Abstract
Our aim is to investigate whether or not the breast cancer metastasis suppressor 1 (BRMS1) gene expression is directly linked to clinico-pathological features of breast cancer. Following a stringent inclusion and exclusion criteria, case–control studies with associations between BRMS1 and breast cancer were selected from articles obtained by way of searches conducted through an electronic database. All statistical analyses were performed with Stata 12.0 (Stata Corp, College Station, TX, U.S.A.). Ultimately, 1,263 patients with breast cancer were found in a meta-analysis retrieved from a total that included 12 studies. Results of our meta-analysis suggested that BRMS1 protein in breast cancer tissues was significantly lower in comparison with normal breast tissues (odds ratio, OR = 0.08, 95% confidence interval (CI) = 0.04–0.15). The BRMS1 protein in metastatic breast cancer tissue was decreased than from that was found in non-metastatic breast cancer tissue (OR = 0.20, 95%CI = 0.13–0.29), and BRMS1 protein in tumor-node-metastasis (TNM) stages 1 and 2 was found to be higher than TNM stages 3 and 4 (OR = 4.62, 95%CI = 2.77–7.70). BRMS1 protein in all three major types of breast cancer was lower than that of control tissues respectively. We also found strong correlations between BRMS1 mRNA levels and TNM stage and tumor size. The results our meta-analysis showed that reduction in BRMS1 expression level was linked directly to clinico-pathological features of breast cancer significantly; therefore, suggesting the loss of expression or reduced levels of BRMS1 is potentially a strong indicator of the metastatic capacity of breast cancer with poor prognosis.
Keywords: BRMS1, Breast cancer, Clinico-pathological features, Meta-analysis, Metastasis, Prognosis
Introduction
Breast cancer accounts for approximately 33% of cancers diagnosed among females in the U.S.A., and is the second leading cause of cancer deaths worldwide [1]. Generally, older women are more commonly diagnosed with breast cancer, with one in 200 women before the age of 40 being diagnosed [2]. Generally, approximately 40% of all patients with breast cancer experience a relapse, of which recurrences 10–20% are locally metastatic and 60–70% are distant metastases [3]. The established risk factors for breast cancer are age, menopausal status, body mass index (BMI), duration of breastfeeding, age at first pregnancy, and postmenopausal hormonal use [4–6]. The predominant cause of death in breast cancer is distant metastasis [7]. Tumor metastasis is a multistep process involving disruption of intercellular adhesions and dispersal of single cells from solid tumor, invasion of blood and lymphatic vessels, immunologic escape in circulation, attachment to endothelial cells, extravasation from blood and lymph vessels and proliferation, and angiogenesis induction [8,9]. This process is assisted by a preferred microenvironment at the primary and metastatic sites [7].
Breast cancer metastasis suppressor 1 (BRMS1), introduced in 2000, inhibits metastasis while having no effect on the growth of primary tumors [10]. The BRMS1 gene is located at 11q13.1-13.2, a region often altered in late-stage breast cancers, while in close proximity to the genomic loci that contains deletions and amplifications commonly observed in progression of breast cancer [11]. BRMS1 is also an inhibitor of metastasis in ovarian cancer, bladder cancer, non-small cell lung cancer, melanoma, and breast cancer. Metastasis mouse models have demonstrated a high capacity for BRMS1 to inhibit metastasis, recording up to 80–90% metastasis inhibition [8,12,13]. The mechanisms by which BRMS1 mediates its anti-metastasis function still remain unknown despite observations of anti-metastatic potential of BRMS1 [7]. Following this context, it is also important that thorough evaluation of the BRMS1 association with metastatic breast cancer is performed, so for this reason performing a meta-analysis based study to test the correlation between BRMS1 expression and the tumor behavior in breast cancers alone was necessary.
Materials and methods
Search strategy
A literature search was performed systematically using PubMed, EBSCO, SpringerLink, Wiley, Ovid, Web of science, Wanfang Database, China National Knowledge Infrastructure (CNKI), and VIP Information databases using MeSH and free text search terms (last updated search on October, 2014). All variants of key search terms: breast cancer and BRMS1 were included. For example, (‘BRMS1 protein, human’ or ‘breast-cancer metastasis suppressor 1’) and (‘breast neoplasms’ or ‘breast cancer’ or ‘breast carcinoma’ or ‘tumors, breast’ or ‘mammary neoplasms, human’ or ‘carcinoma, human mammary’ or ‘mammary cancer’ or ‘malignant neoplasm of breast’ or ‘malignant tumor of breast’ or ‘cancer of the breast’) were selected to retrieve corresponding literatures. Bibliographies regarding the collected trials and review papers were studied and explored manually for potentially relevant and conducive articles.
Criteria for selecting articles included in this meta-analysis
Study articles were incorporated if (1) the study type of selected studies was case–control study; (2) study subjects were patients diagnosed with breast cancer and healthy controls; (3) selected studies provided complete data consisting of sample size, age, ethnicity, gender, pathological types, positive expression rate of BRMS1 protein, expression of BRMS1 mRNA etc.; (4) the extracted studies were published by the same authors, only including the last or complete one.
Studies were omitted if they were unrelated with either BRMS1 expression or breast cancer, the data were incomplete, the study was not published in Chinese and English, or if the article was repeatedly published.
Data extraction
Information was extracted from all included publications systematically by two investigators in adherence with the aforementioned inclusion criteria. The following data were collected from each individual study: first author, country, language, ethnicity, study design, total numbers and mean age of cases and controls, sample size, pathological types etc.
Statistical analysis
All of the meta-analyses performed utilized Stata 12.0 (Stata Corp, College Station, TX, U.S.A.). The standardized mean differences (SMDs), odds ratio (OR), and effect size (ES) with 95% confidence interval (CI) were all used to assess the case–control studies investigating the association between BRMS1 and clinico-pathological features of breast cancer. Moreover, Z-test was applied to determine the significance of pooled SMDs. Cochran’s Q statistic with a significance level of P < 0.05 and the I2 test (0–100%, values of 40% and 75% were considered to indicate moderate and high heterogeneity respectively) were used to assess heterogeneity across studies. If P < 0.05 or I2 > 50%, there was great heterogeneity among studies, thereby implementing use of a random effect model; if no presence of a random effect model, a fixed effect model was performed [14,15].
Results
Literature searching results and baseline characteristics of included studies
Originally through database searches, one hundred and seventy-five articles were identified. Thirty seven papers remained after excluding duplicates (n = 15), animal studies (n = 24), letters, reviews, meta-analyses (n = 2), and unrelated topics (n = 97). After excluding non-case–control or cohort study (n = 11), studies not relevant to BRMS1 (n = 6), studies with no correlation to breast cancer (n = 7), insufficient information in studies (n = 1), 12 articles were finally selected for this meta-analysis [16–27], including 1,263 patients with breast cancer. Among the 12 studies, there were study subjects with nine being performed in Asian trials, two trials in Caucasians, and one trial in Mixed. To break it down further, in compliance with country there were eight studies from China, one from Japan, U.S.A., England, and Italy respectively. The included studies were all published between 2006 and 2014. BRMS1 in different breast tissues, lymph node metastasis (LNM) status, tumor-node-metastasis (TNM) stages, tumor size, histological grades, pathological types, estrogen receptor (ER) status, progesterone receptor (PR) status, overall survival (OS), and relapse free survival (RFS) expressions were all compared in this meta-analysis. The sample size of study subjects ranges from 70 to 200. The marker involved in these studies was either protein or mRNA. The baseline characteristics of included studies are shown in Table 1 respectively.
Table 1. Baseline characteristics of included studies.
| Author | Year | Country | Ethnicity | Language | Disease | Age (years) | Marker | Sample number |
|---|---|---|---|---|---|---|---|---|
| Wang, D.Y. [16] | 2014 | China | Asians | Chinese | Breast neoplasms | 44.5(28-78) | Protein | 160 |
| Wang, L.X. [17] | 2013 | China | Asians | Chinese | Breast neoplasms | 52.0(37-76) | Protein | 65 |
| Han, D.Y. [18] | 2012 | China | Asians | Chinese | Breast neoplasms | 52.0(33-86) | Protein | 75 |
| Wu, Z.Y. [19] | 2011 | China | Asians | Chinese | Breast neoplasms | 49.4(32-74) | Protein | 63 |
| He, X.B. [20] | 2009 | China | Asians | Chinese | Breast neoplasms | 46.2(27-75) | Protein | 78 |
| Cui, M. [22] | 2009 | China | Asians | Chinese | Breast neoplasms | 45.7(29-76) | Protein | 80 |
| Frolova, N. [21] | 2009 | U.S.A. | Mixed | English | Breast neoplasms | 25-89 | mRNA | 174 |
| Tang, L.B. [23] | 2007 | China | Asians | Chinese | Breast neoplasms | 52.4(28-83) | mRNA | 71 |
| Lombardi, G. [24] | 2007 | Italy | Caucasians | English | Breast neoplasms | NR | mRNA | 47 |
| Zhang, Y.L. [26] | 2006 | China | Asians | Chinese | Breast neoplasms | 43.5(31-62) | mRNA | 51 |
| Zhang, Z.H. [25] | 2006 | Japan | Asians | English | Breast neoplasms | 53.0(34-88) | mRNA | 161 |
| Hicks, D.G. [27] | 2006 | U.K. | Caucasians | English | Breast neoplasms | NR | mRNA | 238 |
NR, not reported.
Breast cancer tissues and normal tissues
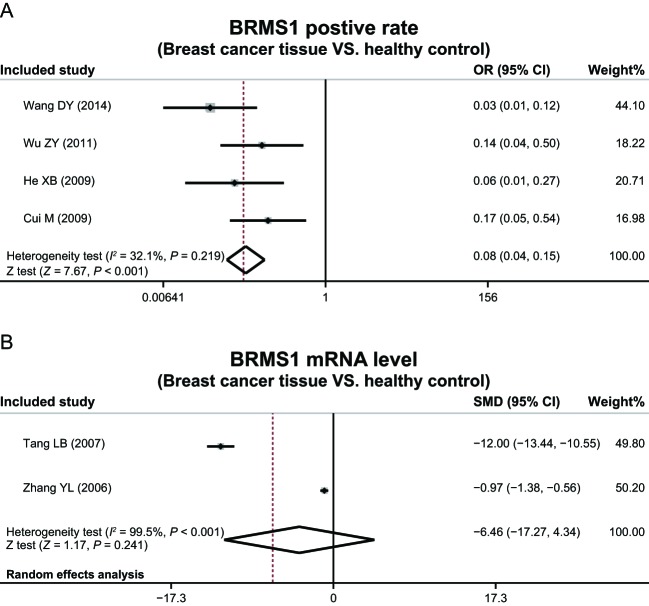
Administration of a heterogeneity test discovered the lack of heterogeneity in the expression of BRMS1 protein between breast cancer tissues and normal tissues, requiring implementation of a random effect model (P = 0.219, I2 = 32.1%). There was heterogeneity in expression of BRMS1 mRNA between breast cancer and normal tissues, thus a fixed-effect model was performed (P < 0.001, I2 = 99.5%).
Results of this meta-analysis suggested that the expression of BRMS1 protein in breast cancer tissues was significantly lower in comparison with normal tissues (OR = 0.08, 95%CI = 0.04–0.15, P < 0.001), while showing no significant difference in expression of BRMS1 mRNA between breast cancer and normal tissues (OR = −6.46, 95%CI = −17.27–4.34, P = 0.241) (Figure 1).
Figure 1. The expressions of BRMS1 in breast cancer and healthy tissues.
Clinico-pathological features of breast cancer
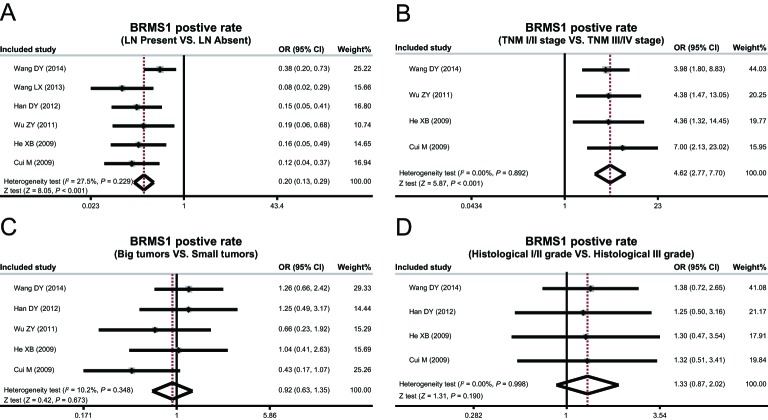
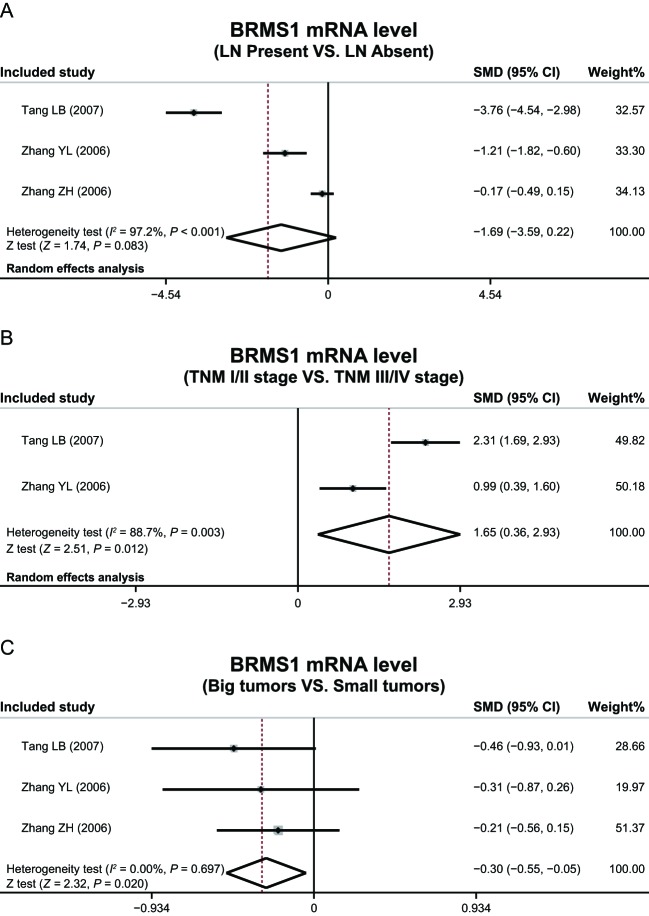
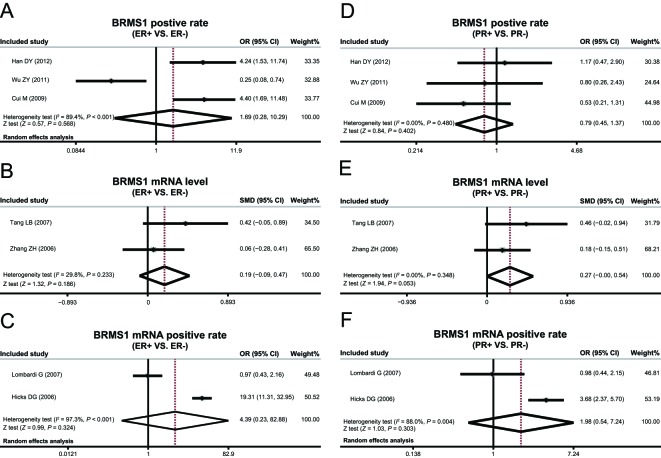
The heterogeneity test revealed absence of heterogeneity in expression of BRMS1 protein in different LNM status, TNM stages, tumor size, histological grades, pathological types, therefore leading to adoption of a fixed-effect model (LNM: P = 0.229, I 2 = 27.5%; TNM stage: P = 0.892, I2 = 0.00%; tumor size: P = 0.348, I2 = 10.2%; histological grade: P = 0.998, I2 = 0.00%; ductal carcinoma: P = 0.466, I2 = 0.00%; lobular carcinoma: P = 0.473, I2 = 0.00%; medullary carcinoma: P = 0.653, I2 = 0.00%) simulating heterogeneity. There was heterogeneity in expression of BRMS1 mRNA in LNM and TNM stage, thus random-effects model was performed (LNM: P < 0.001, I2 = 97.2%; TNM stage: P = 0.003, I2 = 88.7%), while showing no heterogeneity in tumor size, and thereby requiring use of a fixed-effect model (P = 0.697, I2 = 0%).
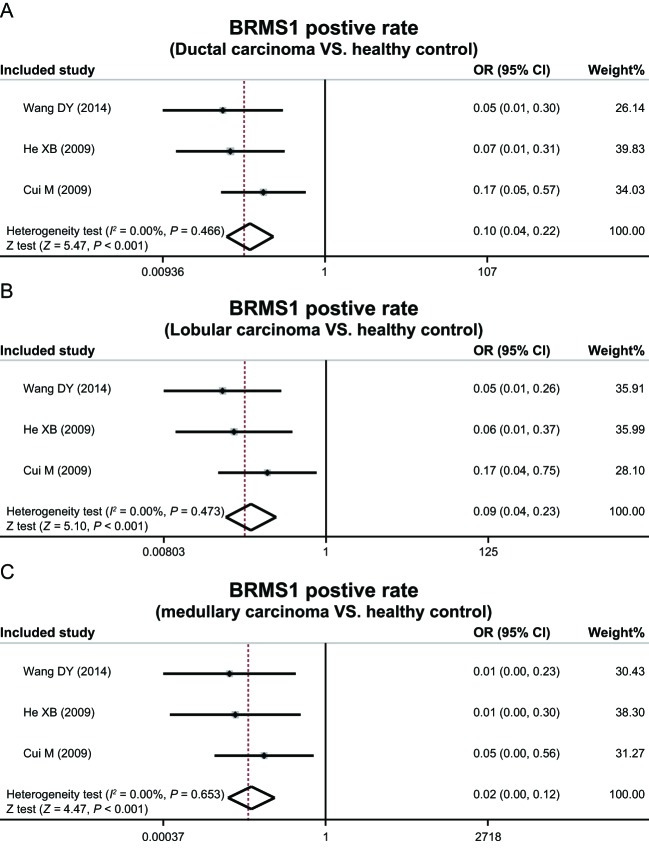
The expression of BRMS1 protein in metastatic breast cancer tissue was found to be lower than that of non-metastatic breast cancer tissue (OR = 0.20, 95%CI = 0.13–0.29, P<0.001), and the expression of BRMS1 protein in TNM stages 1 and 2 was higher than TNM stages 3 and 4 (OR = 4.62, 95%CI = 2.77–7.70, P<0.001) by means of the meta-analysis used. In addition, BRMS1 protein expression level was not considerably related with tumor size and histological grade (tumor size: OR = 0.92, 95%CI = 0.63–1.35, P = 0.673; histological grade: OR = 1.33, 95%CI = 0.87–2.02, P = 0.190) (Figure 2). Additionally, expression of BRMS1 protein in the three types of breast cancer tissues was all lower than the normal tissues (ductal carcinoma: OR = 0.10, 95%CI = 0.04–0.22, P < 0.001; lobular carcinoma: OR = 0.09, 95%CI = 0.04–0.23; P < 0.001; medullary carcinoma: OR = 0.02, 95%CI = 0.00–0.12, P < 0.001) (Figure 3).
Figure 2. Positive expression rates of BRMS1 protein in different LNM status, tumor size, and histological grades of breast cancer.
Figure 3. Positive expression rates of BRMS1 protein in different pathological types of breast cancer.
This meta-analysis also indicated that expression of BRMS1 mRNA was adversely associated with TNM stage and tumor size (TNM stage: OR = 1.65, 95%CI = 0.36–2.93, P=0.012; tumor size: OR = −0.30, 95%CI = −0.55–0.05, P=0.020), while association with LNM status (OR = −1.69, 95%CI = −3.59–0.22, P=0.083) (Figure 4) proved nonexistent.
Figure 4. Expression levels of BRMS1 mRNA in different LNM status, tumor node metastases stages, and tumor size.
Immunohistochemistry
Since heterogeneity was nonexistent among studies exploring the correlation between BRMS1 protein expression and PR status, a fixed-effect model was used (P = 0.480, I2 = 0.00%) substituting the lack thereof. However, with the presence of heterogeneity among studies investigating the associations of BRMS1 protein expression and ER status, application of a random-effect model (P < 0.001, I2 = 89.4%) was allowed. Furthermore, no heterogeneity existed across studies investigating the links between the expression of BRMS1 mRNA with status of ER and PR, and again due to the absence of heterogeneity, a fixed-effect model was put in place (ER: P = 0.233, I2 = 29.8%; PR: P = 0.348, I2 = 0.00%), while heterogeneity existed among studies relative to positive expression rate of BRMS1 mRNA and ER and PR status, a random-effect model was utilized (ER: P < 0.001, I2 = 97.3%; PR: P = 0.004, I2 = 88.0%). The expressions of BRMS1 protein and BRMS1 mRNA were not significant in coordinance with the status of ER and PR in association with the results of this meta-analysis (as shown in Figure 5). The correlation also suggests which test model will be implemented according to presence of heterogeneity.
Figure 5. Correlation of BRMS1 expressions with ER and PR.
Prognosis
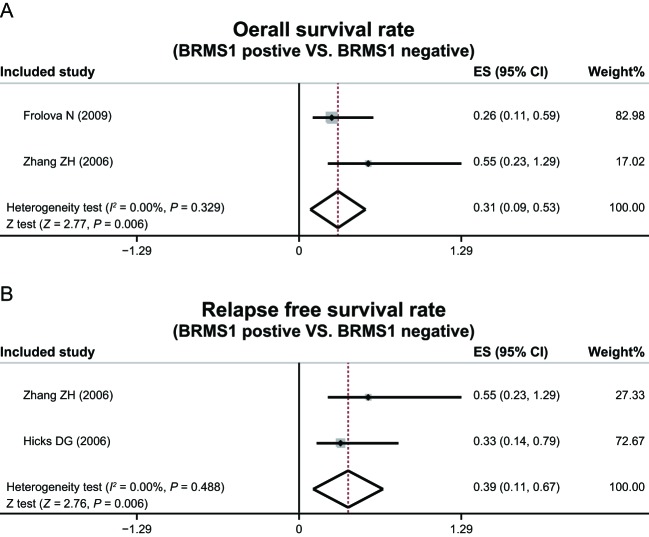
No heterogeneity among studies that investigated the correlation of BRMS1 protein expression with OS and RFS was detected thereby administering performance of a fixed-effect model (OS: P = 0.329, I2 = 0.00%; RFS: P = 0.488, I2 = 0.00%). Had there been heterogeneity, the implication of a random-effect model (ER: P < 0.001, I2 = 97.3%; PR: P = 0.004, I2 = 88.0%) would have been necessary to facilitate lack of expression.
Specifications that there were significant differences in OS and RFS between patients with positive BRMS1 (BRMS1+) and negative BRMS1 (BRMS1−) (OS: ES = 0.31, 95%CI = 0.09–0.53, P=0.006; RFS: ES = 0.39, 95%CI = 0.11–0.67, P=0.006) (Figure 6) were present in this meta-analysis.
Figure 6. OS rates and RFS rates on the correlation between BRMS1 and breast cancer.
Discussion
The present meta-analysis investigated the correlation between BRMS1 and the clinico-pathological features of breast cancer including LNM, TNF stages, tumor size, histological grade, pathological type, ER, PR, OS, and RFS. Aside from involving degradation of extracellular matrix, invasion and metastasis of malignant tumors also include reciprocation between various genes such as oncogenes, tumor suppressor genes, and metastasis-regulating genes [28]. Currently, approximately 30 metastasis suppressor genes have been recognized in multiple carcinomas along with the loss of their function through mutations or gene silencing, facilitating metastatic behavior of tumor cells [8]. BRMS1 is an important nuclear protein providing various functions such as differentially modulating the expression of various genes and inhibiting metastasis with no effects on the primary tumor growth, all while regulating migration of tumor cells [29].
In multiple human tumors, there is either a reduction or complete absence of the expression of the BRMS1 nuclear protein. Current study illustrates that BRMS1 protein expression in breast cancer tissue is significantly lesser than normal tissue. In addition to decreased protein expression, the BRMS1 expression in metastatic breast cancer tissue was lower than non-metastatic breast cancer tissue, suggesting invasion and metastasis of breast cancer might possibly be linked to the reduction or absence of BRMS1 expression. Potential aspects for BRMS1-mediated metastasis suppression in breast cancers might be through altering metastasis-associated microRNA and/or interfering with specific cellular pathways relative to metastasis including: gap junctions, nuclear factor kappa B signaling, phosphoinositide signaling, cell motility and invasion, apoptosis, and tumor cell dissemination, though the exact mechanisms remain elucidated [30–36]. Past studies alluded to reports that BRMS1 reduced lung metastasis in athymic mice when its expression was restored by exogenous expression of BRMS1 in metastatic cell lines from non-small cell lung carcinomas, ovarian, melanoma, and breast [37,38].
In TNM stages 1 and 2, we found that since the expression of BRMS1 protein was drastically higher compared with TNM stages 3 and 4, breast cancer progression might be correlated with low expression of BRMS1 protein. Additionally, another discovery made was finding that the expression levels of BRMS1 mRNA were associated negatively with the TNM stage and tumor size, similar to the result between BRMS1 expression and TNM staging [39]. Contrary to our result which revealed that the expression levels of BRMS1 mRNA were irrelative to LNM, the BRMS1 mRNA expression levels were found to be lower in brain metastasis of breast cancer than in primary tumor while also reduced in breast tumor compared with the expression of that in matched normal breast tissues [40,41]. Limitations in the present study may have led to possible differences in expression levels. In conclusion, a significant difference in the OS and RFS between patients with BRMS1+ and BRMS1−suggests that the BRMS1 protein might be a potential prognostic indicator in breast cancer, which is consistent with a study reported by Hanker et al. which determined that low BRMS1 mRNA expression and poor prognosis of breast cancer were indeed related [42].
Our results might be adversely affected due in part by the small portion size provided by the limitations of the meta-analysis used in this study. Another complication of the limitations of this analysis is the loss of data in several studies, which could have an effect on the final result to a certain extent. Furthermore, our results deemed that there was no association between BRMS1 protein and mRNA expression. The expression of BRMS1 protein in breast cancer tissues was significantly lower compared with normal tissues, while no significant difference in expression of BRMS1 mRNA between breast cancer and normal tissues was found. This was a statistical result and might have been caused by the included studies, those of which might differ in the study of BRMS1 protein and mRNA. According to this, previous clinical studies have revealed that the mRNA and BRMS1 protein expressions were not necessarily conjunctive, and the relative level of mRNA in tumors didn’t necessarily associate with the protein level [21,43,44]. Finally, there is little discussion about the expressions of BRMS1 in in-depth classification of breast cancer, like triple-negative breast cancer (TNBC) and ER+ tumors, a potentially impactful limitation of the analysis that required further investigation.
In summary, the coordinance between reduced or loss of expression of BRMS1 and clinico-pathological features of breast cancer was significant enough to suggest that BRMS1 could be an indicator of the metastatic capacity and low expression of the protein in regards to the poor prognosis surrounding breast cancer.
Acknowledgments
We are obliged to many colleagues who have shared wisdom with us during the writing of this manuscript. We apologized some whose work was not cited though we have strived to be thorough.
Abbreviations
- BMI
body mass index
- BRMS1
breast cancer metastasis suppressor 1
- CI
confidence interval
- ER
estrogen receptor
- ES
effect size
- LNM
lymph node metastasis
- OR
odds ratio
- OS
overall survival
- PR
progesterone receptor
- RFS
relapse free survival
- SMD
standardized mean difference
- TNBC
Triple-negative breast cancer
- TNM
tumor-node-metastasis
Funding
The authors declare that there are no sources of funding to be acknowledged.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
L.-Z.L., M.-G.C., Y.-C.D., Z.-B.Z., F.-F.J., L.-L.S., and Y.P. designed the study. L.-Z.L. and M.-G.C. developed the database; Y.-C.D. and L.-L.S. carried out data analyses; Z.-B.Z. and F.-F.J. produced the initial draft of the manuscript; L.-Z.L., Y.P., and H.-B.S. contributed to drafting the manuscript. All authors have read and approved the final version of the manuscript.
References
- 1.Assi H.A., Khoury K.E., Dbouk H., Khalil L.E., Mouhieddine T.H. and El Saghir N.S. (2013) Epidemiology and prognosis of breast cancer in young women. J. Thorac. Dis. 5, S2–S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munster P.N., Moore A.P., Ismail-Khan R., Cox C.E., Lacevic M., Gross-King M. et al. (2012) Randomized trial using gonadotropin-releasing hormone agonist triptorelin for the preservation of ovarian function during (neo)adjuvant chemotherapy for breast cancer. J. Clin. Oncol. 30, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Wang N., Liu P., Chen Q., Situ H., Xie T. et al. (2014) MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget 5, 7013–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Colditz G.A., Rosner B., Berkey C.S., Collins L.C., Schnitt S.J. et al. (2013) Alcohol intake between menarche and first pregnancy: a prospective study of breast cancer risk. J. Natl. Cancer Inst. 105, 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phipps A.I., Buist D.S., Malone K.E., Barlow W.E., Porter P.L., Kerlikowske K. et al. (2012) Breast density, body mass index, and risk of tumor marker-defined subtypes of breast cancer. Ann. Epidemiol. 22, 340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaghjyan L., Colditz G.A., Rosner B. and Tamimi R.M. (2012) Mammographic breast density and breast cancer risk by menopausal status, postmenopausal hormone use and a family history of breast cancer. Cancer Causes Control 23, 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Ye L., Tan Y., Sun P., Ji K. and Jiang W.G. (2014) Expression of breast cancer metastasis suppressor-1, BRMS-1, in human breast cancer and the biological impact of BRMS-1 on the migration of breast cancer cells. Anticancer Res. 34, 1417–1426 [PubMed] [Google Scholar]
- 8.Nakayama K., Nakayama N., Katagiri H. and Miyazaki K. (2012) Mechanisms of ovarian cancer metastasis: biochemical pathways. Int. J. Mol. Sci. 13, 11705–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tekle C., Nygren M.K., Chen Y.W., Dybsjord I., Nesland J.M., Maelandsmo G.M. et al. (2012) B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int. J. Cancer 130, 2282–2290 [DOI] [PubMed] [Google Scholar]
- 10.Hurst D.R. and Welch D.R. (2011) Unraveling the enigmatic complexities of BRMS1-mediated metastasis suppression. FEBS Lett. 585, 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seraj M.J., Samant R.S., Verderame M.F. and Welch D.R. (2000) Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 60, 2764–2769 [PubMed] [Google Scholar]
- 12.You J., He X., Ding H. and Zhang T. (2015) BRMS1 regulates apoptosis in non-small cell lung cancer cells. Cell Biochem. Biophys. 71, 465–472 [DOI] [PubMed] [Google Scholar]
- 13.Hurst D.R. and Welch D.R. (2011) Metastasis suppressor genes at the interface between the environment and tumor cell growth. Int. Rev. Cell Mol. Biol. 286, 107–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zintzaras E. and Ioannidis J.P. (2005) Heterogeneity testing in meta-analysis of genome searches. Genet. Epidemiol. 28, 123–137 [DOI] [PubMed] [Google Scholar]
- 15.Higgins J.P. and Thompson S.G. (2002) Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 [DOI] [PubMed] [Google Scholar]
- 16.Wang D.Y. (2014) Expression and clinical significance of BRMS1 and Survivin in breast cancer. Hebei Med.J. 342–324 [Google Scholar]
- 17.Wang L.X., Mou Q.J., Mao J., Tang L.Y., Tao Y.J., and Li L.H. and (2013) Significance of MMP9 and BRMSI in metastasis and invasion of breast cancer stem cell, Chinese J. Clin. Exp. Pathol. 45, 943–946 [Google Scholar]
- 18.Han D.Y., et al. (2012) The expression and significance of BRMSI in breast cancer. Shanghai Med. J. 546–548 [Google Scholar]
- 19.Wu Z.Y. and Chen B. (2011) Expression and significance of BRMS1 and VEGF-C protein in breast cancer. J. Prac. Med. 27, 572–574 [Google Scholar]
- 20.He X.B., et al. (2009) Study on expression and clinicopathologic significance of correlative metastasis genes BRMS1 and MMP-9 in breast cancer. Prac. J. Cancer 24, 351–354 [Google Scholar]
- 21.Frolova N., Edmonds M.D., Bodenstine T.M., Seitz R., Johnson M.R., Feng R. et al. (2009) A shift from nuclear to cytoplasmic breast cancer metastasis suppressor 1 expression is associated with highly proliferative estrogen receptor-negative breast cancers. Tumour Biol. 30, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui M., H.X. and Wu H.Y. (2009) Expression and clinical significance of BRMS1 protein in breast cancer. J. Clin. Surg. 17, 756–758 [Google Scholar]
- 23.Tang L.B., et al. (2007) Expression of BRMS1 mRNA in human breast cancer and its clinical significance. Chinese J. Gen. Surg. 16, 58–60 [Google Scholar]
- 24.Lombardi G., Di Cristofano C., Capodanno A., Iorio M.C., Aretini P., Isola P. et al. (2007) High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int. J. Cancer 120, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z., Yamashita H., Toyama T., Yamamoto Y., Kawasoe T. and Iwase H. (2006) Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin. Cancer Res. 12, 6410–6414 [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y.L., Wei R., Liu X.T., Yan J.W., Wang L.X. and Shi H.M. (2006) Expression of BRMS1mRNA in breast carcinoma and its clinical significance. China Cancer 787–789 [Google Scholar]
- 27.Hicks D.G., Yoder B.J., Short S., Tarr S., Prescott N., Crowe J.P. et al. (2006) Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin. Cancer Res. 12, 6702–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L.X., et al. (2013) Significance of MMP9 and BRMS1 in metastasis and invasion of breast cancer stem cells[C]//Control and Automation (ICCA). 2013 10th IEEE Int. Conf. on IEEE 943–946 [Google Scholar]
- 29.Cho W.C. (2010) MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int. J. Biochem. Cell Biol. 42, 1273–1281 [DOI] [PubMed] [Google Scholar]
- 30.Patsialou A., Wyckoff J., Wang Y., Goswami S., Stanley E.R. and Condeelis J.S. (2009) Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 69, 9498–9506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borsig L., Vlodavsky I., Ishai-Michaeli R., Torri G. and Vismara E. (2011) Sulfated hexasaccharides attenuate metastasis by inhibition of P-selectin and heparanase. Neoplasia 13, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finger E.C. and Giaccia A.J. (2010) Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev. 29, 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boimel P.J., Smirnova T., Zhou Z.N., Wyckoff J., Park H., Coniglio S.J. et al. (2012) Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res. 14, R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonda K., Watanabe T.M., Ohuchi N. and Higuchi H. (2010) In vivo nano-imaging of membrane dynamics in metastatic tumor cells using quantum dots. J. Biol. Chem. 285, 2750–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierga J.Y., Hajage D., Bachelot T., Delaloge S., Brain E., Campone M. et al. (2012) High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann. Oncol. 23, 618–624 [DOI] [PubMed] [Google Scholar]
- 36.Hurst D.R., Xie Y., Thomas J.W., Liu J., Edmonds M.D., Stewart M.D. et al. (2013) The C-terminal putative nuclear localization sequence of breast cancer metastasis suppressor 1, BRMS1, is necessary for metastasis suppression. PLoS ONE 8, e55966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith P.W., Liu Y., Siefert S.A., Moskaluk C.A., Petroni G.R. and Jones D.R. (2009) Breast cancer metastasis suppressor 1 (BRMS1) suppresses metastasis and correlates with improved patient survival in non-small cell lung cancer. Cancer Lett. 276, 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng X.J., Zhou D.M., Liu Q., Lou S.Y., Song Q.Y. and Zhou Y.Q. (2014) BRMS1 inhibits expression of NF-kappaB subunit p65, uPA and OPN in ovarian cancer cells. Eur. J. Gynaecol. Oncol. 35, 236–242 [PubMed] [Google Scholar]
- 39.Wang Y., Zhao Z., Chen L., Cong L. and Zhang J. (2011) Expression of BRMS1 gene protein in nasal and paranasal sinus carcinomas. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 25, 920–921 [PubMed] [Google Scholar]
- 40.Bodenstine T.M., Vaidya K.S., Ismail A., Beck B.H., Cook L.M., Diers A.R. et al. (2010) Homotypic gap junctional communication associated with metastasis suppression increases with PKA activity and is unaffected by PI3K inhibition. Cancer Res. 70, 10002–10011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaidya K.S., Harihar S., Phadke P.A., Stafford L.J., Hurst D.R., Hicks D.G. et al. (2008) Breast cancer metastasis suppressor-1 differentially modulates growth factor signaling. J. Biol. Chem. 283, 28354–28360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanker L.C., Karn T., Mavrova-Risteska L., Ruckhaberle E., Gaetje R., Holtrich U. et al. (2011) SATB1 gene expression and breast cancer prognosis. Breast 20, 309–313 [DOI] [PubMed] [Google Scholar]
- 43.Hurst D.R., Xie Y., Edmonds M.D. and Welch D.R. (2009) Multiple forms of BRMS1 are differentially expressed in the MCF10 isogenic breast cancer progression model. Clin. Exp. Metastasis 26, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly L.M., Buggy Y., Hill A., O’Donovan N., Duggan C., McDermott E.W. et al. (2005) Expression of the breast cancer metastasis suppressor gene, BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumour Biol. 26, 213–216 [DOI] [PubMed] [Google Scholar]