomed.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 147.Ramsey CP, Glass CA, Montgomery MB, et al. Expression of Nrf2 in neurodegenerative diseases. Journal of Neuropathology and Experimental Neurology. 2007;66:75–85. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yamamoto N, Sawada H, Izumi Y, et al. Proteasome inhibition induces glutathione synthesis and protects cells from oxidative stress: relevance to Parkinson disease. Journal of Biological Chemistry. 2007;282:4364–4372. doi: 10.1074/jbc.M603712200. [DOI] [PubMed] [Google Scholar]
- 149.Jakel RJ, Townsend JA, Kraft AD, Johnson JA. Nrf2-mediated protection against 6-hydroxydopamine. Brain Research. 2007;1144:192–201. doi: 10.1016/j.brainres.2007.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wruck CJ, Claussen M, Fuhrmann G, et al. Luteolin protects rat PC12 and C6 cells against MPP+ induced toxicity via an ERK dependent Keap1-Nrf2-ARE pathway. Journal of Neural Transmission Suppl. 2007;72:57–67. doi: 10.1007/978-3-211-73574-9_9. [DOI] [PubMed] [Google Scholar]
- 151.Burton NC, Kensler TW, Guilarte TR. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology. 2006;27:1094–1100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 152.Chen PC, Vargas MR, Pani AK, et al. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson’s disease: a critical role for the astrocyte. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bertram L, Tanzi RE. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nature Reviews Neuroscience. 2008;9:768–778. doi: 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- 154.Marshak DR, Pesce SA, Stanley LC, Griffin WS. Increased S100 beta neurotrophic activity in Alzheimer’s disease temporal lobe. Neurobiology of Aging. 1992;13:1–7. doi: 10.1016/0197-4580(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 155.Smith MA, Harris PL, Sayre LM, Perry G. Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. Proc Natl Acad Sci U S A. 1997;94:9866–9868. doi: 10.1073/pnas.94.18.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schipper HM, Bennett DA, Liberman A, et al. Glial heme oxygenase-1 expression in Alzheimer disease and mild cognitive impairment. Neurobiology of Aging. 2006;27:252–261. doi: 10.1016/j.neurobiolaging.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 157.Wang X, Su B, Perry G, et al. Insights into amyloid-beta-induced mitochondrial dysfunction in Alzheimer disease. Free Radical Biology and Medicine. 2007;43:1569–1573. doi: 10.1016/j.freeradbiomed.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 158.Shaftel SS, Griffin WS, O’Banion MK. The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. Journal of Neuroinflammation. 2008;5:7. doi: 10.1186/1742-2094-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raina AK, Templeton DJ, Deak JC, et al. Quinone reductase (NQO1), a sensitive redox indicator, is increased in Alzheimer’s disease. Redox Reports. 1999;4:23–27. doi: 10.1179/135100099101534701. [DOI] [PubMed] [Google Scholar]
- 160.Wang Y, Santa-Cruz K, DeCarli C, Johnson JA. NAD(P)H:quinone oxidoreductase activity is increased in hippocampal pyramidal neurons of patients with Alzheimer’s disease. Neurobiology of Aging. 2000;21:525–531. doi: 10.1016/s0197-4580(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 161.SantaCruz KS, Yazlovitskaya E, Collins J, et al. Regional NAD(P)H:quinone oxidoreductase activity in Alzheimer’s disease. Neurobiology of Aging. 2004;25:63–69. doi: 10.1016/s0197-4580(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 162.Kanninen K, Malm TM, Jyrkkänen HK, et al. Nuclear factor erythroid 2-related factor 2 protects against beta amyloid. Molecular and Cellular Neurosciences. 2008;39:302–313. doi: 10.1016/j.mcn.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 163.Resende R, Moreira PI, Proença T, et al. Brain oxidative stress in a triple-transgenic mouse model of Alzheimer disease. Free Radical Biology and Medicine. 2008;44:2051–2057. doi: 10.1016/j.freeradbiomed.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 164.Wruck CJ, Götz ME, Herdegen T, et al. Kavalactones protect neural cells against amyloid beta peptide-induced neurotoxicity via extracellular signal-regulated kinase 1/2-dependent nuclear factor erythroid 2-related factor 2 activation. Molecular Pharmacology. 2008;73:1785–1795. doi: 10.1124/mol.107.042499. [DOI] [PubMed] [Google Scholar]
- 165.Brennan WA, Jr, Bird ED, Aprille JR. Regional mitochondrial respiratory activity in Huntington’s disease brain. Journal of Neurochemistry. 1985;44:1948–1950. doi: 10.1111/j.1471-4159.1985.tb07192.x. [DOI] [PubMed] [Google Scholar]
- 166.Gu M, Gash MT, Mann VM, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Annals of Neurology. 1996;39:385–389. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 167.Tabrizi SJ, Workman J, Hart PE, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Annals of Neurology. 2000;47:80–86. doi: 10.1002/1531-8249(200001)47:1<80::aid-ana13>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 168.Bogdanov MB, Andreassen OA, et al. Increased oxidative damage to DNA in a transgenic mouse model of Huntington’s disease. Journal of Neurochemistry. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 169.Schapira AH. Oxidative stress and mitochondrial dysfunction in neurodegeneration. Current Opinion in Neurology. 1996;9:260–264. doi: 10.1097/00019052-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 170.Brouillet E, Condé F, Beal MF, Hantraye P. Replicating Huntington’s disease phenotype in experimental animals. Progress in Neurobiology. 1999;59:427–468. doi: 10.1016/s0301-0082(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 171.Shih AY, Imbeault S, Barakauskas V, et al. Induction of the Nrf2-driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. Journal of Biological Chemistry. 2005;280:22925–22936. doi: 10.1074/jbc.M414635200. [DOI] [PubMed] [Google Scholar]
- 172.Calkins MJ, Jakel RJ, Johnson DA, et al. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:244–249. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shih AY, Li P, Murphy TH. A small-molecule-inducible Nrf2-mediated antioxidant response provides effective prophylaxis against cerebral ischemia in vivo. Journal of Neuroscience. 2005;25:10321–10335. doi: 10.1523/JNEUROSCI.4014-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhao J, Kobori N, Aronowski J, Dash PK. Sulforaphane reduces infarct volume following focal cerebral ischemia in rodents. Neuroscience Letters. 2006;393:108–112. doi: 10.1016/j.neulet.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 175.Zhao X, Sun G, Zhang J, et al. Transcription factor Nrf2 protects the brain from damage produced by intracerebral hemorrhage. Stroke. 2007;38:3280–3286. doi: 10.1161/STROKEAHA.107.486506. [DOI] [PubMed] [Google Scholar]
- 176.Wang J, Fields J, Zhao C, et al. Role of Nrf2 in protection against intracerebral hemorrhage injury in mice. Free Radical Biology and Medicine. 2007;43:408–414. doi: 10.1016/j.freeradbiomed.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Satoh T, Okamoto SI, Cui J, et al. Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophilic [correction of electrophillic] phase II inducers. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Satoh T, Kosaka K, Itoh K, et al. Carnosic acid, a catechol-type electrophilic compound, protects neurons both in vitro and in vivo through activation of the Keap1/Nrf2 pathway via S-alkylation of targeted cysteines on Keap1. Journal of Neurochemistry. 2008;104:1116–1131. doi: 10.1111/j.1471-4159.2007.05039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhao J, Moore AN, Redell JB, Dash PK. Enhancing expression of Nrf2-driven genes protects the blood brain barrier after brain injury. Journal of Neuroscience. 2007;27:10240–10248. doi: 10.1523/JNEUROSCI.1683-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhao J, Moore AN, Clifton GL, Dash PK. Sulforaphane enhances aquaporin-4 expression and decreases cerebral edema following traumatic brain injury. Journal of Neuroscience Research. 2005;82:499–506. doi: 10.1002/jnr.20649. [DOI] [PubMed] [Google Scholar]
- 181.Qiang W, Kuang X, Liu J, et al. Astrocytes survive chronic infection and cytopathic effects of the ts1 mutant of the retrovirus Moloney murine leukemia virus by upregulation of antioxidant defenses. Journal of Virology. 2006;80:3273–3284. doi: 10.1128/JVI.80.7.3273-3284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jiang Y, Scofield VL, Yan M, et al. Retrovirus-induced oxidative stress with neuroimmunodegeneration is suppressed by antioxidant treatment with a refined monosodium alpha-luminol (Galavit) Journal of Virology. 2006;80:4557–4569. doi: 10.1128/JVI.80.9.4557-4569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Min KJ, Yang MS, Kim SU, et al. Astrocytes induce hemeoxygenase-1 expression in microglia: a feasible mechanism for preventing excessive brain inflammation. Journal of Neuroscience. 2006;26:1880–1887. doi: 10.1523/JNEUROSCI.3696-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Innamorato NG, Rojo AI, García-Yagüe AJ, et al. The transcription factor Nrf2 is a therapeutic target against brain inflammation. Journal of Immunology. 2008;181:680–689. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
Further reading, resources and contacts
Publications
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Advances in Enzyme Regulation. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. The authors extensively review the available information on Nrf2-Keap1 interaction and the mechanisms of Nrf2 activation. [DOI] [PubMed] [Google Scholar]
- Satoh T, Lipton SA. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends in Neuroscience. 2007;30:37–45. doi: 10.1016/j.tins.2006.11.004. This article reviews the chemical biology of electrophiles and their neuroprotective effects. [DOI] [PubMed] [Google Scholar]
- Patenaude A, Murthy MR, Mirault ME. Emerging roles of thioredoxin cycle enzymes in the central nervous system. Cellular and Molecular Life Sciences. 2005;62:1063–1080. doi: 10.1007/s00018-005-4541-5. This article reviews the importance of this family of enzymes that catalyze the reduction of protein disulfide bonds and its role in neuroprotection. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- The website of the United States National Institute of Neurological Disorders and Stroke provides information for all major neurodegenerative diseases, possible treatments, current clinical trials information and contact information for organisations dedicated to a specific neurodegenerative disease: http://www.ninds.nih.gov/
- Downloadable microarray data from several experimental situations where Nrf2 activation in astrocytes resulted in neuroprotection is available from: http://www.pharmacy.wisc.edu/facstaff/sciences/JohnsonGroup/microdata.cfm