Abstract
Extracellular vesicles (EVs) are membrane particles involved in the exchange of a broad range of bioactive molecules between cells and the microenvironment. While it has been shown that cells can traffic metabolic enzymes via EVs much remains to be elucidated with regard to their intrinsic metabolic activity. Accordingly, herein we assessed the ability of neural stem/progenitor cell (NSC)-derived EVs to consume and produce metabolites. Both our metabolomics and functional analyses revealed that EVs harbour L-asparaginase activity catalysed by the enzyme Asparaginase-like protein 1 (Asrgl1). Critically, we show that Asrgl1 activity is selective for asparagine and is devoid of glutaminase activity. We found that mouse and human NSC-derived EVs traffic ASRGL1. Our results demonstrate for the first time that NSC EVs function as independent, extracellular metabolic units able to modify the concentrations of critical nutrients, with the potential to affect the physiology of their microenvironment.
Introduction
Extracellular vesicles (EVs) are a heterogeneous group of membrane particles secreted by the majority of cell types across all kingdoms of life; they have different mechanisms of biogenesis, structural composition, and functions1. Of these, shedding vesicles, exosomes and apoptotic bodies have been the most studied subtypes of EVs to date2. Several studies aimed at characterizing the content of EVs and went on to demonstrate that a broad range of bioactive molecules, including proteins and different types of nucleic acids, are associated with EVs3,4. Of note, the content of EVs can vary according to the cell type of origin, and/or in response to stimuli from the microenvironment5. We previously demonstrated that neural stem cells (NSCs) secrete EVs containing mRNAs and proteins, whose sorting is regulated by inflammatory cytokines. Our study identified a novel mechanism of intercellular communication regulated by the IFN-γ/Ifngr1 complex on EVs and elucidated its molecular signature and functional relevance to target cells6.
A growing body of evidence supports a key a role for EVs in regulating metabolic homeostasis or associated cellular processes. For instance, glucose deprivation was shown to promote trafficking of glucose transporters and glycolytic enzymes towards EVs in cardiomyocytes7. Prostate-derived, exosome-like prostasomes harbor enzymes involved in adenosine triphosphate (ATP) metabolic turnover that include adenylate kinase, ATPase, 5'-nucleotidase, and hexose transporters8. One quarter of the proteins enriched in prostate cancer-derived EVs large oncosomes (vs. prostate cancer cells) includes enzymes involved in glucose, glutamine and amino acid metabolism9. Furthermore, primary B-cell precursor acute lymphoblastic leukemia cells release a variety of EVs into extracellular fluids (in vitro) and circulation (in vivo) that are internalized by stromal cells, which subsequently undergo a metabolic switch toward a glycolytic phenotype10. Finally, cancer-associated fibroblasts -derived EVs induce central carbon metabolism in target cells and promote tumor growth under nutrient deprivation or nutrient stressed conditions11. It has been recently hypothesized that EVs are involved in the intercellular trafficking of small molecules11. However, whether EVs can act as independent, metabolically active units, capable of perturbing the extracellular milieu is still unknown. In this work, we sought to investigate the enzymatic activities associated with NSC-derived EVs. We found that NSC EVs are metabolically active and, in particular, that they exhibit L-asparaginase activity. We identified asparaginase-like protein 1 (Asrgl1) as the key metabolic enzyme responsible for the L-asparaginase activity of EVs. Finally, we observed that EV-associated Asrgl1 is specific for asparagine (Asn) and is devoid of glutaminase activity. Our study shows for the first time that EVs can behave as autonomous metabolic reactors and are capable of delivering specific and functional L-asparaginase activity to the microenvironment via Asrgl1.
Results
NSCs secrete EVs containing exosomes
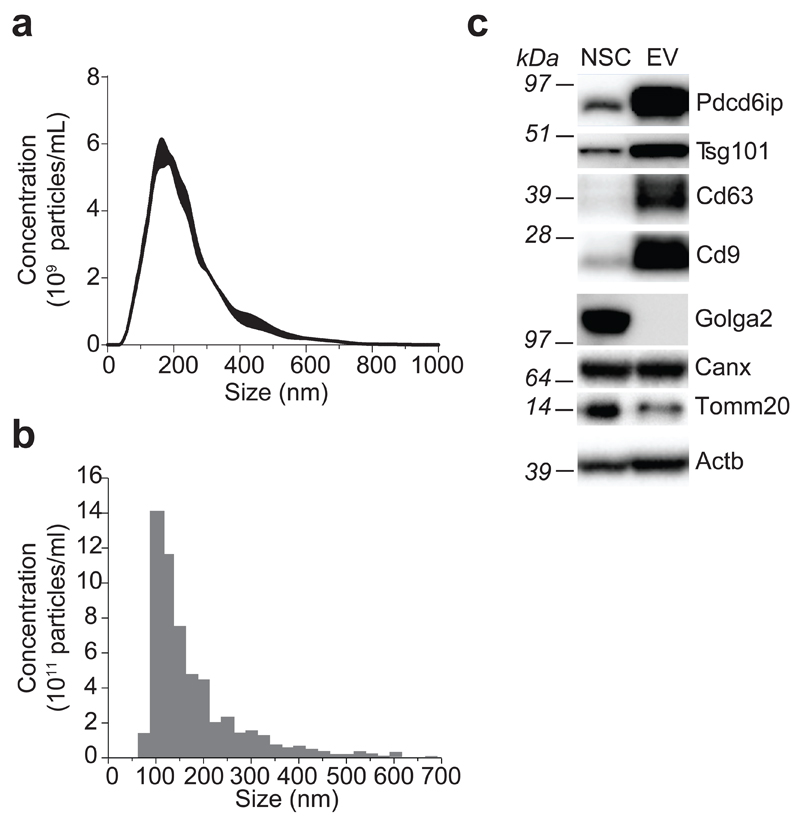
We started our investigation by generating EVs from NSCs. Stably expandable NSCs were derived from the subventricular zone of adult mice, and EVs were purified from NSC supernatants by differential centrifugation (Supplementary Results, Supplementary Fig. 1a), as previously described6. To characterize the population of vesicles secreted by NSCs, we applied a combination of nanoparticle tracking analysis (NTA), tunable resistive pulse sensing (TRPS) technology and Western blot (WB) analysis. NTA identified a major peak (169 ± 3 nm) in the size range of exosomes, and a minor peak (300–500 nm), which is consistent with the dimension of larger shedding vesicles (Fig. 1a). TRPS with a 150 nm nanopore confirmed the presence of small vesicles, with a major peak corresponding to the size described for exosomes (102 ± 9 nm) (Fig. 1b)12. WB analyses further confirmed that protein extracts purified from EVs are specifically enriched in markers associated with exosomes (vs. parental NSC protein extracts), which include Pdcd6ip, Tsg101 and the tetraspanins CD63 and CD9. On the other hand, markers for other cellular compartments, such as Golga2 (Golgi), Calnexin (Endoplasmic reticulum) and Tomm20 (Mitochondria) did not show a similar distribution (Fig. 1c).
Figure 1. NSCs secrete EVs containing exosomes.
(a, b) Particle-size distribution of EVs by NTA (a) and TRPS (b) technologies. The dimension data are expressed as mean values (nm) ± SEM from n= 3 independent experiments. (c) Western blot analysis of markers for exosomes and for different organelles (Golgi, endoplasmic reticulum and mitochondria) in mouse NSCs and EVs. This panel is representative of n= 3 independent protein preparations showing the same trends. Full gels are located in Supplementary Figure 7.
NSC-derived EVs exhibit L-asparaginase activity
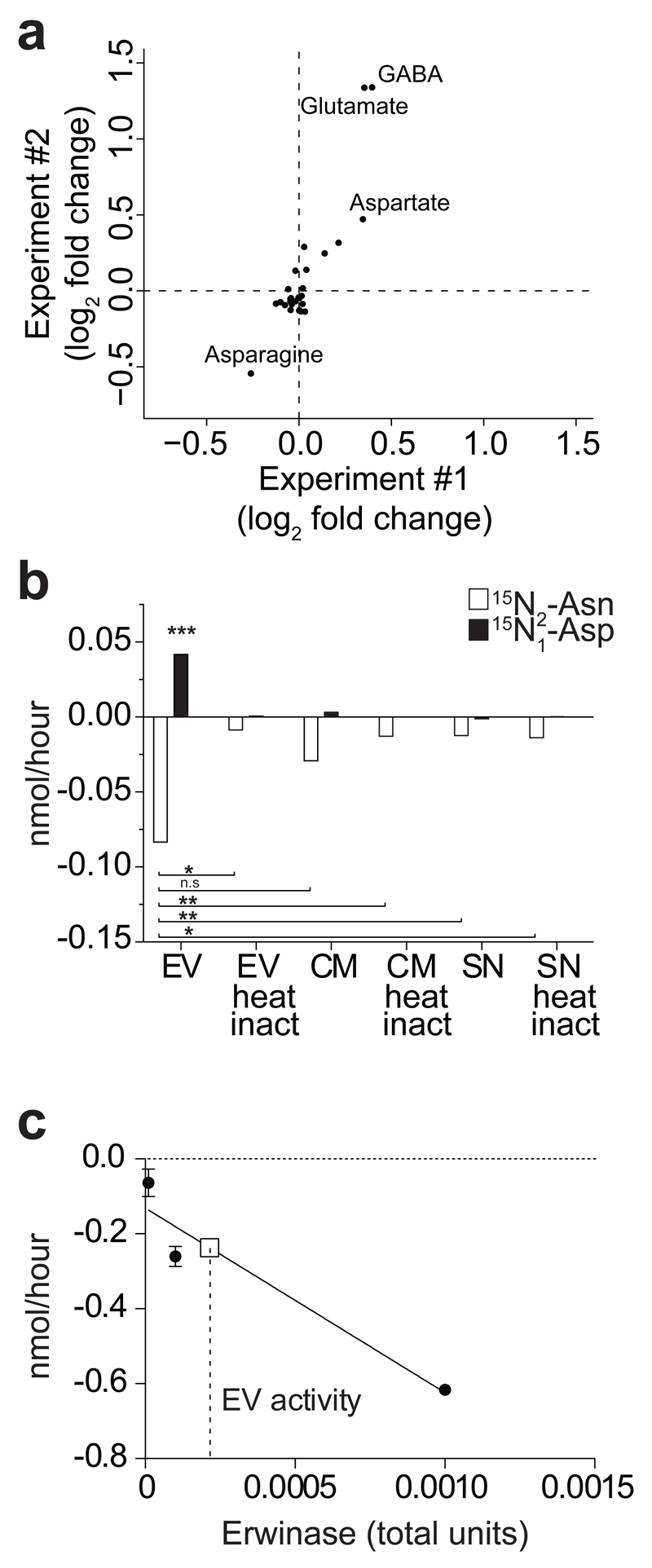
To assess the metabolic activity of NSC EVs, we incubated EVs in commercial NSC medium and analysed the consumption/release of metabolites from/in the medium via liquid chromatography coupled to mass spectrometry (LC-MS) (Supplementary Fig. 1b). We observed that incubation with EVs induced profound changes in the levels of several metabolites within the medium (Supplementary Tab. 1). Importantly, heat inactivation suppressed this metabolic activity of EVs (Supplementary Fig. 2a), suggesting that consumption and release of metabolites is largely due to the intrinsic enzymatic activity of EVs and not to leakage of metabolites. We found that Asn was the most consumed metabolite out of two independent EVs preparations, whilst aspartate (Asp) and glutamate (Glu) were amongst the most abundantly released metabolites (Fig. 2a). Given the significant depletion of Asn coupled with the production of Asp, we hypothesised that EVs harbour L-asparaginase activity. To validate this hypothesis, we incubated EVs in commercial NSC medium supplemented with 15N2-Asn. If EVs carried L-asparaginase activity, 15N-Aspartate should be found in the EV spent medium (Supplementary Fig. 2b). Consistent with this hypothesis, we observed marked consumption of 15N2-Asn coupled with production of 15N-Asp in media containing EVs (Fig. 2b). Of note, EVs exhibited enriched L-asparaginase activity compared to NSC conditioned media (CM) and EV-deprived NSC supernatants (SN). Furthermore, L-asparaginase activity was dampened in heat-inactivated vesicles (Fig. 2b), confirming that this activity depends on intact enzymatic function. Importantly, we found that also NSCs exhibited L-asparaginase activity, as indicated by the uptake of asparagine and intracellular conversion of 15N2-asparagine to 15N-aspartate (Supplementary Fig. 2c). Finally, a calibration curve using clinical-grade L-asparaginase produced by Erwinia chrysanthemi (Erwinase) allowed us to quantify the intrinsic L-asparaginase activity of NSC EVs, giving a value of 2x10–6 U/μg of EV protein (Fig. 2c).
Figure 2. NSC EVs are metabolically active in vitro.
(a) Scatter plot of metabolomics experiment showing log2 fold changes of extracellular metabolites in medium + EVs vs. Vehicle as in a. Positive/negative values indicate production/consumption of metabolites, respectively. Data from two independent experiments are shown. (b) Barplot of the consumption of 15N2-Asn and production of 15N-Asp mediated by EVs, conditioned medium (i.e. CM, medium with EVs) and supernatant (i.e. SN, medium deprived of EVs), with or without heat inactivation (100°C for 10’). Data are mean ± SEM and have been obtained from n= 2 independent experiments. Statistical analysis was performed using one-way ANOVA, followed by Bonferroni’s test correction. * p< 0.05; ** p < 0.01; *** p<0.001. (c) Consumption of Asn with different amounts of clinical-grade L-asparaginase produced by Erwinia chrysanthemi (Erwinase) was used to determine the L-asparaginase activity associated with EVs. One International Unit of L-asparaginase is defined as the amount of enzyme required to generate 1 μmol of ammonia per minute at pH 7.3 and 37°C. Solid line indicates Erwinase calibration curve, while square symbol and dashed line indicate interpolation of the calibration curve with Asn consumption values obtained with EVs.
Mouse and human NSCs traffic Asrgl1 into EVs
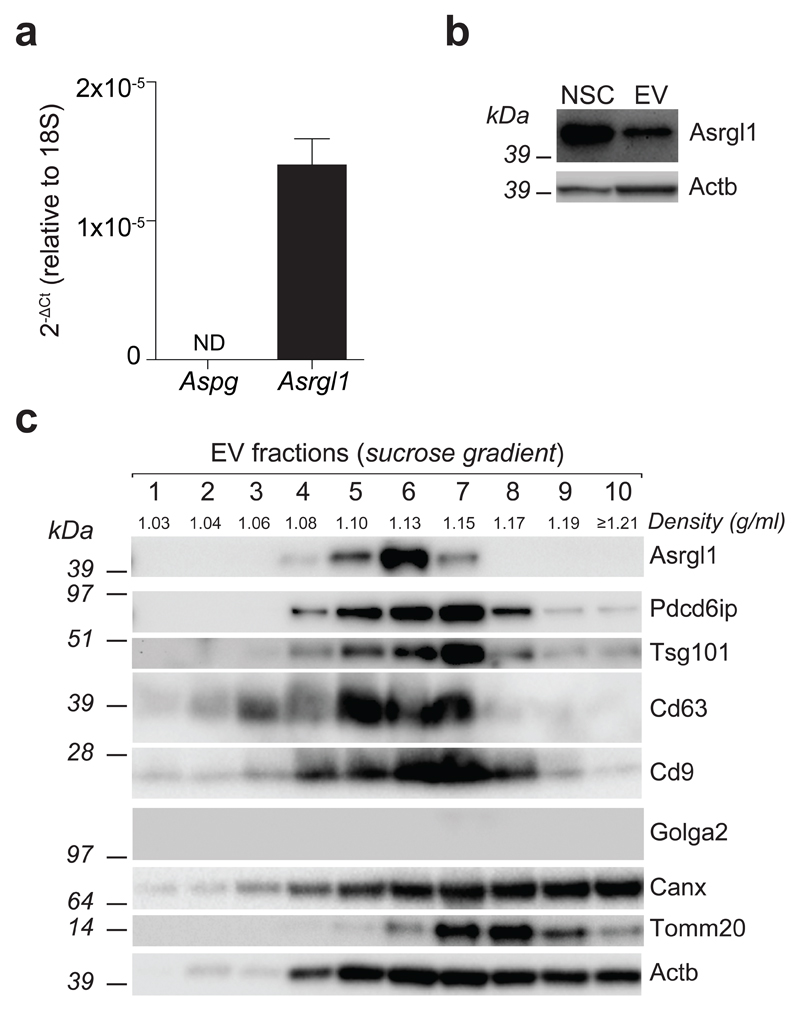
We then investigated the identity of the possible enzymes that might confer such asparaginase activity to NSC-derived EVs. The mouse genome contains two L-asparaginases: a 60 kDa lysophospholipase/L-asparaginase (Aspg) and an isoaspartyl peptidase/L-asparaginase (Asparaginase-like protein 1; Asrgl1) (source http://www.uniprot.org). To assess the expression of Aspg and Asrgl1 in NSCs we took advantage of a comprehensive long RNA-seq analysis that we recently published6, and found that NSCs express Asrgl1 but not Aspg (Supplementary Fig. 3a).
Independent qPCR and WB analyses confirmed the RNA-seq data and showed that EV protein extracts contained Asrgl1 at levels comparable to parental NSCs (Fig. 3a-b). We then assessed the distribution of the enzyme in EVs subjected to sucrose gradient fractionation. We found that Asrgl1 peaks at 1.13 g/ml, overlapping with the exosome fraction, both in terms of markers (i.e. Pdcd6ip, Tsg101, CD63 and CD9) and density (1.13–1.20 g/ml), as described13 (Fig. 3c). We also excluded that Asrgl1 was present in fractions enriched into protein aggregates (density ≥1.21 g/ml)14 (Fig. 3c), thus unambiguously demonstrating that Asrgl1 is genuinely associated with EVs (e.g. shedding vesicles and exosomes).
Figure 3. Mouse NSCs traffic Asrgl1 into EVs.
(a) qPCR and (b) western blot analyses of Asrgl1 expression in mouse NSCs. qPCR data are represented as fold change ± SEM of the Aspg and Asrgl1 mRNA levels in NSCs, and analysed with the 2-ΔCt method over 18S ribosomal RNA, used as housekeeping gene. Both qPCR and WB data have been obtained from n=3 independent experiments. (c) Western blot of Asrgl1 and markers for exosomes and cellular organelles as in Figure 1, after sucrose gradient fractionation of total EVs. This panel is representative of n = 3 independent protein preparations showing the same trends. See also Supplementary Figure 3. Full gels are located in Supplementary Figure 8 and 9.
Next, we extended our analyses to clinical grade human foetal NSCs (hNSCs)15, as well as to EVs collected from hNSC supernatants. Similar to mouse NSCs, hNSCs expressed ASRGL1, but not ASPG, and hNSCs-derived EVs contained ASGRL1 (Supplementary Fig. 3b-c).
Thus, EVs from both mouse and human NSCs are cargoed with Asrgl1.
Asrgl1 catalyses the L-asparaginase activity of EVs
Asrgl1 has been described to be associated to small exosome-like vesicles called prostasomes16, but its metabolic function in EVs remains to be defined.
In vitro experiments indicate that, differently from Aspg, Asgrl1 activity is devoid of enzymatic activity towards glutamine (Gln)17. Therefore, we wanted to investigate substrate specificity of EV-associated Asrgl1. We first tested whether EVs exhibit any glutaminase (Gls) activity. To this aim, we incubated EVs with 13C-Gln and measured the production of 13C-Glu, the product of Gls activity (See Supplementary Fig. 4a for a schematic of the experiment). We could detect Gln consumption by EVs and this was associated with the production of 13C-Glu (Supplementary Fig. 4b). Heat inactivation abolished Gln consumption and Glu production by EVs, indicating that this activity requires the presence of intact enzymes. In line with this finding, we detected protein expression of Gls in EVs (Fig. 4a-b).
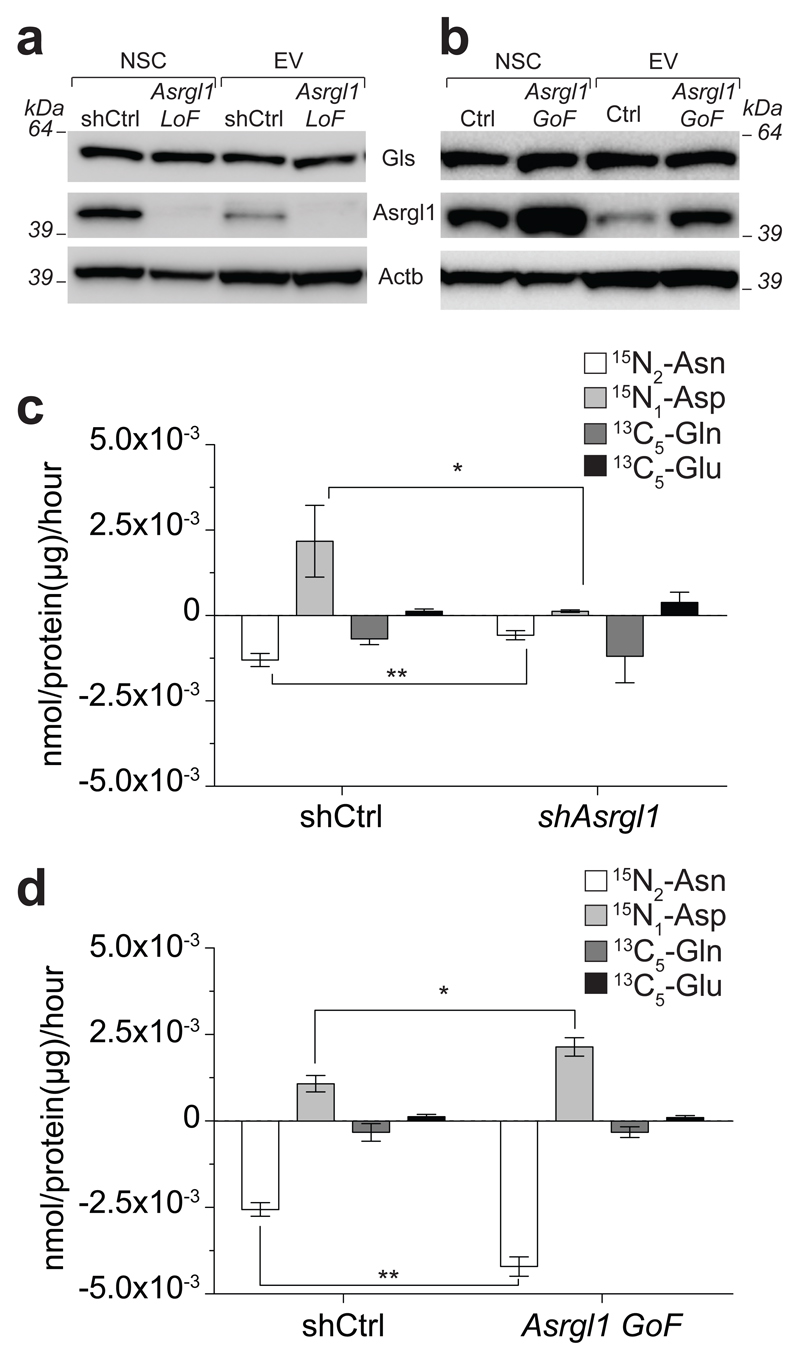
Figure 4. Asrgl1 is responsible for the selective L-asparaginase activity associated with EVs.
(a, b) Western blot analysis of shAsrgl1 (a) and Asrgl1 GoF (b) NSCs and EVs. These panels are representative of n= 3 independent protein preparations showing the same trends. (c, d) Barplot of the consumption of 15N2-Asn and 13C5-Gln followed by production of 15N-Asp and 13C5-Glu mediated by shAsrgl1 (c) and Asrgl1 GoF (d) EVs. Data are mean ± SEM and have been obtained from n≥ 3 independent experiments. Statistical analysis was performed using t-test. * p< 0.05; ** p < 0.01. Full gels are located in Supplementary Figure 11 and 12.
To confirm that Asrgl1 is the L-asparaginase responsible for the EV-mediated hydrolysis of Asn to Asp, and further exclude any possible Asrgl1-related Gls activity, we performed loss- (LoF) and gain-of-function (GoF) experiments by transducing NSCs with either a short hairpin RNA targeting Asrgl1 (shAsrgl1) or with the Asrgl1 coding sequence. NSC transduction with LoF and GoF lentiviral vectors did not affect NSC growth and viability upon serial passaging in vitro (Supplementary Fig. 5). LoF and GoF NSCs and EVs showed effective reductions and increases in the levels of Asrgl1 expression, respectively, as compared to controls. Of note, these changes in the expression of Asrgl1 had no effects on the level of other endogenous enzymes, such as Gls, both in NSC and EVs (Fig. 4a-b).
We then assessed the relative metabolic activity of EV-trafficked Asrgl1 by incubating control, LoF and GoF EVs in cell culture media containing equal amounts of Asn and Gln. Consumption of 15N2-Asn and production of 15N-Asp were significantly reduced in media incubated with shAsrgl1 EVs (vs. shCtrl EVs) (Fig. 4c). Conversely, we observed a significant increase in the consumption of 15N2-Asn and production of 15N-Asp within media incubated with Asrgl1 GoF EVs (vs. Ctrl EVs) (Fig. 4d). Importantly, incubation of media with both shAsrgl1 EVs and Asrgl1 GoF EVs did not alter the consumption of 13C5- Gln or the production of 13C5-Glu (Fig. 4c-d), confirming that Asrgl1 activity is specific for Asn and is devoid of Gls activity. Finally, we compared the L-asparaginase activity of EVs to that of parental cells and of human recombinant ASRGL1. We used the latter as reference to estimate by western blotting the relative abundance of Asgrl1 in NSCs and EVs, both Ctrl and GoF. Interestingly, we found that NSCs and EVs both exhibit much higher Asgrl1-dependent L-asparaginase activity, compared to the recombinant protein (Supplementary Fig. 6).
Discussion
EVs are influential players in intercellular communication where they participate via exchanging lipids, proteins, and nucleic acids. Depending on their origin and cargo molecules, EVs can modulate immune-regulatory processes, set up tumor escape mechanisms and/or mediate regenerative or degenerative processes18. The extent to which EVs interact with and modulate intracellular signaling pathways within target cells is not yet fully understood19.
We have recently shown that NSC EVs carrying receptor proteins within their cargo, including inflammatory cytokine receptors, activate downstream canonical signaling pathways in target cells, a mechanism that grafted stem cells might use to communicate with the host immune system6. More complex mechanisms, however, are emerging. These include direct gene and/or protein delivery, or transfer of non-coding small RNAs (e.g. microRNAs) functioning in RNA silencing and post-transcriptional regulation of gene expression20. The level of complexity in the understanding of signaling properties of EVs increases along with the knowledge that (i) individual cells release different subtypes of EVs simultaneously and that intercellular communication often involves more than two cells; (ii) trafficking of messengers into EVs varies in response to perturbations and stresses, including cytokines and growth factors, relative levels of extra- or intra-cellular amino acids21, hypoxia/normoxia, shear stress and circadian rhythms22,23; and (iii) available techniques to collect, process and analyze EVs and EV-induced cellular responses remain suboptimal24.
Emerging evidence suggests that EVs from different cell types can act as metabolic regulators7–10,25, yet the characterization of EVs intrinsic metabolic activity is still debated. By applying state-of-the-art untargeted and targeted metabolomic analyses, we demonstrated that NSC EVs exhibit enzymatic activities that affect the consumption/release of metabolites in the extracellular compartment. This important observation suggests that EVs are metabolically active and that multiple enzymes are associated with EVs. Unexpectedly, we found that Asn is the most significantly consumed metabolite suggesting a predominant L-asparaginase activity associated with EVs. Of note, this L-asparaginase activity was observed in NSCs, strongly suggesting that EVs do not acquire this metabolic function de novo but, rather, that this activity is transferred from the donor cells. This hypothesis has important implications: whilst L-asparaginase activity in NSCs would result in a possible depletion of asparagine in the extracellular milieu, L-asparaginase activity in EVs has the additional benefit of releasing Asp, a metabolite whose role in supporting cells’ bioenergetics is increasingly recognised26,27. Hence, the biological consequences of Asn-depletion strategies using NSCs or EVs are likely to be very different.
Besides Asn, the levels of several other metabolites with known biological functions were significantly altered by EVs. These include Asp, Glu, lactate, GABA and alanine. For instance, GABA and Glu are well-known neurotransmitters able to shape the activity of brain cells28, as well as the behaviour of other cell types29,30. Lactate is a potent signalling molecule able to affect the proliferation of cancer cells31, promote tumour angiogenesis32, and act as a pro-inflammatory signal to induce T cell migration33. Finally, uptake of Asp has been recently shown to be essential for cells with impaired mitochondrial function both in vitro and in vivo26,27. This evidence supports the hypothesis that, by altering the metabolic composition of the microenvironment, EVs might be able to profoundly shape the behaviour of surrounding cells, with potential important implications for biological functions of different cell types.
Here we identified Asrgl1 as the key metabolic enzyme responsible of the EV-associated L-asparaginase activity. Asrgl1 was initially identified as novel L-asparaginase in the rat and human testis34, and lately found to exhibit specific activity for Asn17. We demonstrated that Asrgl1 is the only metabolic enzyme with known L-asparaginase activity expressed by mouse and human NSCs, which both export towards EVs. Moreover, when equimolar amounts of labelled Asn and Gln are added to medium the relative L-asparaginase activity is higher compared to the Gls activity, further supporting that the L-asparaginase activity is the predominant one in EVs. We also confirmed that Asrgl1 is the enzyme responsible for the Asn-specific L-asparaginase activity mediated by EVs, as indicated by our Asrgl1 Lof and GoF experiments. In fact, when the expression of Asrgl1 was reduced or enhanced we observed that, while Gln and Glu levels were unaltered, only Asn consumption and Asp production were affected.
The finding that EVs-derived Asrgl1 is specific for Asn has important clinical implications. Various bacteria-derived L-asparaginases are currently used to treat acute lymphoblastic leukaemia and malignant lymphoma35. Furthermore, new reports demonstrated the L-asparaginase efficacy also toward glioblastoma cells in vitro36. The residual Gls activity of bacterial asparaginases is thought to be a major drawback of therapy since it leads to secondary immune depression and liver toxicity37. Therefore, the identification of Gls-free functional L-asparaginase into EVs might provide a valid alternative to recombinant L-asparaginases that would limit most of their side effects.
Beyond the characterisation of EV-associated Asrgl1 activity, the mechanism of Asrgl1 action within the EVs compartment remains to be further elucidated. Considering its cytosolic localization, it is tempting to speculate that Asrgl1 may be contained in the lumen of EVs, where it remains functionally active as long as for at least 24 hours, a time frame that is compatible with the experiments here described. Indeed, Asrgl1 enzymatic activity is significantly enriched in EVs, as compared to EV-deprived NSC supernatants, which indicate that extracellular metabolic enzymes are not secreted as free proteins in the extracellular milieu. Furthermore, the observation that the intrinsic Asrgl1 activity is higher in EVs than in NSCs and in the recombinant protein, suggests that Asrgl1 in EVs might be exposed to an activating microenvironment, or subject to post-translational modifications that do not occur in vitro, making EVs ideal vessels to deliver this enzyme.
Applying heavy-light double stable isotope labelling of amino acids in cell culture (SILAC) comparing EVs and exosomes collected from NSCs, we have recently described that NSCs and NSC EVs express a few members of the solute carrier (SLC) family of membrane transporters, which include the high affinity Glu and Asp transporter (Slc1a3) and the two system A family members the Sodium-coupled neutral amino acid transporter 2 (Slc38a2), and the Sodium-coupled neutral amino acid transporter 3 (Slc38a3)6. It would be then plausible to hypothesize that metabolically active EVs uptake extracellular Asn via sodium–coupled neutral amino acid (system N/A) transporters (SNATs), hydrolyze Asn to Asp into Asrgl1-coargoed EVs and then export Asp via EV-associated Slc1a3 in the extracellular milieu. Further studies are necessary to validate this hypothesis, as well as to identify the possible mechanisms by which metabolic enzymes are loaded into (or sampled by) EVs.
In conclusion, our work shows for the first time the intrinsic ability of NSCs to deliver functional L-asparaginase activity in the microenvironment via Asrgl1 in EVs. Our work further highlights a surprising novel role for stem cell-derived EVs, with a potential role in the propagation of specific metabolic signals to the surrounding cells and to the microenvironment.
Online methods
Cell culture preparations
NSCs were prepared from the subventricular zone of 7- to 12-week-old SJL mice, as described1. Good Manufacturing Practice-grade, foetal human NSCs from natural in utero death (hNSCs) were prepared as described15. Both mouse and human NSCs were tested for Mycoplasma contamination, several times throughout the study, and always resulted Mycoplasma-free.
Purification of EVs from media and sucrose gradient fractionation
Mouse and human NSC EVs were collected and characterized as described5.
Nanoparticle Tracking Analysis (NTA)
Samples were diluted 1:1000 with PBS, vortexed and analysed by Nanoparticle Tracking Analysis (NTA) instrument (NS500, NanoSight ltd) fitted with an Electron Multiplication Charge-Couple Device camera and a 532 nm laser. The temperature was maintained within the range 20-25°C and resultant liquid viscosity calculated by the instrument. 3 repeated measurements of 60 seconds were recorded for each of the 3 independent samples (9 analysis in total). Fresh sample was loaded into the chamber prior to each video capture. Static mode (no flow) was used for analysis and detection threshold was adjusted to 5. Automatic settings were set for blur size, minimum track length and minimum particle size. The movement of each particle in the field of view was measured to generate the average displacement (in terms of x and y) of each particle per unit time. From this measurement, the particle diffusion coefficient was estimated and the hydrodynamic diameter calculated through application of the Stokes-Einstein equation.
Tunable resistive pulse sensing (TRPS)
The concentration and size distribution of EVs was analysed with TRPS (qNano, Izon Science Ltd) using a NP150 nanopore at 0.5 V with 47 mm stretch. The concentration of particles was standardized using 100 nm calibration beads (CPC 100) at a concentration of 1x1010 particles/mL.
Protein purification and Western blot analysis
Whole NSCs, EVs and sucrose gradient fraction extracts were processed as described5. Due to the limited amount of material obtained from sucrose gradient fractions, the correspondent WBs were performed using independent EV preparations starting from the same NSC batch. The following primary antibodies were used: mouse monoclonal anti-Pdcd6ip (BD transduction lab and Cell Signaling Technology); rat monoclonal and mouse monoclonal anti-CD9 (BD transduction lab and Life Technologies, respectively); goat polyclonal anti-TSG101 (Santa Cruz); rat monoclonal and mouse monoclonal anti-CD63 (MBL and Life Technologies, respectively); rabbit polyclonal anti-Asrgl1 (Proteintech); mouse monoclonal anti-GM-130 (BD transduction lab); rabbit polyclonal anti-calnexin (Abcam); rabbit polyclonal anti-Tom-20 (Santa Cruz); rabbit monoclonal anti-Gls (Abcam); mouse monoclonal anti-beta-Actin (Sigma). Molecular weight marker: SeeBlue Plus2 (Invitrogen). WB for mouse and human Cd63 and Cd9 were run under non reducing conditions, i.e sample buffer used for electrophoresis was devoid of reducing agent.
RNA extraction, RT and qPCR
RNAs from mouse and human NSCs were obtained using the miRCURY™ RNA Isolation Kit - Cell & Plant (Exiqon). Total RNA quantity and purity were assessed with the NanoDrop 2000c instrument (Thermo Scientific). cDNA synthesis was performed using SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen), with random hexamers as primers. qPCRwas performed with the TaqMan® Universal PCR Master Mix (Applied Biosystems) and TaqMan® Gene Expression Assays for FAM™ Aspg, Asrgl1, ASPG and ASRGL1 and VIC® euk 18S rRNA probes. Samples were tested in triplicate on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems) and analysed with the 2-ΔCt method over 18S ribosomal RNA, used as housekeeping gene.
Vector production, titration and NSC infection
Lentiviral particles were obtained and used as described (Viral manipulation of neural stem/precursor cells, ISSN: 08932336, DOI: 10.1007/978-1-62703-610-8_14). shAsrgl1 NSCs (Asrgl1 LoF) were obtained using the pLKO.1-puro third generation vector (Sigma) with the shRNA sequence TRCN32311 (CCGGCCAGAGTTCAACGCAGGTTATCTCGAGATAACCTGCGTTGAACTCTGGTTTTTG) targeting mouse Asrgl1. Asrgl1 GoF NSCs were obtained using the pCDH-EF1-MCS-T2A-GFP third generation vector (System Biosciences) with the coding sequence expressing the mouse Asrgl1. A Multiplicity of Infection (MOI) of 30 was used for both shAsrgl1 and Asrgl1 GoF NSCs.
LC-MS metabolomic analysis
For the LC separation, column used was the Sequant ZIC-HILIC (150mm × 4.6 mm, particle size 5 μm) with a guard column (20 mm × 2.1 mm, 5 µm) from Merck Millipore (HiChrom, Reading, UK). Mobile phase A: 0.1% formic acid v/v in water. Mobile B: 0.1% formic acid v/v in acetonitrile. The flow rate was kept at 300 μL/min and gradient was as follows: 0 minutes 80% of B, 12 minutes 50% of B, 26 minutes 50% of B, 28 minutes 20% of B, 36 minutes 20% of B, 37-45 minutes 80% of B. The mass spectrometer (Thermo Q-Exactive Orbitrap) was operated in a polarity-switching mode. Samples were randomised to avoid bias due to machine drift. Chromatographic peaks were integrated using XCalibur Quan software (Thermo).
Untargeted metabolomics and metabolic tracing analyses
DMEM high glucose (Life Technology) was incubated for 24 hours at 37°C and 5% CO2 with or without isolated EVs, corresponding to 100 µg of protein extract. To compare metabolic activities of EVs, conditioned medium (CM) and supernatant (SN), medium was supplemented with 50 μM 15N2-L-Asn and 2.5mM 13C5-Gln and incubated for 24 hours at 37°C and 5% CO2 with or without isolated EVs. For all other isotope tracing experiments medium was supplemented with equimolar concentrations of 15N2-L-Asn and 13C5-Gln (50 μM). To assess NSC L-asparaginase activity 1.5*10e6 Ctrl or Asrgl1 GoF cells were seeded in 6-well plates and incubated at 37°C with medium containing 50 μM 15N2-L-Asn for 24 hours. At the end of incubation period cells or EVs were harvested and centrifuged for 5 minutes at 300g or 30 minutes at 100000g, respectively. Supernatants were collected and NPC pellets were washed three times in PBS. Extracellular metabolites were extracted by diluting 50 μL of supernatant in 750 μL extraction buffer (EB, MeOH:ACCN:H2O, 50:30:20). NSC intracellular metabolites were extracted by re-suspending washed cellular pellets in EB (1mL/10e6 cells). Extracted samples were mixed at 4°C for 15 minutes and proteins were precipitated by centrifugation at 16000g at 4°C for 10 minutes. Supernatant was collected and submitted to LC-MS analysis. To investigate extracellular metabolic activity LC-MS peak areas of fresh medium compared to EVs spent medium were compared. Absolute quantification of metabolites was obtained by comparing external standard curves composed of at least 4 dilution points.
Quantification of Asrgl1 activity in intact NSCs and EVs was performed by quantifying Asrgl1 protein abundance in the different fractions. Image J was used to quantify the signal of the active form (alpha-chain) of ASRGL1 and to extrapolate the amount of Asrgl1 in NSCs and EVs. 15N-Asparagine consumption from NSCs, EVs, and recombinant protein was measured using LCMS and normalised to Asrgl1 content.
Statistical analysis
For untargeted metabolomics analysis statistical analysis was performed with the R package “muma” (https://cran.r-project.org/) by comparing levels of metabolites in fresh and EVs conditioned medium via student’s t test. Benjamini-Hochberg correction for multiple testing and FDR=0.05 were applied to determine statistical significance. For isotope tracing experiments statistical analysis was performed with Prism Graphpad and tests were applied as indicated in figure legends.
Supplementary Material
Acknowledgments
The authors thank Francesco Dazzi, Claudio Mauro, Jayden Smith, and Aviva Tolkovsky for critically discussing the article. We are grateful to Julien Muzard (iZON) for his help with the qNano. We acknowledge the technical assistance of Iacopo Bicci, Matthew Davis, Florian Gessler, Giovanni Pluchino, and Beatriz Vega-Blanco. This work has received support from the Italian Multiple Sclerosis Association (AISM, grant 2010/R/31 and grant 2014/PMS/4 to SP), the Italian Ministry of Health (GR08-7 to SP), the European Research Council (ERC) under the ERC-2010-StG Grant agreement n° 260511-SEM_SEM, the Medical Research Council, the Engineering and Physical Sciences Research Council, and the Biotechnology and Biological Sciences Research Council UK Regenerative Medicine Platform Hub “Acellular Approaches for Therapeutic Delivery” (MR/K026682/1 to SP), The Evelyn Trust (RG 69865 to SP), The Bascule Charitable Trust (RG 75149 to SP) and core support grant from the Wellcome Trust and Medical Research Council to the Wellcome Trust – MRC Cambridge Stem Cell Institute. N.I. was supported by a FEBS long-term fellowship. C.F., A.S.H., and E.G. were funded by the Medical Research Council, Core Fund SKAG006.
Footnotes
Author contributions
NI, EG, CF, and SP conceived the study and designed the experiments; NI, TL, CC, LPJ, JDB elucidated the trafficking of Asrgl1 into NSC-EVs, and performed experiments including cell culture preparations, EV purification from media, tunable sensitive pulse sensing (TRPS), western blots, RT and qPRC, vector production, and generation of gain-of-function and loss-of-function tools; NI, CB and NF performed Nanoparticle Tracking Analysis (NTA); ASHC and EG performed the LC-MS metabolomic analyses; ALV, LGO, CF, and SP provided key reagents and resources; NI, EG, ASHC, TL, AJE, CF, and SP analyzed the data, interpreted, and discussed the results; NI, EG, CF, and SP prepared the figures, wrote, and edited the manuscript; CF and SP supervised the research.
Competing Financial Interests
NI, EG, TL, CF and SP are listed as inventors in a patent application on related to the technology described in this work (European patent application No. 16189525.5).
Data Availability Statement
Request for materials, associated protocols, and other supporting data should be sent to S. Pluchino (spp24@cam.ac.uk) or C. Frezza (cf366@MRC-CU.cam.ac.uk).
References
- 1.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80:1948–57. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DK, Lee J, Simpson RJ, Lotvall J, Gho YS. EVpedia: A community web resource for prokaryotic and eukaryotic extracellular vesicles research. Semin Cell Dev Biol. 2015;40:4–7. doi: 10.1016/j.semcdb.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Kalra H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 6.Cossetti C, et al. Extracellular vesicles from neural stem cells transfer IFN-gamma via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell. 2014;56:193–204. doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia NA, Moncayo-Arlandi J, Sepulveda P, Diez-Juan A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc Res. 2016;109:397–408. doi: 10.1093/cvr/cvv260. [DOI] [PubMed] [Google Scholar]
- 8.Ronquist KG, Ek B, Stavreus-Evers A, Larsson A, Ronquist G. Human prostasomes express glycolytic enzymes with capacity for ATP production. Am J Physiol Endocrinol Metab. 2013;304:E576–82. doi: 10.1152/ajpendo.00511.2012. [DOI] [PubMed] [Google Scholar]
- 9.Minciacchi VR, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6:11327–41. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson SM, et al. Metabolic reprogramming of bone marrow stromal cells by leukemic extracellular vesicles in acute lymphoblastic leukemia. Blood. 2016;128:453–6. doi: 10.1182/blood-2015-12-688051. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5 doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobb RJ, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thery C, et al. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr. 2000;56:791–4. doi: 10.1107/s090744490000679x. [DOI] [PubMed] [Google Scholar]
- 15.Mazzini L, et al. Human neural stem cell transplantation in ALS: initial results from a phase I trial. J Transl Med. 2015;13:17. doi: 10.1186/s12967-014-0371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliakov A, Spilman M, Dokland T, Amling CL, Mobley JA. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate. 2009;69:159–67. doi: 10.1002/pros.20860. [DOI] [PubMed] [Google Scholar]
- 17.Cantor JR, Stone EM, Chantranupong L, Georgiou G. The human asparaginase-like protein 1 hASRGL1 is an Ntn hydrolase with beta-aspartyl peptidase activity. Biochemistry. 2009;48:11026–31. doi: 10.1021/bi901397h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 19.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228:1713–9. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- 21.Fafournoux P, Bruhat A, Jousse C. Amino acid regulation of gene expression. Biochem J. 2000;351:1–12. doi: 10.1042/0264-6021:3510001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braicu C, et al. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22:34–45. doi: 10.1038/cdd.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christie GR, Hyde R, Hundal HS. Regulation of amino acid transporters by amino acid availability. Curr Opin Clin Nutr Metab Care. 2001;4:425–31. doi: 10.1097/00075197-200109000-00014. [DOI] [PubMed] [Google Scholar]
- 24.Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–51. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan LB, et al. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–63. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petroff OA. GABA and glutamate in the human brain. Neuroscientist. 2002;8:562–73. doi: 10.1177/1073858402238515. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe M, et al. Gamma-aminobutyric acid (GABA) and cell proliferation: focus on cancer cells. Histol Histopathol. 2006;21:1135–41. doi: 10.14670/HH-21.1135. [DOI] [PubMed] [Google Scholar]
- 30.Stepulak A, Rola R, Polberg K, Ikonomidou C. Glutamate and its receptors in cancer. J Neural Transm (Vienna) 2014;121:933–44. doi: 10.1007/s00702-014-1182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 32.Sonveaux P, et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7:e33418. doi: 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haas R, et al. Lactate Regulates Metabolic and Pro-inflammatory Circuits in Control of T Cell Migration and Effector Functions. PLoS Biol. 2015;13:e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bush LA, et al. A novel asparaginase-like protein is a sperm autoantigen in rats. Mol Reprod Dev. 2002;62:233–47. doi: 10.1002/mrd.10092. [DOI] [PubMed] [Google Scholar]
- 35.Duval M, et al. Comparison of Escherichia coli-asparaginase with Erwinia-asparaginase in the treatment of childhood lymphoid malignancies: results of a randomized European Organisation for Research and Treatment of Cancer-Children's Leukemia Group phase 3 trial. Blood. 2002;99:2734–9. doi: 10.1182/blood.v99.8.2734. [DOI] [PubMed] [Google Scholar]
- 36.Karpel-Massler G, et al. Metabolic reprogramming of glioblastoma cells by L-asparaginase sensitizes for apoptosis in vitro and in vivo. Oncotarget. 2016 doi: 10.18632/oncotarget.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ollenschlager G, et al. Asparaginase-induced derangements of glutamine metabolism: the pathogenetic basis for some drug-related side-effects. Eur J Clin Invest. 1988;18:512–6. doi: 10.1111/j.1365-2362.1988.tb01049.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.