Abstract
The proto-oncogene PTTG and its binding partner PBF have been widely studied in multiple cancer types, particularly thyroid and colorectal, but their combined role in tumourigenesis is uncharacterised. Here, we show for the first time that together PTTG and PBF significantly modulate DNA damage response (DDR) genes, including p53 target genes, required to maintain genomic integrity in thyroid cells. Critically, DDR genes were extensively repressed in primary thyrocytes from a bitransgenic murine model (Bi-Tg) of thyroid-specific PBF and PTTG overexpression. Irradiation exposure to amplify p53 levels further induced significant repression of DDR genes in Bi-Tg thyrocytes (P=2.4x10-4) compared to either PBF- (P=1.5x10-3) or PTTG-expressing thyrocytes (P=NS). Consistent with this, genetic instability was greatest in Bi-Tg thyrocytes (mean GI index of 35.8±2.6%), as well as significant induction of gross chromosomal aberrations in thyroidal TPC-1 cells following overexpression of PBF and PTTG. We extended our findings to human thyroid cancer using TCGA datasets (n=322) and found striking correlations with PBF and PTTG expression in well-characterised DDR gene panel RNA-seq data. In addition, genetic associations and transient transfection identified PBF as a downstream target of the RTK-BRAF signalling pathway, emphasising a role for PBF as a novel component in a pathway well-described to drive neoplastic growth. We also showed that overall survival (P=1.91x10-5) and disease-free survival (P=4.9x10-5) was poorer for TCGA patients with elevated tumoural PBF/PTTG expression and mutationally activated BRAF. Together our findings indicate that PBF and PTTG have a critical role in promoting thyroid cancer that is predictive of poorer patient outcome.
Keywords: DNA repair, p53, thyroid, PTTG1IP, DNA damage
Introduction
The world-wide incidence of thyroid cancer has risen sharply in the past decade1 with an estimated 637,115 people living with the disease in the US in 2013. The 5-year survival rate of papillary thyroid carcinomas (PTCs) is high at 98.1%; however PTCs can differentiate into more aggressive and lethal thyroid cancers especially in individuals aged >45 years.2 In addition, recurrence of thyroid cancer has been estimated at up to nearly 30% of cases.3 Recent progress in large scale multi-platform studies has identified 96.5% of oncogenic drivers in PTCs.4 The emerging complexity in molecular subtypes of PTCs however, with four newly-identified subgroups of BRAF-mutant tumours,4 represents a significant challenge in developing targeted therapies based on genomic insight. A better understanding will be needed of interactions between key effectors of tumourigenesis that influence molecular subgroups to enable development of effective treatments.
Previous studies by our group5,6 and others7 implicated the proto-oncogene PTTG and its binding partner PBF in thyroid cancer but their precise roles in tumourigenesis remain relatively undefined. PTTG is the better characterised proto-oncogene and has been studied recently in a broad range of tumour types such as breast,8,9 colorectal10 and ovarian.11 In particular, PTTG is a complex, multifunctional protein involved in a wide range of processes such as cell cycle regulation, invasion, genetic instability, senescence and metabolism.8,10–14 Also known as the human securin, PTTG ensures the meticulous segregation of chromosomes during mitosis,15 and both over- and under-expression cause genetic instability.16,17 The central functions of PTTG require its presence in the nucleus and yeast 2 hybrid screening initially identified PBF, which contains a bipartite nuclear localisation signal, as a specific interacting partner of PTTG.18 Thus PBF became an obligate model of subcellular shuttling for PTTG, facilitating its nuclear transport.
Also known as PTTG1IP, PBF had not been studied functionally before it was identified as a binding partner for PTTG. However, similar to PTTG, PBF demonstrates transforming ability and is overexpressed in a growing number of tumour types.6,19,20 PBF was recently identified as a central driver gene in human cancer as discerned across 34 tumour types via DOTS-Finder bioinformatic analysis.21 We initially highlighted a role for PBF in tumourigenesis as subcutaneous PBF overexpression in fibroblasts induced high-grade malignant tumour formation in athymic nude mice.6 However, a thyroid-specific transgenic model (PBF-Tg) showed significant hyperplastic thyroid growth but did not exhibit frequent tumours.22
Inactivation of the tumour suppressor protein p53 is a critical event in the pathogenesis of most cancers.23 Loss of p53 function, for instance, has been linked to genomic instability, insensitivity to apoptotic signals, invasiveness and altered cellular metabolism.24 The role of p53 in differentiated thyroid cancer (DTC) is unclear due to a very low prevalence (0.8%) of TP53 mutations.4 PTTG has been reported to bind directly to p53 and reduce the activity of p53-specific promoters,25 including p53-regulated transcription of PTTG-targeting miRNAs in pituitary cells.26 More recently, we identified PBF as a negative regulator of p53 in thyroid27 and colorectal cancer,28 with implications for impairing the DNA damage response (DDR) and increased tumour invasiveness. Given these observations, the relationship between PBF and PTTG warrants further investigation in vivo to determine the extent to which they interact to modulate the DDR, including the role of p53 target genes, and thus promote thyroid cancer progression.
Our study now suggests that PBF and PTTG modulate DDR genes to significantly increase genomic instability - an underlying mechanism implicated in thyroid cancer progression. In particular, we provide extensive insight into DNA repair genes affected by PBF and PTTG in bi-transgenic mice, as well as in human thyroid cancer TCGA data. We further characterise genetic alterations associated with PBF and PTTG, which implicates PBF as a downstream target of the RTK-BRAF signalling pathway that drives neoplastic growth in multiple different cancer types. These findings have implications for using PBF and PTTG to identify patients with more aggressive thyroid cancer.
Results
Enlarged thyroids in a murine model of targeted induction of PBF and PTTG
To investigate PBF and PTTG in thyroid cells, we constructed a bitransgenic model (Bi-Tg) of thyroid-specific PBF and PTTG over-expression by crossing two FVB/N murine models of human PBF (PBF-Tg) and PTTG (PTTG-Tg) transgenes under the control of the bovine thyroglobulin promoter (Figure 1a). Greater expression of PTTG did not appear to increase PBF protein levels and vice versa (Figure 1b). In keeping with previous data, expression of both proto-oncogenes was confined to the thyroid gland (Supplementary Figure S1a). Examination of body weight did not reveal any significant differences between Bi-Tg and wild-type (WT) mice (Supplementary Figure S1b).
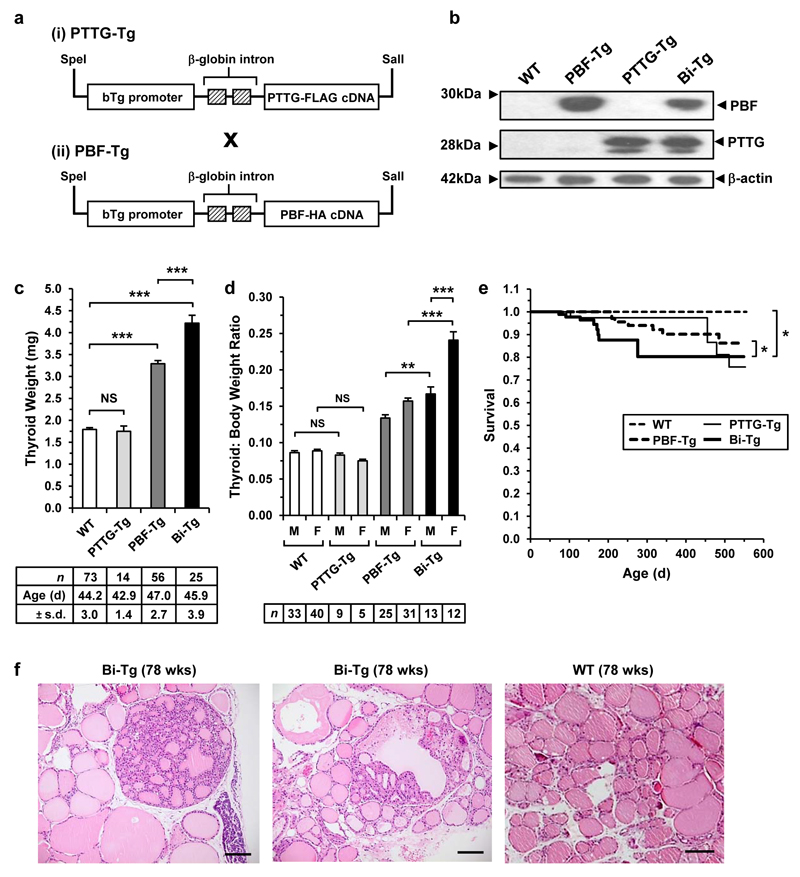
Figure 1. Enlargement of thyroid glands in Bi-Tg mice.
(a) Schematic of the bovine Tg-PTTG-FLAG and Tg-PBF-HA transgenes. (b) Detection of PBF and PTTG expression by Western blot analysis of WT and transgenic thyroids. (c) Thyroid weight of WT and transgenic mice (mean±s.d., number (n) and mean age (d) of mice are indicated, unpaired two-tailed t-test) (NS, not significant; ***P<0.001). (d) Thyroid: body weight ratios of male (M) and female (F) transgenic mice at 7 weeks of age (mean±s.d., number (n) of mice are indicated, unpaired two-tailed t-test) (NS, not significant; **P<0.01; ***P<0.001). (e) Kaplan-Meier survival curves for WT (n=30), PTTG-Tg (n=129), PBF-Tg (n=82) and Bi-Tg (n=89) mice. P-values were determined using the log-rank test (*P<0.05). (f) Representative H&E stained images of hyperplastic lesions in Bi-Tg thyroids from 78-week old mice. WT thyroid follicles (right) are shown for comparison. Scale bar, 100 μm.
A striking phenotype of Bi-Tg mice was a significantly greater thyroid weight than in all other genotypes (Figure 1c). Bi-Tg thyroid glands, for instance, were up to 2.4-fold heavier than those of either PTTG-Tg or WT littermates (Figure 1c; P<0.001), and >50% heavier than in age-matched female PBF-Tg mice (Figure 1d; P<0.001). Indeed, simultaneous induction of both proto-oncogenes exceeded the additive effect expected from combined over-expression since the thyroid weight of PTTG-Tg mice was not significantly different compared to WT (Figure 1c and d). A distinctive characteristic of Bi-Tg mice was significantly reduced survival compared to WT (P<0.05), PBF-Tg (P<0.05) and PTTG-Tg (P<0.05) littermates up to 400 days of age (Figure 1e). No consistent physiological causes of this increased mortality were identified.
In terms of thyroid function, the only significant alteration in serum TSH across genotypes and genders was in female PTTG-Tg mice compared to age- and sex-matched WT (Supplementary Figure S1c; P<0.05). Total T3 and T4 were induced in Bi-Tg and PTTG-Tg males (Supplementary Figures S1d and e), and total T3 was marginally higher in Bi-Tg females than WT females (Supplementary Figure S1d). However, the significantly altered thyroid sizes apparent in Bi-Tg mice were not explained by consistent changes in TSH, T3 and T4 levels. Similar to observations in PBF-Tg thyroids,22 microscopic examination of Bi-Tg thyroids (Figure 1f and Supplementary Figures S2a-e) demonstrated a high occurrence of hyperplastic lesions by 78 weeks of age, especially in females (>80% occurrence) compared with sex-matched WT (Supplementary Figure S2f; P<0.05). In contrast, PTTG-Tg thyroids in aged mice retained normal cellular architecture (Supplementary Figure S2g). Positive immunostaining with the proliferation markers cyclin D1 (Supplementary Figure S3) and cyclin A (Supplementary Figure S4) further demonstrated that PBF and PTTG co-expression resulted in increased thyroid cell proliferation in vivo.
Taken together our results show that thyroidal co-expression of PBF and PTTG in vivo gives rise to mice phenotypically more similar to PBF-Tg than PTTG-Tg mice, but with significant differences in thyroid gland size and overall survival.
Modulation of DDR gene expression by raised PBF and PTTG
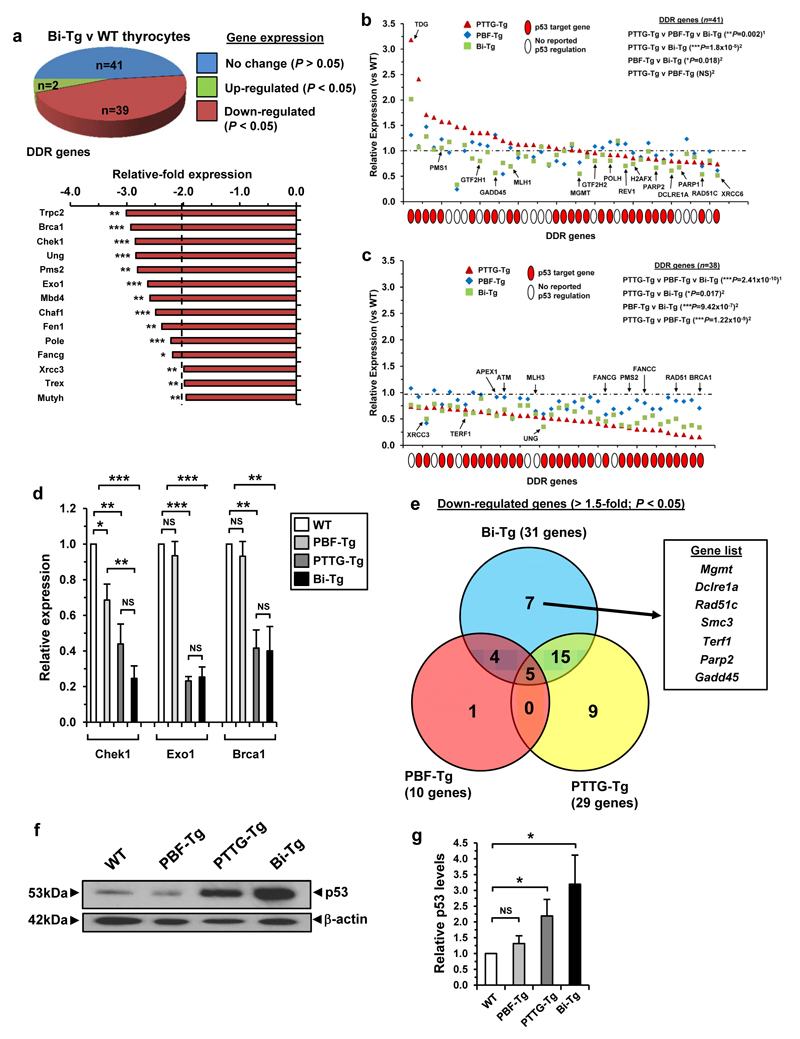
Separate studies have implicated PBF and PTTG in tumourigenesis due to their ability to regulate p53 genes involved in DDR, cell-cycle and apoptotic pathways.27,29 Having established the Bi-Tg murine model, we next characterised the impact of simultaneous induction of these proto-oncogenes on DDR genes in primary thyrocytes. Importantly, significant changes in gene expression occurred in Bi-Tg thyrocytes in response to raised PTTG and PBF compared to WT (Figure 2a; n=41/82 genes; P<0.05). The majority of genes were down-regulated >1.5-fold (Supplementary Figure S5a and Figure 2b and c; n=31), including those known to maintain genomic integrity, such as Brca1 (2.9 ± 0.1 fold; P<0.001) and Pms2 (2.8 ± 0.1 fold; P<0.01). Overall the DDR transcriptional signature in Bi-Tg thyrocytes was distinct compared to either PTTG-Tg or PBF-Tg (Supplementary Figure S5b). In particular, there was significant repression for a subset of 41 DDR genes with relative expression >0.8 (range 0.8-3.2 in PTTG-Tg) between Bi-Tg and PTTG-Tg thyrocytes (Figure 2b and Supplementary Figure S5c; P=1.8x10-5). Further experiments confirmed significant changes in DDR genes Brca1, Chek1 and Exo1 between genotypes (Figure 2d). Seven genes with >1.5-fold reduction were identified solely in Bi-Tg thyrocytes (Figure 2e), of which six were p53 target genes (Supplementary Figure S6 and Supplementary Table S1) such as Mgmt that has been associated with increased genomic instability in thyroid carcinoma.30
Figure 2. Impaired DDR gene expression in Bi-Tg thyroids.
(a) Pie chart summarizes number of DDR gene expression changes between thyrocytes from male Bi-Tg and WT mice (n=3 arrays). Graph (below) shows DDR genes with mRNA repressed ≥2.0-fold in Bi-Tg thyrocytes (mean, n=3, unpaired two-tailed t-test) (*P<0.05; **P<0.01; ***P<0.001). (b) Transcriptional signature of subset of DDR genes in PTTG-Tg (triangles), PBF-Tg (diamonds) and Bi-Tg (squares) thyrocytes with relative expression >0.8 (range 0.8-3.2 in PTTG-Tg) (mean, n=3 arrays, 1Kruskal-Wallis test, 2Mann-Whitney test) (NS, not significant; *P=0.018; **P=0.002; ***P=1.8x10-5). Red filled circle indicates p53 target gene. SRD5A2, TNP1 and RBBP4 from (a) were not included due to lack of expression in all 4 genotypes. (c) Same as (b) except using subset of DDR genes with relative expression <0.8 (range 0.15-0.8 in PTTG-Tg) (*P=0.017; ***P values are shown). (d) qPCR analysis of Chek1, Exo1 and Brca1 expression in thyrocytes of indicated genotypes (mean±s.e.m., n=4, unpaired two-tailed t-test) (NS, not significant; *P<0.05; **P<0.01; ***P<0.001). (e) Venn diagram of DDR genes with repressed expression (>1.5-fold; *P<0.05) for thyrocytes of indicated genotype compared to WT (n=3). Genes listed are unique to Bi-Tg thyrocytes (box). (f) Western blot of p53 levels in thyroid gland lysates for each genotype as indicated. Blot shown is representative from 4 independent experiments. (g) Quantification of mean p53 protein levels relative to β-actin (mean±s.e.m., n=4, unpaired two-tailed t-test) (NS, not significant; *P<0.05).
Western blot analysis indicated that p53 protein levels were elevated in PTTG-Tg (2.2±0.5-fold; P<0.05) and Bi-Tg (3.2±0.9-fold; P<0.05) thyroids which might account for some transcriptional differences (Figure 2f and g). An intense pattern of pChek1(Ser 345) expression in Bi-Tg thyrocytes implicated replication stress as a potential explanation for p53 activation (Supplementary Figure S7). Interestingly, p53 protein was not altered in PBF-Tg thyroids (Figure 2f).
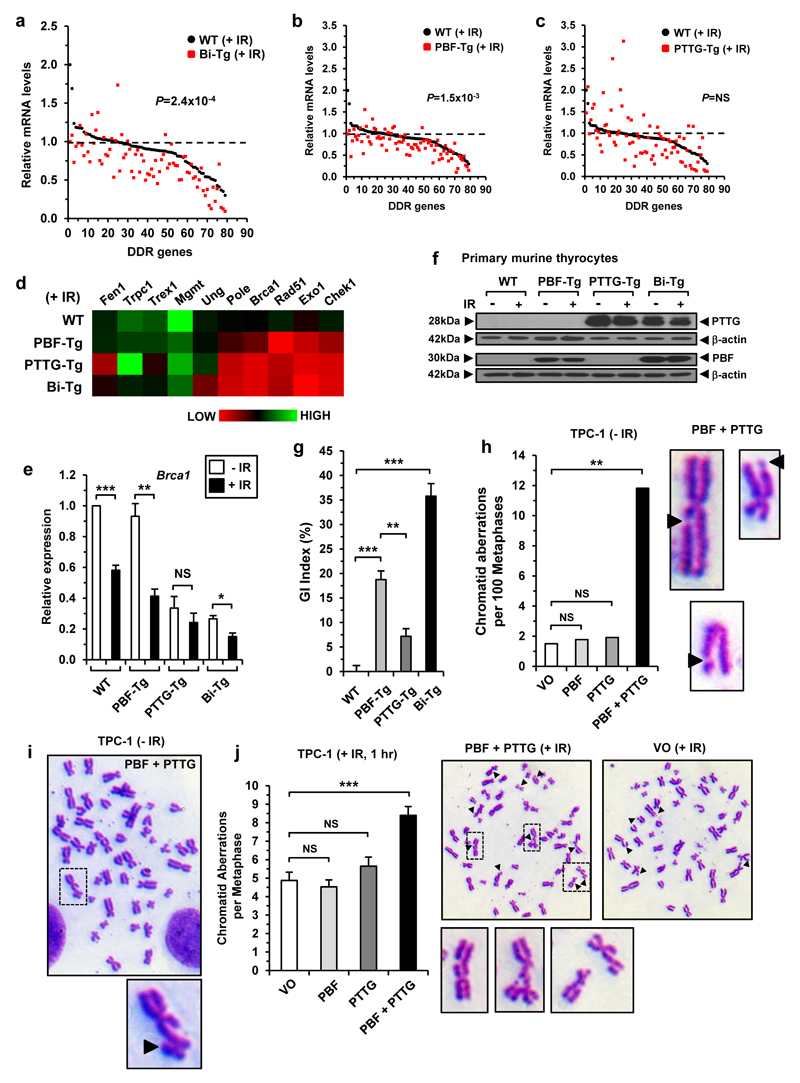
To examine the influence of p53 further we irradiated thyrocytes to amplify p53 levels in response to DNA damage. The most significant overall reduction for DDR genes following DNA damage was between irradiated Bi-Tg and WT thyrocytes (P=2.4x10-4) compared with either PBF-Tg (P=1.5x10-3) or PTTG-Tg thyrocytes (P=NS) (Figure 3a-d). Importantly, subsequent validation confirmed that DDR genes Chek1, Brca1, Rad51 and Exo1 were most highly suppressed in irradiated Bi-Tg thyrocytes (Figure 3e and Supplementary Figure S8). In control experiments irradiation did not alter protein expression of PBF and PTTG (Figure 3f).
Figure 3. Transcriptional signature of DDR genes in irradiated murine thyrocytes.
(a) DDR gene expression profile of irradiated (+IR) Bi-Tg and WT thyrocytes (mean, n=3 arrays, Mann-Whitney test, ***P=2.4x10-4). (b) Same as (a) except DDR expression profile shown between irradiated PBF-Tg and WT thyrocytes. (c) Same as (a) except DDR expression profile shown between irradiated PTTG-Tg and WT thyrocytes. (d) Heatmap showing relative expression levels for 10 DDR genes between indicated genotypes. (e) Relative fold change in Brca1 expression in irradiated (+IR) thyrocytes of each genotype versus non-irradiated (-IR) controls (mean±s.e.m., n=4, unpaired two-tailed t-test) (NS, not significant; *P<0.05; **P<0.01; ***P<0.001). (f) Representative Western blot of PTTG and PBF in irradiated (+IR) thyrocytes of each genotype relative to untreated controls (-IR). (g) Quantification of genetic instability (GI) in murine primary thyrocytes for each genotype (mean±s.e.m., n=5, unpaired two-tailed t-test) (**P<0.01; ***P<0.001). (h) Chromosomal aberrations in TPC-1 cells at 48 h post-transfection (VO, n=69; PBF, n=56; PTTG, n=52; PBF+PTTG, n=93; Fisher’s exact test). Representative images of PBF+PTTG-induced chromatid gaps are shown (right, black arrowheads) (i) Representative metaphase spread of TPC-1 cells transfected with PBF+PTTG. Boxed area is magnified showing chromatid break (black arrowhead). (j) Chromatid aberrations in TPC-1 cells irradiated with 1 Gy dose at 48 h post-transfection (mean±s.e.m., VO, n=32; PBF, n=34; PTTG, n=31; PBF+PTTG, n=47; unpaired two-tailed t-test) (NS, not significant; ***P<0.001). Representative metaphase spreads for irradiated TPC-1 cells transfected with PBF+PTTG or VO are shown. Boxed areas are magnified and show representative chromatid aberrations (i.e. gaps, exchanges and breaks respectively). Black arrowheads indicate chromatid aberrations.
Given the extent of suppression of DDR genes, we next used fluorescent inter simple sequence repeat (FISSR) PCR to determine whether there was any associated impact on genetic instability with PBF and PTTG.16,17 In comparison to primary thyrocytes derived from age- and sex-matched WT mice (arbitrarily assigned a GI of 0%), Bi-Tg thyrocytes demonstrated the most disrupted genome, with a mean GI index of 35.8±2.6% (Figure 3g; P<0.001). This level of GI was significantly greater than in the genomes for PBF-Tg (18.8±1.8%; P<0.01) and PTTG-Tg (7.2±1.6%; P<0.001) thyrocytes. To determine whether PBF and PTTG might also contribute to gross chromosomal aberrations we next examined metaphases of thyroidal TPC-1 cells. Strikingly, co-expression of PBF and PTTG produced >6-fold increase in the number of chromatid aberrations compared to PBF-, PTTG- or VO- control cells in the absence of irradiation (Figure 3h-i and Supplementary Figure S9a; P<0.01). Similarly, chromosomal aberrations in PBF+PTTG-overexpressing TPC-1 cells irradiated with a dose of 1 Gy were significantly greater than in all controls (Figure 3j; P<0.001). Further studies confirmed that PBF and PTTG suppressed DDR genes in TPC-1 cells (Supplementary Figure S9b).
Collectively, these results provide evidence for the ability of PBF and PTTG together to drive genomic instability in thyroid cells that may be due to alterations in the regulation of DDR genes required for normal replication or impaired p53-dependent signalling.
Elevated PTTG and PBF are associated with DDR genes in DTC
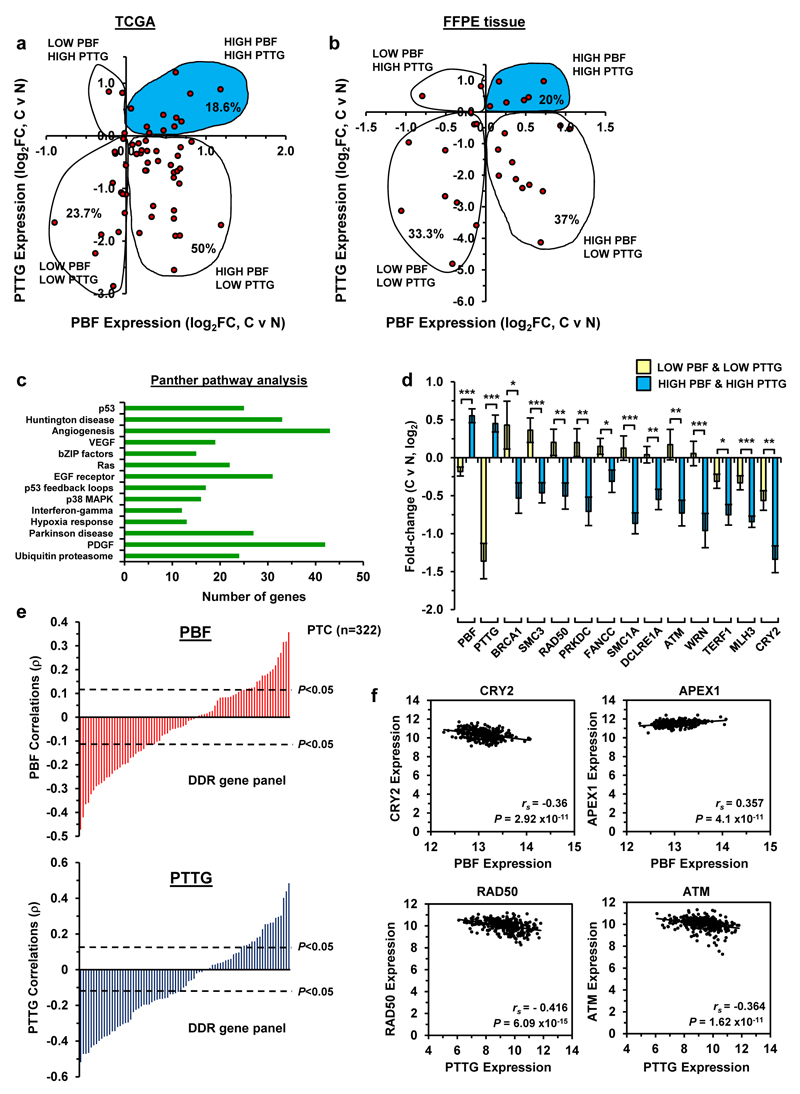
Whilst PBF and PTTG have been reported individually to be overexpressed in human DTC, there is a lack of information regarding their co-expression in tumours. To extend our findings to human DTC, we examined PBF and PTTG expression by RNA-seq (TCGA) in 59 matched thyroid tumour and normal samples.4 We found that PBF and PTTG expression were simultaneously up-regulated in 18.6% of DTC cases (Figure 4a; n=11/59), while PBF was higher in ~70% of tumours (n=41/59) compared to 24% with positive PTTG expression (Figure 4a; n=14/59). The subsequent challenging of our findings in our in-house matched FFPE tumour and normal DTC demonstrated a comparable number of tumours (n=6/30; 20%) with positive PBF and PTTG expression (Figure 4b).
Figure 4. Significant association between PBF and PTTG with DDR genes.
(a) Scatterplot showing fold-change (FC) in PBF and PTTG expression in DTC versus matched normal samples (log2, n=59, TCGA dataset). (b) Same as (a) except PBF and PTTG expression in matched DTC and normal samples from FFPE tissue (log2, n=30). Shaded areas in (a) and (b) indicate DTC with high PBF/PTTG expression. (c) DAVID analysis of differentially expressed genes in matched DTC and normal samples with low (n=15) versus high (n=11) PBF/PTTG-expressing tumours. P<0.05 for all subgroups (modified Fisher’s exact test). (d) Fold-change (FC) in expression of DDR genes repressed in DTC with high PBF/PTTG (n=11) versus DTC with low PBF/PTTG (n=15) (mean log2FC±s.e.m., Mann-Whitney test) (*P<0.05; **P<0.01; ***P<0.001; see Supplementary Figure S12 for additional data). (e) Correlation values (ρ) for PBF (top) and PTTG (bottom) in a panel of DDR genes (n=82) using thyroid TCGA RNA-seq data (n=322). P and ρ values were calculated using Spearman’s correlation tests. (f) Correlation of PBF and PTTG expression with indicated DDR genes in human thyroid cancer TCGA dataset (n=322, Spearman’s correlation test).
Having identified a subset of DTC with elevated PBF and PTTG expression, we investigated their relationship with DDR and p53 target genes. DAVID analysis31,32 of differentially expressed genes confirmed the involvement of genes in DNA repair and p53 pathways by comparing DTC with either low or high PBF/PTTG expression (Figure 4c and Supplementary Figures S10 and S11). Importantly, DDR genes previously identified in Bi-Tg thyrocytes (Figure 2a) were altered significantly between DTC subsets (Figure 4d and Supplementary Figure S12; n=18/41 genes). Further evaluation revealed significant correlations between DDR genes with PBF (Supplementary Figure S13a; n=13/18 genes) and PTTG alone (Supplementary Figure S13b; n=16/18 genes). In addition, there were striking correlations with PBF and PTTG in well-characterised DDR and p53 target gene panels (>60% of genes; 81-95 genes per panel; P<0.05) using unmatched TCGA samples (Figure 4e and Supplementary Figures S14 and S15; n=322). The most significant correlations with PBF and PTTG, for instance, were observed in DTC with Cry2, Apex1, Rad50 and Atm (Figure 4f). A list of differentially expressed genes in DTC with high PBF/PTTG is provided (Supplementary Table S2). Altogether these findings demonstrate significant associations between PBF and PTTG with DDR and p53 target genes (Supplementary Table S1) in human DTC, as well as in the murine Bi-Tg thyroid model.
Association of genetic abnormalities with PBF and PTTG
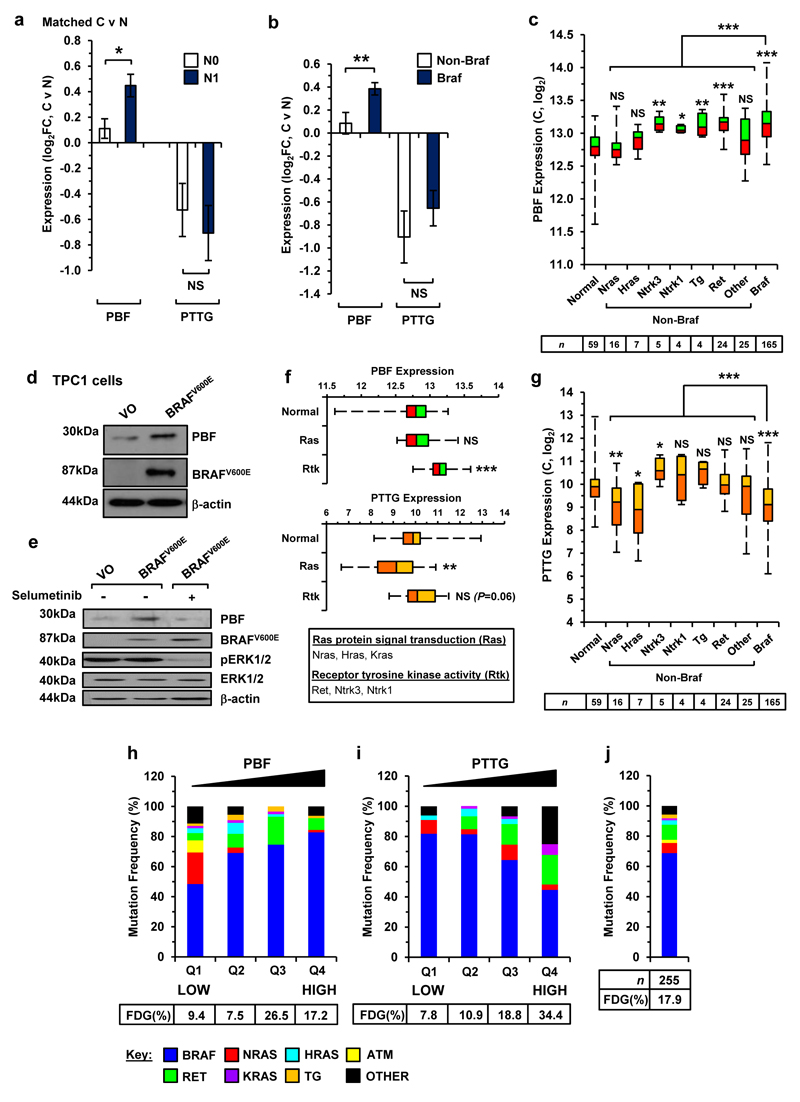
Genomic profiling has revealed extensive diversity in mutations and fusion driver genes associated with thyroid cancer.4 To gain insights into alterations that might influence PBF and PTTG in the genetically diverse tumour environment, we next correlated their expression with invasive properties and mutational status in DTC. We found that PBF expression was significantly greater in regional metastatic DTC (N1) for matched tumour and normal samples in TCGA (Figure 5a; P<0.05) and FFPE thyroid specimens (Supplementary Figure S16; P<0.05) than in non-metastatic DTC (N0). Of particular significance, PBF expression was higher in BRAF-mutant DTC compared to non-BRAF variants (Figure 5b; P<0.01). Similarly, PBF expression was elevated in BRAF-mutant DTC in a larger cohort of unmatched TCGA cases (Figure 5c; P<0.001 vs non-BRAF; n=255). These findings paralleled our in vitro studies in which BRAFV600E over-expression induced PBF in TPC-1 cells (Figure 5d). In agreement with this, the addition of the MEK inhibitor selumetinib effectively blocked induction of PBF protein by BRAFV600E (Figure 5e). Control experiments confirmed that PBF and PTTG overexpression did not activate MEK/ERK signalling (Supplementary Figures S9b and S17a).
Figure 5. Genetic alterations associated with PBF and PTTG.
(a) Fold-change (FC) in PBF and PTTG expression in metastatic (N1) and non-metastatic (N0) DTC relative to matched normal tissue (N) (mean±s.e.m., N0, n=23; N1, n=21; Mann-Whitney test) (NS, not significant; *P<0.05). (b) Fold-change in PBF and PTTG expression in Braf-mutant and non-Braf mutant DTC relative to matched normal tissue (mean±s.e.m., non-Braf, n=21; Braf, n=32; Mann-Whitney test) (NS, not significant; **P<0.01). (c) Box-whisper plots of PBF expression (log2) in unmatched DTC with the indicated genetic alteration and number of samples (below) (NS, not significant; *P<0.05; **P<0.01; ***P<0.001; Mann-Whitney test). (d) Western blot analysis of PBF and BRAFV600E in TPC-1 cells transfected with either VO or BRAFV600E. (e) Western blot analysis of PBF, BRAFV600E, pERK1/2, and ERK1/2 in TPC-1 cells transfected with either VO or BRAFV600E and then incubated with 200 nM selumetinib. (f) Expression of PBF (top) and PTTG (bottom) in unmatched DTC with the indicated subgroup of genetic alteration (Normal, n=59; Ras, n=25; Rtk, n=33; Mann-Whitney test) (NS, not significant; **P<0.01; ***P<0.001). (g) Same as (c) but instead showing PTTG expression in unmatched DTC samples. (h) Frequency (%) of indicated genetic alterations in unmatched DTC with different quartiles (Q1-Q4) of PBF expression. Frequency of fusion driver genes (FDG) in each quartile is shown (below) (Q1, Q2 and Q4, n=64; Q3, n=63). P-values were determined by Fisher’s exact test. (i) Same as (h) but instead showing mutation frequency (%) present in DTC with different quartiles of PTTG expression. (j) Frequency of mutations and FDG in all 255 unmatched DTC cases.
We also observed that higher PBF was associated with the receptor tyrosine kinase (RTK) genes NTRK1, NTRK3 and RET (Figure 5f; P<0.001) but remained unchanged for RAS genes (Figure 5f; P=NS). In contrast, PTTG expression was lower in both RAS- (Figure 5f; P<0.01) and BRAF-mutant DTC (Figure 5g; P<0.001). The relative levels of PTTG however tended to be raised in RTK-mutant DTC (Figure 5f), which reached significance for NTRK3 fusion genes (Figure 5g; P<0.05). In keeping with this, the highest frequency of BRAF-mutation in DTC (82.8%; n=53/64) corresponded with the top quartile (Q4) of PBF expression compared to 48.4% for DTC with the bottom quartile Q1 (Figure 5h; P<0.0001; n=31/64). By comparison, DTC with top quartile PTTG expression (Figure 5i) had the lowest frequency of BRAF-mutation (44.6% in Q4 vs 81.8% in Q1; P<0.001) and highest frequency of RET alterations (19.6% in Q4 v 0% in Q1; P<0.001). Overall, fusion driver genes (FDG) were ~2-fold more prevalent in DTC with high PTTG expression (Figure 5i; top quartile; 34.4%) compared to all 255 cases of DTC (Figure 5j; 17.9%; P<0.01).
These results demonstrate for the first time that the underlying genetic alteration in DTC is related to distinct patterns of PBF and PTTG expression. Furthermore, PBF overexpression was associated with regional metastatic DTC.
High expression of PBF and PTTG in DTC is associated with poor patient survival
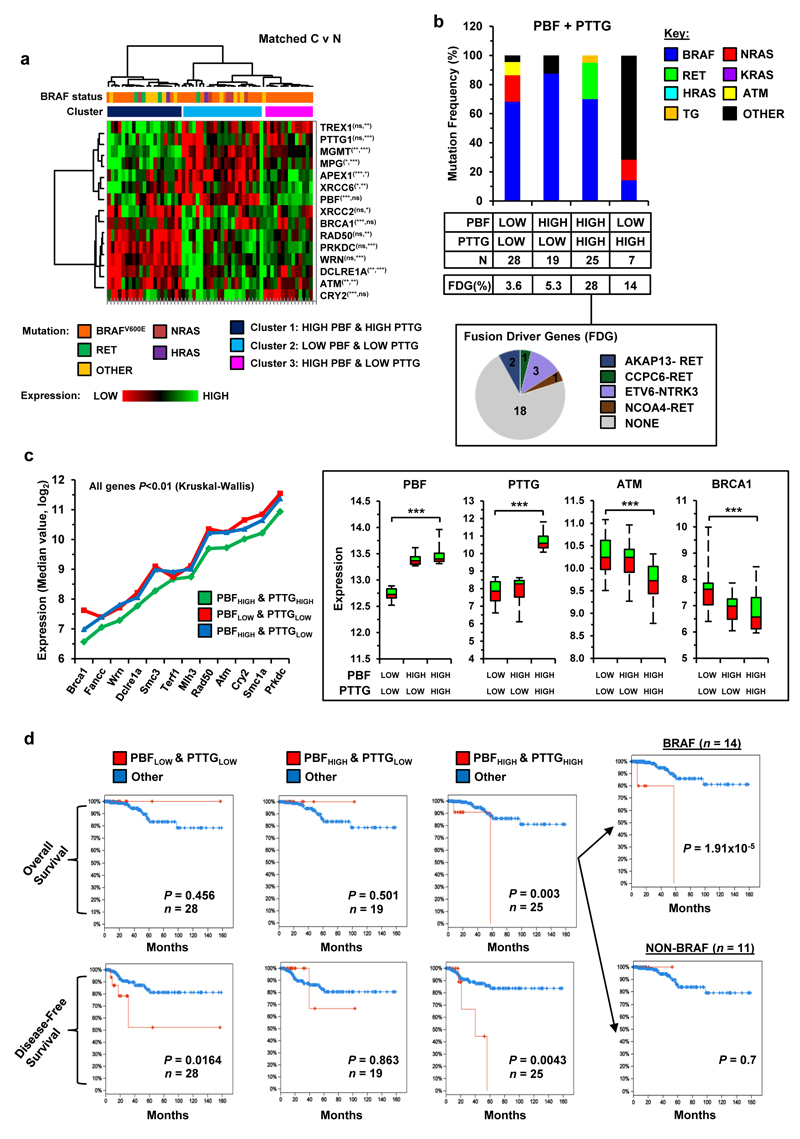
To further define the molecular subtypes of DTC associated with high expression of both PBF and PTTG, we applied hierarchical clustering to matched tumour/normal DTC data using a panel of 13 DDR genes and identified 3 main clusters (Figure 6a; n=59). BRAF (n=10/21) and RET (n=3/21) were frequent alterations in cluster 1 (i.e. elevated PBF and PTTG), while mutated BRAF was present in all cases of DTC in cluster 3 (Figure 6a; i.e. high PBF and low PTTG; n=14/14). The unmatched DTC dataset (n=255) supported these observations with BRAF (70%) and RET (25%) the most common alterations in DTC with elevated PBF and PTTG (Figure 6b; top quartile; n=25), while BRAF was more prominent in DTC (87.5%) with high PBF and low PTTG expression (Figure 6b; n=19). Overall the occurrence of fusion driver genes was greatest in DTC with elevated PBF and PTTG (Figure 6b; 28%), especially the ETV6-NTRK3 rearrangement (n=3/7) which is associated with exposure to 131I.33 In contrast, mutated NRAS was more common in DTC with low PBF and PTTG expression in matched (Figure 6a; cluster 2; n=4/22) and unmatched (Figure 6b; 18.2%; n=28) DTC. Importantly, the expression of DDR genes in unmatched DTC samples was also significantly altered (P<0.05) in DTC with high PBF and PTTG (top quartile) compared to DTC in other expression subsets (Figure 6c and Supplementary Figures S18 and S19), as well as demonstrating unique correlations in a panel of DDR genes (Supplementary Figure S20).
Figure 6. Poor patient survival with high PBF/PTTG-expressing tumours and mutant BRAF.
(a) Hierarchical cluster analysis of matched DTC and normal samples (n=59) based on expression of PBF and PTTG, as well as a panel of 13 DDR genes. Significance of correlation between DDR genes with PBF (left asterisk) and PTTG (right asterisk) are shown (NS, not significant; *P<0.05; **P<0.01; ***P<0.001; Spearman’s correlation test). (b) Frequency (%) of genetic alterations in unmatched DTC with different subsets of PBF and PTTG expression. Number of DTC samples and frequency of fusion driver genes (FDG) per subset are shown. Pie chart (below) summarizes the proportion of FDG in DTC with high PBF/PTTG expression. (c) Reduced expression of a panel of DDR genes in DTC with high PBF/PTTG expression. P-values were determined by the Kruskal-Wallis test (median, PBF/PTTG expression subsets: low/low, n=28; high/low, n=19; high/high, n=25). Example box-whisper plots for PBF, PTTG, ATM and BRCA1 in expression subsets are shown (right) (**P<0.01; ***P<0.001; see Supplementary Figures S18-S20 for additional data). (d) TCGA clinical data showing overall survival (upper) and disease-free survival (lower) curves for DTC of PBF and PTTG expression subsets compared to all DTC cases (n=255). Overall survival curves for BRAF-mutant and non-BRAF mutant DTC with high PBF/PTTG expression are shown (right) (Number (n) of DTC samples per subset are indicated, P-values were determined using the log-rank test).
Having observed strong repression of DDR genes in DTC with elevated PBF and PTTG, we next evaluated whether this correlated with tumour aggressiveness and patient survival. The TCGA clinical dataset showed significantly poorer survival (P=0.003) and disease-free survival (P=0.043) for patients (n=25) with high tumoural PBF and PTTG expression than for all other patients (Figure 6d; n=255). In particular, poor overall survival was a characteristic for patients with BRAF-mutant DTC (P=1.91x10-5) and not non-BRAF mutant DTC (Figure 6d and Supplementary Figure S21; P=NS). Further analysis of BRAF-mutant DTC alone confirmed these observations and showed that PBF and PTTG status were both required to identify aggressive tumours associated with poor patient survival, as well as a higher incidence of stage IVA disease (Supplementary Figure S22a and b). Potential high-risk modifiers (e.g. USP9X) in aggressive BRAF-mutant DTC with elevated PBF and PTTG were also identified (Supplementary Figure S22c). By comparison, high tumoural expression of a panel of proliferation markers was not associated with diminished overall patient survival (Supplementary Figure S23).
Taken together our study reveals that expression patterns of PBF and PTTG in thyroid cancer cells are intimately linked to specific genetic alterations and may promote oncogenic characteristics such as modulating the DNA damage response. We therefore propose that PBF and PTTG have a critical role in promoting thyroid cancer that correlates with poorer patient survival.
Discussion
Differentiated thyroid cancer is the most rapidly increasing cancer in the UK and US, with ~300,000 new cases reported worldwide annually.34 Extensive genomic profiling has recently identified new driver mutations and fusion driver genes in thyroid tumours.4 However, a better understanding of contributory factors that promote aggressive tumours is urgently needed to reduce the >38,000 worldwide deaths from thyroid cancer per annum. In separate studies we previously implicated elevated expression of the proto-oncogene PTTG and its binding partner PBF in thyroid cancer.5,6 Our current findings now demonstrate for the first time a role for PBF and PTTG together in modulating genes associated with the DNA damage response that is predictive of poor survival in thyroid cancer.
Disruption of p53 activity has a critical role in promoting tumourigenesis for most human cancers. This role is relatively undefined in DTC however due to a very low incidence of p53 mutations,4 although a recent study detected p53 mutations in 73% of aggressive anaplastic thyroid carcinomas (ATC).35 In the present study a comparative approach was used in a novel bi-transgenic model and human TCGA data to examine the impact of PBF and PTTG on a panel of DDR genes of which the majority were p53 target genes and known to be impaired in cancer cells.36 A remarkable finding was that many DDR genes were highly repressed in both murine thyrocytes and human thyroid tumours in the presence of raised PBF and PTTG. Our findings therefore provide strong evidence of a model in which overexpression of PBF and PTTG impairs DDR gene activity thus promoting tumour progression.
The association between PTTG and DDR pathways has been described in multiple studies,14 with DDR genes up-regulated in β-cells in PTTG-null mice,29 as well as in PTTG-deficient bone marrow stem cells.37 Therefore, our result of extensive repression of DDR genes in PTTG-Tg thyrocytes mirrors these earlier observations, and provides support for a pivotal role for PTTG in the inhibition of DDR pathways in vivo. Individually, PBF and PTTG both regulate p53,27,38 which plays a key role in eliciting many cellular responses to DNA damage. Here, we found significant induction of p53 protein in PTTG-Tg and Bi-Tg thyroids, which was consistent with greater DDR gene changes and correlated with in vitro studies demonstrating p53 induction by PTTG in transformed cell lines38 and human fibroblasts.13
In contrast, the impact of PBF on p53 and DDR pathways is less well understood due to a lack of mouse knockout models. Here we demonstrated in murine thyrocytes that DDR genes were more highly repressed in the presence of raised PBF and PTTG following irradiation than with either PBF or PTTG alone. In addition, expression of well-characterised panels of DDR and p53 target genes were highly correlated with PBF and PTTG levels in human DTC. The underlying mechanism might share a common pathway especially as both proto-oncogenes bind specifically to p53 and block transcriptional activity.25,27,28 PBF however also interacts with PTTG18 and the ability of PBF to facilitate the translocation of PTTG to the nucleus18 might augment interaction with p53 to repress DDR genes, especially in cells stressed by irradiation with increased nuclear p53. In addition, our study and others39 have implicated PTTG in promoting replication stress which is associated with activation of p53.40 The exact mechanism for PBF regulation of p53-responsive DDR genes in combination with PTTG is therefore likely to be multifaceted and remains to be fully clarified. In particular, it will be important to determine whether PTTG influences the ability of PBF to inhibit p53 transcriptional activity or diminish p53 stability by enhancing ubiquitination27, as well as better defining the contribution of p53-independent pathways on DDR gene modulation.
Inhibition of DDR genes in the tumour environment contributes to genomic instability and promotes cancer progression.41 Given the extent of repression of DDR genes in transgenic thyroids, we examined genetic instability which has previously been reported in thyroid cancer.16,42,43 An important finding was that genetic instability in Bi-Tg thyrocytes was greater than in either PTTG-Tg or PBF-Tg thyrocytes. However, tumour induction was absent in Bi-Tg thyroids, despite extensive hyperplasia, repressed DDR genes and elevated genetic instability. Previously, a PTTG transgenic mouse was crossbred with a p53(+/-) mouse to effectively induce ovarian tumours,44 which infers a contributory role for PTTG. To further define the oncogenic roles of PBF and PTTG in the Bi-Tg mouse it will therefore be necessary to cross-breed with p53-null mice to model ATC or transgenic mice with DTC driver mutations such as BRAFV600E.45
In this study we aimed to correlate our findings in the Bi-Tg mouse model to human DTC. An important consideration was to determine which genetic alterations were associated with high PBF and PTTG expression. In particular, genetic drivers in PTC have distinct signalling consequences and have been categorised into BRAFV600E-like (BVL) and RAS-like (RL) PTCs according to distinct gene signatures.4 In this study high PBF expression was associated predominately with alterations that were highly (i.e. BRAF mutations) or weakly BVL (i.e. RET fusions), as well as a few potentially neutral BVL (i.e. NTRK1/3 fusions). Critically, there was no association with the RL phenotype and RAS mutations. Downstream signalling events in BVL-PTCs are therefore likely to be responsible for increased PBF expression, especially as the top quartile of PBF expression corresponded with a majority of BRAF (82.8%) and RET (7.8%) alterations. Importantly, we were able to confirm that overexpression of BRAFV600E in thyroid cancer cells led to increased PBF protein levels. Together our findings reveal a novel role for PBF as an integral downstream target of the BRAF signalling pathway to promote thyroid tumourigenesis.
In contrast, genetic alterations linked to raised PTTG were less clear, especially as high PTTG expression was detected in just ~25% of DTC compared to almost 70% for PBF. Fusion genes such as ETV6-NTRK3 were associated with high PTTG expression in a small subset of DTC. Low PTTG expression was however predominately associated with mutations that were highly BVL (i.e. BRAF) and RL (i.e. RAS). Importantly, our cluster and quartile expression data analysis showed that BRAF and RET were the main driving events in PTCs with high PBF and PTTG expression. It will be important to establish whether this subset of DTC has a unique molecular profile or corresponds to one of the four distinct classifications of BRAF-mutant PTC.4 In addition, a distinguishing feature was the high prevalence of fusion driver genes in DTC with raised PBF and PTTG compared to other DTC expression subsets, which most likely reflects an increased number of oncogenic chromosomal translocations due to greater disruption of DNA repair processes.46 In support of this, we demonstrated significantly greater levels of gross chromosomal aberrations in thyroid TPC-1 cells by overexpressing PBF and PTTG.
The immediate challenges in thyroid cancer are to identify new biomarkers and molecular targets for small subsets of DTC that are more aggressive and more likely to recur. Here, both the overall survival and disease-free survival rates were significantly reduced in a subset of DTC with high PBF and PTTG expression. These findings therefore suggest that the status of PBF and PTTG should together be regarded as an important clinical indicator for aggressive thyroid disease. In particular, diminished patient survival was a characteristic of BRAF-mutant DTC and not tumours with RET fusion genes. Further evidence linking DDR genes and tumour aggressiveness was revealed by a panel of DDR genes identified in murine Bi-Tg thyroids, including cancer-associated genes such as Brca1 and Atm, which were also significantly repressed in human DTC with raised PBF and PTTG.
In summary, our study based on comparative analysis of mice and human data demonstrates that PBF and PTTG have a pivotal role in modulating DDR genes in thyroid cells, as well as altering genomic instability, both characteristics that expedite malignant initiation and progression.24,41 Indeed, these results raise the possibility that abrogation of DDR genes by PBF and PTTG might also impact on emerging oncogenic roles for DDR genes such as spontaneous epithelial to mesenchymal transition observed in BRCA1-depleted cells.47 Further, we have defined the genetic alterations in DTC correlated with PBF and PTTG, thereby providing novel insights into the integral role of these proto-oncogenes in thyroid cancer pathways. We therefore propose that high tumoural expression of PBF and PTTG contributes to significant disruption of DDR regulated activity that is associated with poorer clinical outcome.
Materials and Methods
Human tissue, cell culture and transfection
Human thyroid samples were obtained with local ethics committee approval (Birmingham Clinical Research Office, UK) and informed patient consent. Thyroid TPC-1 cells were kindly provided by Dr Rebecca Schweppe (University of Colorado, Denver, CO, USA) and maintained in RPMI 1640 (Life Technologies, Paisley, UK) supplemented with 10% fetal bovine serum, penicillin (105 U/l), and streptomycin (100 mg/l). Cells were cultured as recommended at low passage and authenticated by short tandem repeat analysis (DNA Diagnostics Centre, London, UK) and tested for mycoplasma contamination (EZ-PCR kit; Geneflow, Lichfield, UK). pEFP BRAFV600E containing BRAFV600E cDNA was kindly provided by Jim Fagin (Memorial Sloan-Kettering Cancer Center, New York, USA). Cells were transfected using TransIT LT1 (Mirus Bio LLC, Madison, WI, USA) according to manufacturer's protocol.
Transgenic mice and primary thyrocytes
Wild-type (WT) and transgenic FVB/N mice were bred at the Transgenic Mouse Facility (University of Birmingham, UK) and all experiments performed in accordance with U.K. Home Office regulations. Generation of PTTG-Tg and PBF-Tg transgenic mice have been described previously.22,48 The bitransgenic line (Bi-Tg) was generated by crossbreeding PBF-Tg and PTTG-Tg mice. Mice were genotyped using tail DNA and PCR protocols as described.22 Equivalent numbers of male and female mice were used per group unless otherwise stated. Power calculations (biomath.info/power/ttest.htm) showed that at least 6 mice were sufficient to detect a 30% change in thyroid size (power=0.8, alpha=0.05). No blinding or randomisation of mice allocation was performed. Thyroid glands were removed and primary thyrocytes cultured as described previously.27 Seven days after seeding DNA damage was induced by Caesium-137 irradiation at 20 Gy dose (IBL 437C type H unit, CIS Bio international, Gif-Sur-Yvette, France).
RNA analysis
Total RNA was extracted using the RNeasy Micro Kit (Qiagen, Manchester, UK) prior to reverse transcription using the Reverse Transcription System (Promega, Madison, USA). Expression of specific mRNAs was determined on a 7500 Real-time PCR system (Applied Biosystems) using primers listed in Supplementary Table S3. Total RNA was extracted from human thyroids using the RNeasy FFPE kit (Qiagen) as per manufacturer’s instructions and expression determined using the QuantiTect Probe RT-PCR kit (Qiagen). Relative expression was calculated using the 2−ΔΔCt method. Gene expression was also analysed using the DNA damage signalling pathway-focused RT2 Profiler PCR Array (Qiagen) according to manufacturer’s instructions.
Thyroid function
Total T3 and total T4 in the serum of WT and transgenic mice were measured after centrifugation of clotted blood samples using RIA kits (MP Biomedicals, Solon, OH, USA). Mouse serum concentrations were determined by Prof Samuel Refetoff (University of Chicago, Chicago, IL, USA).
Western blotting and Immunohistochemistry
Western blot and immunohistochemical analyses were performed as described previously.20,49 Primary antibodies against PBF (custom20,49), HA (MMS-101P; BioLegend, San Diego, CA, USA), hPTTG (700791; Invitrogen, Paisley, UK), phospho-p44/42 MAPK (Erk1/2) (9101), p44/42 MAPK (Erk1/2) (4695), Myc-Tag (2276; Cell Signaling Technology, Danvars, MA, USA), p53 (sc-126; Santa Cruz Biotechnology, Dallas, TX, USA), cyclin D1 (ab16663), cyclin A2 (ab181591; Abcam, Cambridge, UK), Chk1 (GTX50463), Brca1 (GTX50557; GeneTex, Irvine, CA, USA), phospho-CHK1(Ser345) (PA5-34625; Thermofisher Scientific, Waltham, MA, USA) and β-actin (A5441; Sigma-Aldrich, Dorset, UK) were used. Densitometry was performed on blots scanned into Photoshop (Adobe Systems) and analysed using ImageJ as described.27
Analysis of genomic instability
Fluorescent inter simple sequence repeat (FISSR)-PCR amplications were performed as described previously17,27 using a 5’-6-carboxyfluorescein-labeled primer (CA)8RG with 5 ng genomic DNA. PCR products were electrophoresed on an ABI3730 capillary sequencer (Applied Biosystems, Foster City, CA, USA), and data analysed using Peak Scanner software. Five replicate experiments were performed to verify the reproducibility of the assay. The degree of genetic instability was determined as described50 to generate the GI index, which represents the standard measure of GI with ISSR-PCR analysis. Chromosomal aberrations were scored in Giemsa stained metaphase spreads as described previously51 (Supplementary Information).
Datasets
Normalized gene expression data generated using the Illuminia RNA-seq platform and clinical information was downloaded from cBioPortal.52,53 Gene expression values were transformed as X=log2(X+1) where X represents the normalized fragments per kilobase transcript per million mapped reads (FPKM) values. For matched normal (N) and tumour (C) pairs relative fold-changes (FC) were transformed as log2FC = log2(C) – log2(N) where C and N represent normalized FPKM values. Transcriptome datasets were selected for tumours with TNM staging of T1-3N0 (non-metastatic) or T1-3N1 (lymph node metastasis). In total RNAseq data for 322 unmatched DTC and 59 matched DTC/normal samples were used. Non-parametric Mann-Whitney, Kruskal-Wallis and Spearman’s correlation tests were performed as expression levels of PBF and PTTG in TCGA datasets were not normally distributed.
Statistical analysis
All results were obtained from triplicate experiments unless otherwise indicated. Data were analysed using IBM SPSS Statistics (version 22) and Microsoft Excel. The biological number of samples and corresponding statistical test and significance level is indicated in each figure legend. Normal distribution and homogeneity of variance has been tested using the D’Agostino-Pearson test of normality and the F-test. P<0.05 were considered significant.
Supplementary Material
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Acknowledgments
This work was supported by the Medical Research Council (MR/J001414/1), Wellcome Trust, Cancer Research UK, Get Ahead Charitable Trust and AMEND. Results are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol. 2013;2013:965212. doi: 10.1155/2013/965212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haymart MR. Understanding the relationship between age and thyroid cancer. Oncologist. 2009;14:216–221. doi: 10.1634/theoncologist.2008-0194. [DOI] [PubMed] [Google Scholar]
- 3.Grogan RH, Kaplan SP, Cao H, Weiss RE, Degroot LJ, Simon CA, et al. A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up. Surgery. 2013;154:1436–1446. doi: 10.1016/j.surg.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boelaert K, McCabe CJ, Tannahill LA, Gittoes NJ, Holder RL, Watkinson JC, et al. Pituitary tumor transforming gene and fibroblast growth factor-2 expression: potential prognostic indicators in differentiated thyroid cancer. J Clin Endocrinol Metab. 2003;88:2341–2347. doi: 10.1210/jc.2002-021113. [DOI] [PubMed] [Google Scholar]
- 6.Stratford AL, Boelaert K, Tannahill LA, Kim DS, Warfield A, Eggo MC, et al. Pituitary tumor transforming gene binding factor: a novel transforming gene in thyroid tumorigenesis. J Clin Endocrinol Metab. 2005;90:4341–4349. doi: 10.1210/jc.2005-0523. [DOI] [PubMed] [Google Scholar]
- 7.Heaney AP, Nelson V, Fernando M, Horwitz G. Transforming events in thyroid tumorigenesis and their association with follicular lesions. J Clin Endocrinol Metab. 2001;86:5025–5032. doi: 10.1210/jcem.86.10.7886. [DOI] [PubMed] [Google Scholar]
- 8.Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ, Yoo KC, et al. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population. J Biol Chem. 2012;287:19516–19527. doi: 10.1074/jbc.M111.337428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurvits N, Repo H, Loyttyniemi E, Nykanen M, Anttinen J, Kuopio T, et al. Prognostic implications of securin expression and sub-cellular localization in human breast cancer. Cell Oncol (Dordr) 2016;39:319–313. doi: 10.1007/s13402-016-0277-5. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, Guo J, Zhou J, Lu J, Chen Q, Zhang C, et al. FoxM1 transactivates PTTG1 and promotes colorectal cancer cell migration and invasion. BMC Med Genomics. 2015;8:49. doi: 10.1186/s12920-015-0126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Duan W, Li X, Liu J, Li D, Ye L, et al. PTTG regulates the metabolic switch of ovarian cancer cells via the c-myc pathway. Oncotarget. 2015;6:40959–40969. doi: 10.18632/oncotarget.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao H, Zhong F, Xie J, Peng J, Han Z. PTTG promotes invasion in human breast cancer cell line by upregulating EMMPRIN via FAK/Akt/mTOR signaling. Am J Cancer Res. 2016;6:425–439. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Hsu YH, Liao LJ, Yu CH, Chiang CP, Jhan JR, Chang LC, et al. Overexpression of the pituitary tumor transforming gene induces p53-dependent senescence through activating DNA damage response pathway in normal human fibroblasts. J Biol Chem. 2010;285:22630–22638. doi: 10.1074/jbc.M109.096255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salehi F, Kovacs K, Scheithauer BW, Lloyd RV, Cusimano M. Pituitary tumor-transforming gene in endocrine and other neoplasms: a review and update. Endocr Relat Cancer. 2008;15:721–743. doi: 10.1677/ERC-08-0012. [DOI] [PubMed] [Google Scholar]
- 15.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, Pemberton H, Stratford AL, Buelaert K, Watkinson JC, Lopes V, et al. Pituitary tumour transforming gene (PTTG) induces genetic instability in thyroid cells. Oncogene. 2005;24:4861–4866. doi: 10.1038/sj.onc.1208659. [DOI] [PubMed] [Google Scholar]
- 17.Kim DS, Franklyn JA, Smith VE, Stratford AL, Pemberton HN, Warfield A, et al. Securin induces genetic instability in colorectal cancer by inhibiting double-stranded DNA repair activity. Carcinogenesis. 2007;28:749–759. doi: 10.1093/carcin/bgl202. [DOI] [PubMed] [Google Scholar]
- 18.Chien W, Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J Biol Chem. 2000;275:19422–19427. doi: 10.1074/jbc.M910105199. [DOI] [PubMed] [Google Scholar]
- 19.McCabe CJ, Khaira JS, Boelaert K, Heaney AP, Tannahill LA, Hussain S, et al. Expression of pituitary tumour transforming gene (PTTG) and fibroblast growth factor-2 (FGF-2) in human pituitary adenomas: relationships to clinical tumour behaviour. Clin Endocrinol(Oxf) 2003;58:141–150. doi: 10.1046/j.1365-2265.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 20.Watkins RJ, Read ML, Smith VE, Sharma N, Reynolds GM, Buckley L, et al. Pituitary Tumor Transforming Gene Binding Factor: a New Gene in Breast Cancer. Cancer Res. 2010;70:3739–3749. doi: 10.1158/0008-5472.CAN-09-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melloni GE, Ogier AG, de Pretis S, Mazzarella L, Pelizzola M, Pelicci PG, et al. DOTS-Finder: a comprehensive tool for assessing driver genes in cancer genomes. Genome Med. 2014;6:44. doi: 10.1186/gm563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Read ML, lewy gd, Fong JC, Sharma N, Seed RI, Smith VE, et al. Proto-oncogene PBF/PTTG1IP regulates thyroid cell growth and represses radioiodide treatment. Cancer Res. 2011;71:6153–6164. doi: 10.1158/0008-5472.CAN-11-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, et al. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat Genet. 2002;32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- 26.Liang HQ, Wang RJ, Diao CF, Li JW, Su JL, Zhang S. The PTTG1-targeting miRNAs miR-329, miR-300, miR-381, and miR-655 inhibit pituitary tumor cell tumorigenesis and are involved in a p53/PTTG1 regulation feedback loop. Oncotarget. 2015;6:29413–29427. doi: 10.18632/oncotarget.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Read ML, Seed RI, Fong JC, Modasia B, Ryan GA, Watkins RJ, et al. The PTTG1-binding factor (PBF/PTTG1IP) regulates p53 activity in thyroid cells. Endocrinology. 2014;155:1222–1234. doi: 10.1210/en.2013-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read ML, Seed RI, Modasia B, Kwan PP, Sharma N, Smith VE, et al. The proto-oncogene PBF binds p53 and is associated with prognostic features in colorectal cancer. Mol Carcinog. 2016;55:15–26. doi: 10.1002/mc.22254. [DOI] [PubMed] [Google Scholar]
- 29.Chesnokova V, Wong C, Zonis S, Gruszka A, Wawrowsky K, Ren SG, et al. Diminished pancreatic beta-cell mass in securin-null mice is caused by beta-cell apoptosis and senescence. Endocrinology. 2009;150:2603–2610. doi: 10.1210/en.2008-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santos JC, Bastos AU, Cerutti JM, Ribeiro ML. Correlation of MLH1 and MGMT expression and promoter methylation with genomic instability in patients with thyroid carcinoma. BMC Cancer. 2013;13:79. doi: 10.1186/1471-2407-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeman-Neill RJ, Kelly LM, Liu P, Brenner AV, Little MP, Bogdanova TI, et al. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer. 2014;120:799–807. doi: 10.1002/cncr.28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, Levi F, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer. 2015;136:2187–2195. doi: 10.1002/ijc.29251. [DOI] [PubMed] [Google Scholar]
- 35.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubinek T, Chesnokova V, Wolf I, Wawrowsky K, Vlotides G, Melmed S. Discordant proliferation and differentiation in pituitary tumor-transforming gene-null bone marrow stem cells. Am J Physiol Cell Physiol. 2007;293:C1082–C1092. doi: 10.1152/ajpcell.00145.2007. [DOI] [PubMed] [Google Scholar]
- 38.Hamid T, Kakar SS. PTTG/securin activates expression of p53 and modulates its function. Mol Cancer. 2004;3:18. doi: 10.1186/1476-4598-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan JW, Liao YC, Lua I, Li MH, Hsu CY, Chen JH. Human pituitary tumor-transforming gene 1 overexpression reinforces oncogene-induced senescence through CXCR2/p21 signaling in breast cancer cells. Breast Cancer Res. 2012;14:R106. doi: 10.1186/bcr3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 41.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 42.Mitmaker E, Alvarado C, Begin LR, Trifiro M. Microsatellite instability in benign and malignant thyroid neoplasms. J Surg Res. 2008;150:40–48. doi: 10.1016/j.jss.2007.12.760. [DOI] [PubMed] [Google Scholar]
- 43.Vaish M, Mishra A, Kaushal M, Mishra SK, Mittal B. Microsatellite instability and its correlation with clinicopathological features in a series of thyroid tumors prevalent in iodine deficient areas. Exp Mol Med. 2004;36:122–129. doi: 10.1038/emm.2004.18. [DOI] [PubMed] [Google Scholar]
- 44.Fong MY, Farghaly H, Kakar SS. Tumorigenic potential of pituitary tumor transforming gene (PTTG) in vivo investigated using a transgenic mouse model, and effects of cross breeding with p53 (+/-) transgenic mice. BMC Cancer. 2012;12:532. doi: 10.1186/1471-2407-12-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knauf JA, Ma X, Smith EP, Zhang L, Mitsutake N, Liao XH, et al. Targeted expression of BRAFV600E in thyroid cells of transgenic mice results in papillary thyroid cancers that undergo dedifferentiation. Cancer Res. 2005;65:4238–4245. doi: 10.1158/0008-5472.CAN-05-0047. [DOI] [PubMed] [Google Scholar]
- 46.Byrne M, Wray J, Reinert B, Wu Y, Nickoloff J, Lee SH, et al. Mechanisms of oncogenic chromosomal translocations. Ann N Y Acad Sci. 2014;1310:89–97. doi: 10.1111/nyas.12370. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Bierie B, Li AG, Pathania S, Toomire K, Dimitrov SD, et al. BRCA1/FANCD2/BRG1-Driven DNA Repair Stabilizes the Differentiation State of Human Mammary Epithelial Cells. Mol Cell. 2016;63:277–292. doi: 10.1016/j.molcel.2016.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewy GD, Ryan GA, Read ML, Fong JC, Poole V, Seed RI, et al. Regulation of pituitary tumor transforming gene (PTTG) expression and phosphorylation in thyroid cells. Endocrinology. 2013;154:4408–4422. doi: 10.1210/en.2012-2156. [DOI] [PubMed] [Google Scholar]
- 49.Smith VE, Read ML, Turnell AS, Watkins RJ, Watkinson JC, lewy gd, et al. A novel mechanism of sodium iodide symporter repression in differentiated thyroid cancer. J Cell Sci. 2009;122:3393–3402. doi: 10.1242/jcs.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basik M, Stoler DL, Kontzoglou KC, Rodriguez-Bigas MA, Petrelli NJ, Anderson GR. Genomic instability in sporadic colorectal cancer quantitated by inter-simple sequence repeat PCR analysis. Genes Chromosomes Cancer. 1997;18:19–29. [PubMed] [Google Scholar]
- 51.Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, et al. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol Cell. 2015;59:462–477. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.