Abstract
Aflatoxin B1 (AFB1) is a potent human and animal hepatocarcinogen. To investigate the effects of aflatoxin on miRNA expression during the initiation phase of carcinogenesis, next-generation sequencing was used to analyze liver tissues from F344 rats exposed to 200 μg/kg per day AFB1 for 4 weeks. A panel of miRNAs was identified that was upregulated with AFB1 treatment compared to controls: rno-miR-434-3p, rno-miR-411-5p, rno-miR-221-3p, rno-miR-127-3p, rno-miR-205, rno-miR-429, rno-miR-34a-5p, rno-miR-181c-3p, rno-miR-200b-3p, and rno-miR-541-5p. Analysis of rat livers exposed to AFB1 plus the chemopreventive triterpenoid CDDO-Im revealed a striking abrogation of this upregulation. These changes were validated by real-time PCR. We also explored the temporal variation in expression of the candidate miRNAs during the 4-week dosing period. Most of the candidate miRNAs were upregulated at week 1 and increased for the duration of AFB1 dosing over the 4-week period. Treatment with CDDO-Im ameliorated these effects at all time points. All candidate miRNAs were detectable in serum from aflatoxin treated animals; however, there was no significant difference in expression for 7 of the 11 miRNAs examined. Exposure to AFB1 upregulated miR-122-5p (5 fold), 34a-5p (13 fold), and 181c-3p (170 fold) compared with controls. The findings from this study give insight into epigenetic changes induced by aflatoxin taking place during the initial step of carcinogenesis.
Keywords: miRNA, aflatoxin B1, CDDO-Im, RNA sequencing, hepatocellular carcinoma
Introduction
Aflatoxin is a potent human and animal carcinogen produced by the fungi Aspergillus flavus and parasiticus that flourish on many staple food crops during environmentally stressful conditions. Ingestion of this compound leads to the development of hepatocellular carcinoma (HCC), the most common form of liver cancer [1]. HCC accounts for at least 750,000 deaths across the globe each year [2]. It is estimated by formal risk assessment that aflatoxin may singularly play a causal role in 4.6–28.2% of all HCC cases, with the population attributable risk estimated to be 17% in high exposure areas [3,4]. Thus, the public health impact of aflatoxin is substantial, spanning diverse populations, and warrants expanded effort in exposure mitigation and HCC prevention and early detection in these areas [5,6].
Mechanistic studies of aflatoxin have been accelerated through the development of a quantitative rat model that affords the opportunity to examine the biological impact of this carcinogen across the life course. Many studies to date have shown that rats are highly susceptible to this toxin, develop preneoplastic foci and HCC, and utilize metabolic activation pathways yielding DNA adduct formation that are qualitatively and quantitatively very similar to humans [6,7]. This quantitative rat model has also been used to evaluate a number of cancer chemoprotective agents, including the potent synthetic triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)- dien-28-oyl]imidazole (CDDO-Im)[8,9]. In a recent bioassay, we observed that co-treatment of AFB1 with CDDO-Im during a 28 day dosing period evoked complete protection against aflatoxin-induced hepatocarcinogenesis [9].
The power of this validated, quantitative model is being harnessed to gain insight into emerging sensitive biomarkers that can serve as a reliable method for predicting increased risk for the development of HCC. Mature microRNAs (miRNAs, miRs) are small non-coding RNAs (19–25 nts) that control gene expression post-transcriptionally by binding the 3′ untranslated region (UTR) of messenger RNAs [10]. Previous studies have shown that miRNAs are functional in developmental and physiological processes [11]. In addition, miRNAs are dysregulated in various diseases, especially cancer, and are being explored as biomarkers for disease risk and prognosis [11–14]. Most studies to date analyzing the abnormal expression of miRNAs in HCC have focused on end stage disease [15,16]. There are very few studies that examine miRNA changes due to aflatoxin exposure, especially throughout the initiation and post-initiation phases of carcinogenesis. In this study, we utilized next-generation sequencing to investigate the global miRNA profile of animals subchronically dosed with aflatoxin. We also examined the impact of the chemopreventive agent CDDO-Im on miRNA expression. From these data, we have identified a panel of miRNAs that could be used to predict the trajectory to HCC following aflatoxin exposure.
Materials & Methods
Biological Sample Selection
Tissue samples were obtained from a 4-week and a lifetime bioassay assessing burdens of DNA adducts and hepatic preneoplastic lesions that has been previously described [9]. Rats were randomly assigned to two treatment groups: 200 μg/kg AFB1 daily, or 200 μg/kg plus 30 μmol/kg rat CDDO-Im as shown in Figure 1. There were three additional animals on control diet maintained during the 4 weeks of the dosing regimen. After 7, 14, 21 or 28 days of dosing animals were euthanized and necropsied with hepatic tissue and serum stored. These samples were analyzed using RNA-seq (tissue) and RT-qPCR (tissue & serum). Sera from pairs of AFB1 only and AFB1 plus CDDO-Im treated animals sacrificed at 70, 75, 80, and 85 weeks of age were analyzed by RT-qPCR. All experiments were approved by The Johns Hopkins University Animal Care and Use Committee.
Figure 1.
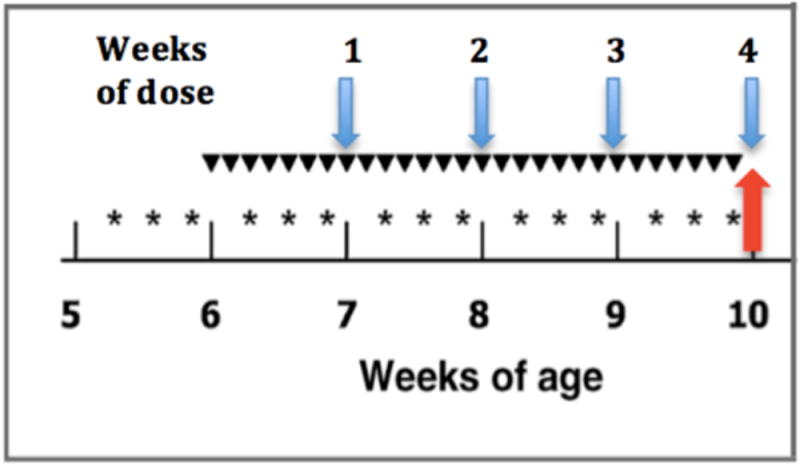
Schematic of dosing protocol. Time of treatment with (*) 30 μmol CDDO-Im, p.o., (▼) 200 μg AFB1/kg BW, p.o., and total weeks of administration. Arrows indicate sampling time points for sequencing (red) and time course (blue) experiments.
RNA Isolation
Rat liver tissue
Total RNA was isolated from approximately 10 mg liver tissue using the miRCURY Isolation tissue kit (Exiqon, Vedbaek, Denmark) according to the manufacturer’s protocol. RNA was eluted with 50 μL RNase-free water.
Rat serum samples
Small RNAs (<1000 nts) were isolated from 50μL serum using the miRCURY miRNA Isolation kit for biofluids (Exiqon, Vedbaek, Denmark) according to the manufacturer’s protocol. RNA was eluted with 50μL RNase-free water.
Small RNA Sequencing
Samples with an RNA Integrity Number (RIN) greater than 7 identified using the 2100 Bioanalyzer (Agilent, Santa Clara, CA) were used for subsequent library preparation and sequencing. The TruSeq Small RNA Sample Preparation kit (Illumina, San Diego, CA) was used for generating small RNA libraries following the manufacturer’s protocol, using a total amount of 1μg total RNA. Samples were prepared with a unique index and sequenced across one lane with single-end 50 bp reads. After library prep and cluster generation, the cDNA libraries were sequenced using the HiSeq 2000 platform (Illumina, San Diego, CA).
Analysis of RNA sequencing data
From the total output reads, contaminant reads with length less than 15 bp and a phred score less than 20 were discarded. The processed reads were aligned to miRNA sequences in the rat genome by conducting a BLAST search (NCBI, v.2.2.30) against a built database of rat miRNA sequences downloaded from miRBase v. 21 (www.mirbase.org)[17]. Only hits with 100% identity and matching the complete length of the miRNA were counted. Normalization of the count data was calculated using estimateSizeFactors. The variance estimation was calculated using estimateDispersions. Count data was averaged across all biological replicates for each treatment group and normalized to reads per million miRNA reads for each miRNA across all treatment groups (Equation 1). Bioconductor’s DESeq package was used for differential expression analysis.
| Equation 1) |
Candidate microRNA selection
Count data was primarily used for candidate selection. After calculating the fold change for all miRNAs with reference to the control group, a list was created containing the miRNAs that had greater than 10-fold change between the AFB1 treated and control groups. From that list, multiple candidate lists were generated that contained the top dysregulated miRNAs as well as functional information. A finalized panel of 10 candidate miRNAs was determined that included the most dysregulated miRNAs with known biological significance.
MicroRNA expression analysis
Nine ng total eluted RNA from tissue was reverse transcribed (RT) using the TaqMan miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers following the manufacturer’s instructions (Applied Biosystems, Foster City, CA). The expression level of the candidate miRNAs was determined by quantitative real-time PCR (qRT-PCR) using TaqMan miRNA assays (Applied Biosystems) and the StepOne Plus System (Applied Biosystems). All assays were analyzed in triplicate. The relative amount of each miRNA was calculated using the 2−ΔΔCt method [18].
Statistical Analysis
Graphs represent the mean ± SD. All statistical analyses were performed using GraphPad Prism 7.0 software. Analysis of variance (ANOVA) tests were used for statistical analysis of differences among the three treatment groups followed by Tukey’s multiple comparisons test. The level of significance was set at p<0.05; *p < 0.05; **p<0.005; ***p<0.0005.
Clinical Chemistry Measurements
Levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured in serum samples by the VetACE Chemistry Analyzer (Alfa Wasserman, West Caldwell, NJ). AST normal range: 192–262 U/L; ALT normal range: 16–89 U/L.
Results
Perturbation of the global miRNA profile in rat liver by AFB1 and its modulation by the triterpenoid, CDDO-Im
Initial experiments were performed to quantitatively characterize the global miRNA expression profiles in rat liver using RNA sequencing. Samples were obtained from both animals that had been fed a control diet and rats that had been dosed for 28 consecutive days with AFB1, a cancer-inducing regimen. The AFB1 exposed animals had been further divided into two experimental groups; one of which received additionally the chemoprotective triterpenoid CDDO-Im during the dosing protocol (Figure 1). All of these rat liver samples had been obtained as part of a liver cancer study that demonstrated the nearly complete protection (96%) against aflatoxin-induced hepatocarcinogenesis by CDDO-Im [9]. Thus, this study explored the patterns of miRNAs during the critical initiation phase of aflatoxin-induced carcinogenesis that establishes the trajectory to disease outcome over the lifetime of the rat.
A total of nine liver samples from the 4-week protocol; three control diet, three AFB1, and three AFB1 plus CDDO-Im were sequenced for miRNA using the Illumina HiSeq 2000 platform yielding an average of 14,132,166 clean reads per sample in the control group, 20,674,284 clean reads per sample in the AFB1 only group, and 19,568,578 clean reads per sample in the AFB1 plus CDDO-Im treatment group (Figure S1). These reads were then explored using the miRNA database miRBase (www.mirbase.org), identifying a total of 579 known rat miRNAs. Of these identified miRNAs, 173 miRNAs were expressed to levels with statistical significance as adjudged by abundances above 50 reads per million aligned miRNA reads. As shown in figure 2, the significantly expressed miRNAs varied in distribution among the control and treatment groups. The majority of identified miRNAs (75%, 130 miRNAs) were expressed in the livers of animals from all three experimental groups. There were 29 miRNAs (17%) that were exclusively expressed in the AFB1 treatment group (figure 2, Table 1). Seven miRNAs (4%) were significantly expressed across the AFB1 only and AFB1 plus CDDO-Im regimens. Three miRNAs (2%) were shared between the control and AFB1 groups. The miRNAs rno-miR-26b-3p and rno-miR-3577 were predominantly expressed in the control group, while rno-miR-29b-3p and rno-miR-96-5p were only expressed in the AFB1 plus CDDO-Im test group (Table 1). These RNA-seq data were further examined by pairwise comparison analysis revealing a total of 96 miRNAs differentially expressed between control and AFB1 treatment groups, and 34 miRNAs differentially expressed between control and AFB1 plus CDDO-Im treatment groups. Finally, there are 135 miRNAs differentially expressed between the AFB1 and CDDO-Im intervention groups. Collectively, these initial analyses demonstrated that the carcinogenic 28-day dosing by AFB1 affected the expression of multiple miRNAs in the liver and that co-treatment with CDDO-Im abrogated the changed expression of many of them
Figure 2.

Venn diagram of significantly expressed miRNAs identified by RNA-seq [61]. Number of miRNAs with greater than 50 aligned reads per million miRNA reads in expression for each biological replicate in control, aflatoxin (AFB1), and aflatoxin plus CDDO-Im (AFB1+CDDO-Im) treatment groups.
Table 1.
Distribution of shared and exclusively expressed miRNAs in treatment groups
| Treatment Group | MicroRNAs |
|---|---|
| AF* only | 409a-3p, 541-5p, 410-3p, 127-3p, 136-3p, 411-5p, 205, 434-3p, 429, 204-5p, 141-3p, 181c-3p, 92b-3p, 130b-3p, 181d-5p, 221-5p, 200c-3p, 221-3p, 222-3p, 455-5p, 421-3p, 455-3p, 10b-5p, 374-5p, 1247-5p, 155-5p, 582-3p, 351-5p, 195-3p |
| AF & AF+CDDO-Im† | 200a-3p, 214-3p, 181a-1-3p, 24-2-5p, 19a-3p, 542-3p, 450b-5p |
| AF & Control | 342-3p, 652-3p, 361-3p |
| AF+CDDO-Im only | 29b-3p, 96-5p |
| Control only | 26b-3p, 3577 |
| AF+CDDO-Im | – |
Ordered by decreasing abundance;
AF, Aflatoxin;
CDDO-Im, 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole
Validation of sequencing data by RT-qPCR
To independently validate the small RNA sequencing data set described above, candidate miRNAs were examined in detail by RT-qPCR. Qualitatively, the results from the RT-qPCR analyses proved to be completely consistent with the RNA sequencing data. This prompted a more detailed set of quantitative studies using RT-qPCR. The criteria for selecting candidate miRNAs included biological significance in the literature and a focus on miRNAs that displayed greater than a 10-fold change in hepatic expression between control and AFB1 treatment groups, as these might exhibit sufficient dynamic range as to make them suitable as biomarkers. The miRNAs rno-miR-541-5p, 34a-5p, 127-3p, 205, 434-3p, 429, 411-5p, 181c-3p, 200b-3p, 221-3p were selected because RNA-seq data showed that they were all upregulated in the AFB1-treated samples compared to controls (Table 2). These miRNAs are either identical to human miRNAs or have human homologs. In addition, the putative liver specific rno-miR-122 was also examined. Prior studies have shown this miRNA to be the most abundant in the liver amounting to 52% percent of the total miRNAs in the human liver [19,20].
Table 2.
List of candidate miRNAs and expression abundances by RNA sequencing
| Normalized to reads per million miRNA reads | Average Count* | Ratio | ||||||
|---|---|---|---|---|---|---|---|---|
| miRNA | CTL* | AF† | CD‡ | % reduction by CDDO-Im | Overall CTL:AF:CD | AF:CTL | CD:CTL | AF:CD |
| 122-5p | 36761 | 32422 | 21258 | 34.4 | 1:1:1 | 1:1 | 1:1 | 2:1 |
| 434-3p | 7 | 246 | 2 | 99.3 | 1:33:0 | 33:1 | 0:1 | 154:1 |
| 411-5p | 4 | 228 | 2 | 99.0 | 1:65:1 | 65:1 | 1:1 | 104:1 |
| 221-3p | 35 | 281 | 28 | 90.1 | 1:8:1 | 8:1 | 1:1 | 10:1 |
| 127-3p | 24 | 1941 | 8 | 99.6 | 1:82:0 | 82:1 | 0:1 | 231:1 |
| 205 | 2 | 83 | 1 | 98.4 | 1:40:1 | 40:1 | 1:1 | 64:1 |
| 429 | 18 | 481 | 43 | 91.1 | 1:27:2 | 27:1 | 2:1 | 11:1 |
| 34a-5p | 33 | 2675 | 252 | 90.6 | 1:81:8 | 81:1 | 8:1 | 11:1 |
| 181c-3p | 4 | 94 | 5 | 95.1 | 1:22:1 | 22:1 | 1:1 | 21:1 |
| 200b-3p | 138 | 3005 | 562 | 81.3 | 1:22:4 | 22:1 | 4:1 | 5:1 |
| 541-5p | 1 | 136 | 0 | 99.9 | 1:136:0 | 136:1 | 0:1 | 680:1 |
Average read count for 3 biological replicates within treatment group;
CTL, Control;
AF, Aflatoxin;
CD, Aflatoxin and CDDO-Im co-treatment
Figure 3 depicts the miRNA findings from the RT-qPCR experiments. ANOVA statistical tests revealed that all of the candidate miRNAs were significantly upregulated in AFB1 treated samples, with rno-miR-541 having the largest increase compared to control (~1000 fold). Treatment with CDDO-Im reduced the expression levels of all candidate miRNAs; even shifting expression levels for rno-miR-411-5p and rno-miR-127-3p to below control levels (Figure 3). Analysis of clinical chemistries in serum samples at the 4-week dosing point was also performed in parallel to miR-122 expression levels. Studies have shown miR-122 as an indication of liver toxicity and its potential use as a biomarker of acute liver injury in humans [21–25]. Results from this study revealed no increase in serum ALT and AST levels, suggesting a compensating mechanism occurring in the livers of these animals despite being given 28 doses of AFB1 (Table S2). For AST, all animals had levels below 210 U/L except for one animal. All animals had ALT levels below 80 U/L.
Figure 3.
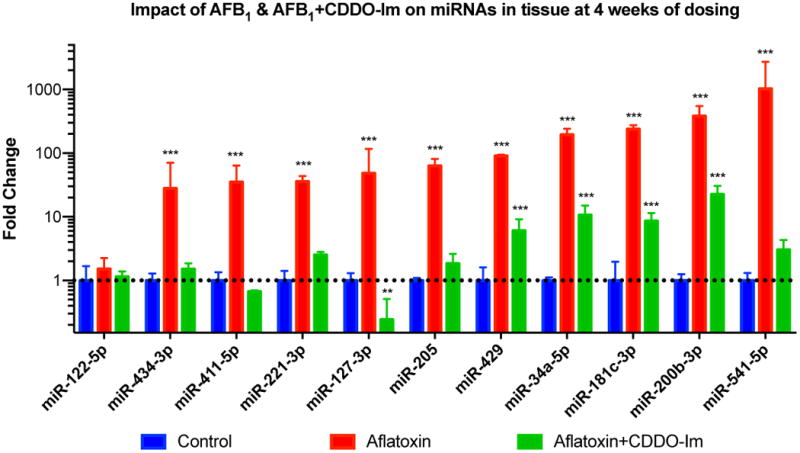
Real-time PCR analysis of candidate miRNAs in liver tissue after 4 weeks of dosing (mean ± SD). Fold change calculated using the 2−ΔΔCt method. (*) indicates significant difference (p<0.05) when compared to control for that miR, (**) indicates significant difference (p<0.01) when compared to control for that miR, (***) indicates significant difference (p<0.001) when compared to control for that miR.
Temporal trends in miRNA expression during chronic dosing of AFB1 and its modulation by CDDO-Im
Most studies in the current literature evaluating miRNA expression during disease focus on one specific time point, particularly at end stage disease diagnosis. To further understand the effect of AFB1 on miRNA expression, we analyzed candidate miRNA expression levels during the 4-week carcinogenic dosing regimen by RT-qPCR (Figure 1, blue arrows). MicroRNA expression increased over time with exposure to aflatoxin compared to controls. Changes in expression for some miRNAs did not occur until 21 doses of aflatoxin (e.g. 411-5p, 205, 127-3p, 541-5p). rno-miR-34a showed a marked increase after just 7 doses of AFB1 and maintained increased expression throughout the dosing period (Figure 4a). The miRNAs 181c-3p and 541-5p displayed the most dramatic change in expression between the first and last week of treatment. Co-treatment with CDDO-Im attenuated these effects with expression levels of all candidate miRNAs not exhibiting statistically significant changes over the dosing period (Figure 4b, d). Thus, the selected candidate miRNAs were all upregulated and tracked with aflatoxin dosing over time. This effect was reversed by chemoprotective intervention with CDDO-Im, further highlighting the possible role of these miRNAs as candidate biomarkers of risk of developing HCC.
Figure 4.
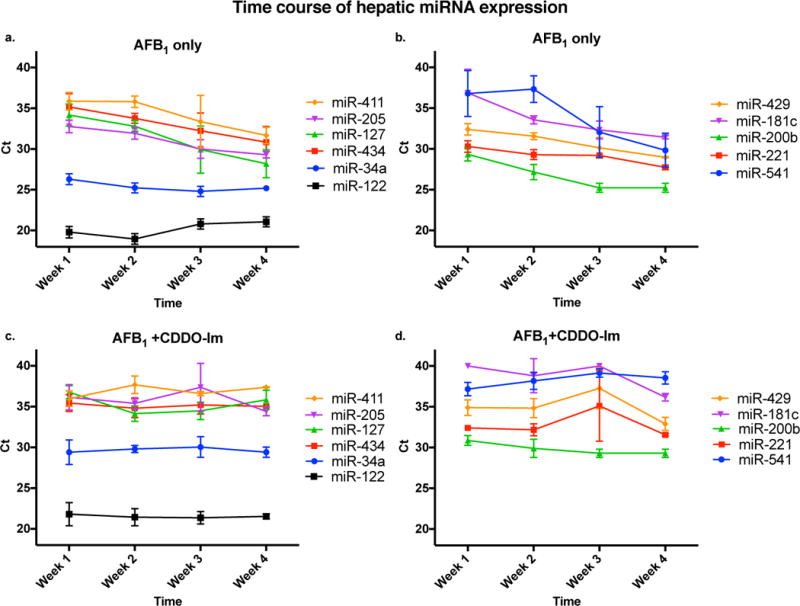
Time course of candidate miRNA expression in liver tissue during the 4 week dosing period measured by RT-qPCR in a-b) aflatoxin only treated samples; majority of miRNAs (except 34a, 122, 429, and 221) have a significant increase in expression from week 1 to week 3; majority of miRNAs (except 34a and 122) are significantly increased in expression from week 1 to week 4 of aflatoxin treatment; and c–d) aflatoxin plus CDDO-Im treated samples; no significant change in expression from week 1 to week 4 for all miRNAs except miR-181c; significant decrease in expression of miRs 429 and 221 from week 1 to week 3.
Detection of candidate miRNAs in serum samples
It is known from the literature that miRNAs are highly stable in the circulation, found in extracellular vesicles or bound to proteins [26,27]. To examine the potential of the candidate miRNAs identified in this study to be reliable blood-based biomarkers, the miRNAs described above were explored in serum by RT-qPCR at the end of the 4-week dosing period. These experiments showed that all candidate miRNAs were detectable in aflatoxin treated samples; however, there was no significant difference in expression for 7 of the 11 miRNAs examined (Figure 5). Exposure to AFB1 significantly upregulated miR-122-5p (5 fold), 34a-5p (13 fold), and 181c-3p (170 fold) compared with controls. Interestingly for miR-541, treatment with AFB1 dramatically decreased the expression of this miRNA, though not statistically significant. Co-treatment with CDDO-Im eliminated these changes. Serum samples from animals diagnosed with HCC (only exposed to aflatoxin) were also examined and compared to sera obtained from animals at the same age dosed with CDDO-Im or controls. According to preliminary data, the miRNAs 34a-5p and 181c-3p were detectable in all serum samples, but miRs-122-5p and 541 were not detectable in the control samples. The expression levels of these four miRNAs were found to be identical in the aflatoxin and aflatoxin plus CDDO-Im treatment groups. Further studies tracking patterns of serum miRNA across time-course after the dosing regimen and leading to cancer will need to be done to validate any of the other candidate miRNAs as biomarkers of HCC.
Figure 5.
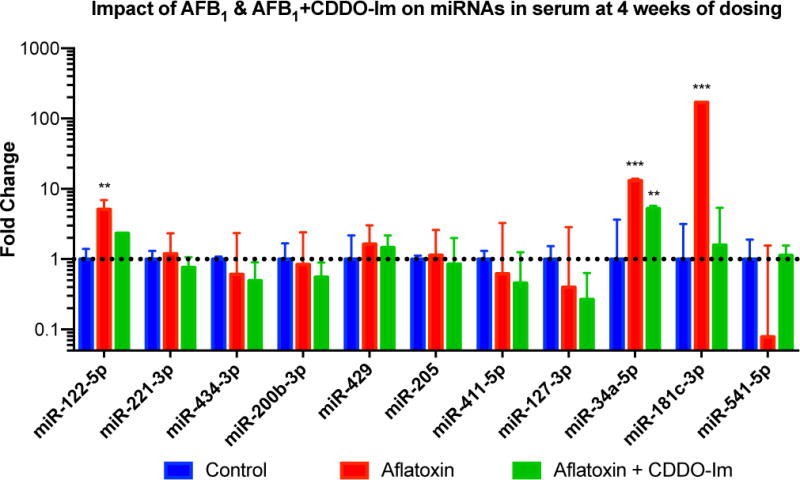
Real-time PCR analysis of candidate miRNAs in serum after 4 weeks of dosing (mean ± SD). Fold change calculated using the 2−ΔΔCt method. Large variability in rno-miR-541 expression in aflatoxin group due to an animal outlier. (*) indicates significant difference (p<0.05) when compared to control for that miR, (**) indicates significant difference (p<0.01) when compared to control for that miR, (***) indicates significant difference (p<0.001) when compared to control for that miR.
Discussion
MicroRNAs play an important role in biological processes such as embryogenesis, cell growth and differentiation, and tumorigenesis. The altered expression of miRNAs due to disease or environmental exposures has been shown in multiple organisms, but little research has been done during cancer initiation with environmental carcinogens such AFB1. Some miRNAs have been identified as potential disease biomarkers for liver cancer based on miRNA targets and functionality, such as miR-122-5p and miR-429 [28–30]. In such studies, however, the focus is on end stage disease [31–35]. Few studies have examined how aflatoxin affects miRNA expression [36–40]. The present study used a quantitative rat model to explore the dysregulation of miRNAs during cancer initiating exposure to aflatoxin but before the development of clinical disease. Global miRNA profiles were agnostically obtained using next-generation RNA sequencing that identified a subset of miRNAs for validation.
In this study, rats dosed with aflatoxin over four consecutive weeks had 96 differentially expressed hepatic miRNAs compared to controls, with the majority being upregulated. From that expression abundance data, further analysis yielded a candidate list of 10 miRNAs, which were all significantly upregulated compared to controls. This effect was reduced by co-exposure to CDDO-Im. Only two in vivo studies to date have explored the dysregulation of miRNAs due to aflatoxin at this early stage of disease. Yang et al., 2014 found that a high acute dose (1.5 mg/kg) of AFB1 caused significant changes in the hepatic expression of miRNAs such as miR-34a and 92a [39]. A study by Liu et al., 2015 also showed the upregulation of rno-miR-34a and the abnormal hepatic expression of other miRs (rno-miR-200b, rno-miR-429, rno-miR-542, rno-miR-130a-3p) after four weeks of a relatively low dose of AFB1 (100 μg/kg or 200 μg/kg every two days) [36]. The present study found rno-miR-34a, 429, and 200b-3p to be upregulated, as well as rno-miRs- 541, 181c-3p, 205, 434-3p, 411-5p, 127-3p and 221-3p after 4-week exposure to 200 μg/kg AFB1 (daily). Frequently dysregulated in different cancers, miR-34a has been shown to regulate many processes such as apoptosis and cell proliferation [41]. This miRNA is typically downregulated in HCC, but studies have found miR-34a to be increased in response to genotoxic agent exposure [42,43]. In this investigation miR-34a could be upregulated at such an early time point in response to aflatoxin exposure (Figure 4a). Also dysregulated in many cancers, miR-429 (a member of the miR-200 family) has been shown to be involved in metastasis in HCC and correlated with higher burden of aflatoxin-DNA adducts [30,44]. The mir-181c-3p was one of many surprising candidate miRNAs found in this study. This miRNA is thought to be protective in leukemia and glioblastoma [45,46]. Little information is known about this miR in the context of HCC except a study showing subsequent increase in expression after Wnt/β-catenin signaling activation [47]. We also found several miRNAs to be exclusively induced by aflatoxin (Table 1). Many of those were chosen as candidate miRNAs in a separate analysis. Additional standouts from this exclusive list include rno-miR-204-5p and rno-miR-410-3p. Important for eye development and adipogenesis, miR-204 is reported to be a tumor suppressor in HCC [48,49]. miR-410 is clustered with miR-541, a candidate miRNA we identified in this study, and is shown to be overexpressed in clinical liver cancers [50]. This miRNA has also been found to be upregulated in human liver cells after exposure to aflatoxin [38].
The majority of microRNAs are stable molecules with a subset that are very dynamic with turnover rates ranging from hours to days [51]. To see how the hepatic expression of the candidate miRNAs change over the course of the four week dosing period, we analyzed their expression by RT-qPCR after 7, 14, 21, and 28 doses of AFB1. Most of the candidate miRNAs increased in expression and tracked with aflatoxin exposure over the dosing period, except rno-miR-122-5p and rno-miR-34a-5p which had constant high expression. miR-122 is the most abundant miRNA in the liver, so its high expression is expected. Additionally, the human miR-122 gene has been shown to contain the binding site for hepatocyte nuclear factor 4 alpha (HNF4α), a transcription factor involved in liver development, which can upregulate miR-122 expression in human Huh7 and HepaRG cells [38,52,53]. Therefore, the dysregulation of rno-miR-122 in this study could also be explained by the modulation of HNF4α by aflatoxin. Recent evidence has shown that miR-34a is transcriptionally regulated by p53 and involved in liver regeneration [36,54]. We hypothesize that the high constant expression of miR-34a during the dosing period could be due to the role of this miR in these processes. Rno-miR-541-5p had the largest significant change in expression between the first and last weeks of dosing. Little is known about this miR in the literature. In cancer, studies have found the human analogue of rno-miR-541 to be a tumor suppressor by regulating telomerase expression and targeting TGIF2 in non-small cell lung cancer [55,56]. Another microRNA found to be significantly increased during the dosing period was rno-miR-200b-3p. Related to rno-miR-429, the expression of this miRNA has been found to be reduced in methyl-deficient diet-induced HCC in rats and in HepG2 cells[57,58]. A study by Li et al., 2016 has also found the human analogue of miR-200b to be downregulated in tumor tissue directly targeting DNA methyltransferase 3a (DNMT3a) expression [57]. Co-treatment with CDDO-Im resulted in no significant change in the hepatic expression of any of the candidate miRNAs over the 4-week dosing period.
Circulating miRNAs are very attractive as a minimally invasive biomarker: they are detectable in serum and plasma, resistant to RNase activity, and stable at different temperatures and pHs [59,60]. We examined the plausibility of the candidate miRNAs, which were highly dysregulated in liver tissue, to be reliable blood-based biomarkers that predict the trajectory to developing HCC. Analysis of serum samples revealed significantly increased circulating levels of rno-miRs 122-5p, 34a-5p, and 181c-3p in animals after 28 doses of aflatoxin. Co-treatment CDDO-Im attenuated this effect, as seen in other experiments throughout this study. We also wanted to see if the miRs 34a, 181c-3p, and 541 were dysregulated in the sera of animals during end-stage disease since they were the most dysregulated miRNAs in tissue and serum during the initiation phase of the disease. Preliminary data revealed that these miRNAs were not useful at these late time points, most likely due to the complex tumor biology taking place. They may be more advantageous in indicating early biological changes occurring in the cancer initiation phase during and following exposure to aflatoxin.
In conclusion using a quantitative rat model, we have shown modifications in the global hepatic miRNA profile as well as identified a panel of miRNAs that are significantly upregulated with subchronic exposure to aflatoxin. Co-treatment with the chemopreventive agent CDDO-Im reduced the increase in expression of these miRs. Analysis of levels during the 4-week dosing period revealed these miRNAs tracked with aflatoxin exposure. These changes were diminished by CDDO-Im as well. To our knowledge, this is the first study to show a time course of miRNA expression affected by aflatoxin and effective reduction by a chemopreventive agent. Further validation of this list of miRNAs could yield a powerful biomarker of increased risk of developing liver cancer after exposure to aflatoxin.
Supplementary Material
Acknowledgments
We thank the Next Generation Sequencing Center (Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins School of Medicine) for assistance with RNA sequencing and data analysis. We thank the Phenotyping and Pathology Core (Johns Hopkins School of Medicine) for technical assistance in measuring the clinical chemistries.
Funding Support:
This research supported by NIH grants T32 ES007141, P30 CA006973, and R35 CA197222.
Abbreviations
- AFB1
Aflatoxin B1
- CDDO-Im
1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole
- miRNA or miR
microRNA
- HCC
hepatocellular carcinoma
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Wu F. Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ Health Perspect. 2010;118(6):818–824. doi: 10.1289/ehp.0901388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Chang CC, Marsh GM, Wu F. Population attributable risk of aflatoxin-related liver cancer: systematic review and meta-analysis. Eur J Cancer (Oxford, England : 1990) 2012;48(14):2125–2136. doi: 10.1016/j.ejca.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JG, Egner PA, Ng D, et al. Reduced aflatoxin exposure presages decline in liver cancer mortality in an endemic region of China. Cancer Prev Res. 2013;6(10):1038–1045. doi: 10.1158/1940-6207.CAPR-13-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler TW, Roebuck BD, Wogan GN, Groopman JD. Aflatoxin: a 50-year odyssey of mechanistic and translational toxicology. Toxicol Sci. 2011;120(Suppl 1):S28–48. doi: 10.1093/toxsci/kfq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kensler TW, Groopman JD, Roebuck BD. Use of aflatoxin adducts as intermediate endpoints to assess the efficacy of chemopreventive interventions in animals and man. Mutat Res. 1998;402(1–2):165–172. doi: 10.1016/s0027-5107(97)00294-7. [DOI] [PubMed] [Google Scholar]
- 8.Yates MS, Kwak MK, Egner PA, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66(4):2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 9.Johnson NM, Egner PA, Baxter VK, et al. Complete Protection against Aflatoxin B1-Induced Liver Cancer with a Triterpenoid: DNA Adduct Dosimetry, Molecular Signature, and Genotoxicity Threshold. Cancer Prev Res. 2014;7(7):658–665. doi: 10.1158/1940-6207.CAPR-13-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. MicroRNA: function, detection, and bioanalysis. Chem Rev. 2013;113(8):6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 11.Valencia-Quintana R, Sanchez-Alarcon J, Tenorio-Arvide MG, et al. The microRNAs as potential biomarkers for predicting the onset of aflatoxin exposure in human beings: a review. Front Microbiol. 2014;5:102. doi: 10.3389/fmicb.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allegra A, Alonci A, Campo S, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41(6):1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 13.Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56(6):1371–1383. doi: 10.1016/j.jhep.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes CN, Chayama K. MicroRNAs as Biomarkers for Liver Disease and Hepatocellular Carcinoma. Int J Mol Sci. 2016;17(3):280. doi: 10.3390/ijms17030280. doi: 210.3390/ijms17030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afonso MB, Rodrigues PM, Simao AL, Castro RE. Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. J Clin Med. 2016;5(3) doi: 10.3390/jcm5030030. (pii):E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 20.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122 – A key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Pro Natl Acad of Sci. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bala S, Petrasek J, Mundkur S, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56(5):1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starckx S, Batheja A, Verheyen GR, et al. Evaluation of miR-122 and other biomarkers in distinct acute liver injury in rats. Toxicol Pathol. 2013;41(5):795–804. doi: 10.1177/0192623312464436. [DOI] [PubMed] [Google Scholar]
- 24.Farid WRR, Pan Q, van der Meer AJP, et al. Hepatocyte-derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18(3):290–297. doi: 10.1002/lt.22438. [DOI] [PubMed] [Google Scholar]
- 25.Antoine DJ, Dear JW, Lewis PS, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58(2):777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–U672. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.El-Hefnawy T, Raja S, Kelly L, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin Chem. 2004;50(3):564–573. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 28.Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 30.Tang J, Li L, Huang W, et al. MiR-429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial-mesenchymal transition. Cancer Lett. 2015;364(1):33–43. doi: 10.1016/j.canlet.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Leung WK, He M, Chan AW, Law PT, Wong N. Wnt/beta-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015;362(1):97–105. doi: 10.1016/j.canlet.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC) Biomed Pharmacother. 2014;68(2):143–148. doi: 10.1016/j.biopha.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto Y, Kosaka N, Tanaka M, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14(7):529–538. doi: 10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Li J, Shen J, Wang C, Yang L, Zhang X. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. doi: 10.1186/1471-2407-12-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi KQ, Lin Z, Chen XJ, et al. Hepatocellular carcinoma associated microRNA expression signature: integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget. 2015;6(28):25093–108. doi: 10.18632/oncotarget.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Yu H, Zhang Y, et al. Upregulation of miR-34a-5p antagonizes AFB1-induced genotoxicity in F344 rat liver. Toxicon. 2015;106:46–56. doi: 10.1016/j.toxicon.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 37.Zhu L, Gao J, Huang K, Luo Y, Zhang B, Xu W. miR-34a screened by miRNA profiling negatively regulates Wnt/beta-catenin signaling pathway in Aflatoxin B1 induced hepatotoxicity. Sci Rep. 2015;5:16732. doi: 10.1038/srep16732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marrone AK, Tryndyak V, Beland FA, Pogribny IP. MicroRNA Responses to the Genotoxic Carcinogens Aflatoxin B1 and Benzo[a]pyrene in Human HepaRG Cells. Toxicol Sci. 2016;149(2):496–502. doi: 10.1093/toxsci/kfv253. [DOI] [PubMed] [Google Scholar]
- 39.Yang W, Lian J, Feng Y, et al. Genome-wide miRNA-profiling of aflatoxin B1-induced hepatic injury using deep sequencing. Toxicol Lett. 2014;226(2):140–149. doi: 10.1016/j.toxlet.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Liu YX, Long XD, Xi ZF, et al. MicroRNA-24 Modulates Aflatoxin B1-Related Hepatocellular Carcinoma Prognosis and Tumorigenesis. Biomed Res Int. 2014;2014:482926. doi: 10.1155/2014/482926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Koufaris C, Wright J, Currie RA, Gooderham NJ. Hepatic microRNA profiles offer predictive and mechanistic insights after exposure to genotoxic and epigenetic hepatocarcinogens. Toxicol Sci. 2012;128(2):532–543. doi: 10.1093/toxsci/kfs170. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Branham WS, Dial SL, et al. Genomic analysis of microRNA time-course expression in liver of mice treated with genotoxic carcinogen N-ethyl-N-nitrosourea. BMC genomics. 2010;11:609. doi: 10.1186/1471-2164-11-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang X-Y, Yao J-G, Huang H-D, et al. MicroRNA-429 Modulates Hepatocellular Carcinoma Prognosis and Tumorigenesis. Gastroenterol Res Pract. 2013;2013:10. doi: 10.1155/2013/804128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao L, Li Y, Song X, et al. Upregulation of miR-181c inhibits chemoresistance by targeting ST8SIA4 in chronic myelocytic leukemia. Oncotarget. 2016 doi: 10.18632/oncotarget.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruan J, Lou S, Dai Q, Mao D, Ji J, Sun X. Tumor suppressor miR-181c attenuates proliferation, invasion, and self-renewal abilities in glioblastoma. Neuroreport. 2015;26(2):66–73. doi: 10.1097/WNR.0000000000000302. [DOI] [PubMed] [Google Scholar]
- 47.Ji J, Yamashita T, Wang XW. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011;1(1):1–8. doi: 10.1186/2045-3701-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37(9):11667. doi: 10.1007/s13277-016-5144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge Y, Yan X, Jin Y, et al. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015;11(12):e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Fu J, Jiang M, et al. MiR-410 is overexpressed in liver and colorectal tumors and enhances tumor cell growth by silencing FHL1 via a direct/indirect mechanism. PloS One. 2014;9(10):e108708. doi: 10.1371/journal.pone.0108708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y, Liu J, Elfenbein SJ, et al. Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res. 2015;43(4):2326–2341. doi: 10.1093/nar/gkv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, He JH, Xiao ZD, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52(4):1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 53.Li ZY, Xi Y, Zhu WN, et al. Positive regulation of hepatic miR-122 expression by HNF4alpha. J Hepatol. 2011;55(3):602–611. doi: 10.1016/j.jhep.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 54.Chen H, Sun Y, Dong R, et al. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PloS One. 2011;6(5):e20238. doi: 10.1371/journal.pone.0020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hrdlickova R, Nehyba J, Bargmann W, Bose HR., Jr Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PloS One. 2014;9(2):e86990. doi: 10.1371/journal.pone.0086990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu YJ, Liu RY, Hu K, Wang Y. MiR-541–3p reverses cancer progression by directly targeting TGIF2 in non-small cell lung cancer. Tumour Biol. 2016;37(9):12685–95. doi: 10.1007/s13277-016-5241-5. [DOI] [PubMed] [Google Scholar]
- 57.Li XY, Feng XZ, Tang JZ, et al. MicroRNA-200b inhibits the proliferation of hepatocellular carcinoma by targeting DNA methyltransferase 3a. Mol Med Rep. 2016;13(5):3929–35. doi: 10.3892/mmr.2016.4995. [DOI] [PubMed] [Google Scholar]
- 58.Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48(6):479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Pro Natl Acad Sci. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveros JC. Venny. An interactive tool for comparing lists with Venn’s diagrams. 2007–2015 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


