Abstract
Heterologous protection against swine influenza viruses (SwIVs) of different lineages is an important concern for the pig industry. Cross-protection between ‘avian-like’ H1N1 and 2009 pandemic H1N1 lineages has been observed previously, indicating the involvement of cross-reacting T-cells. Here, reverse vaccinology was applied to identify cross-reacting MHC class I T-cell epitopes from two different SwIV H1 lineages in pigs. In silico prediction followed by in vitro and in vivo testing was used to identify SLA-1*0702 T-cell epitopes in heterologous SwIV-infected pigs. Following viral infection, tetramer specific T-cell populations were identified. The majority of the identified T-cell epitopes were conserved between the examined lineages, suggesting that targeting cross-reactive T-cell epitopes could be used to improve vaccines against SwIV in SLA-1*0702-positive pigs.
Keywords: swine influenza virus, reverse vaccinology, MHC class I T cell epitopes, cross-reactive epitopes
Abbreviations
d.p.b., days post boost; d.p.i., days post primary infection; IV, influenza virus; PSCPL, positional scanning combinatorial peptide library; SwIV, swine influenza virus.
Full-Text
Swine influenza virus (SwIV) is considered to be an important pathogen in pig herds. Different subtypes and lineages are circulating in pigs [1] and the involvement of both arms of the immune system is considered to be necessary for an effective immune response [2]. Although commercially available vaccines were designed to confer homologous protection, the control of heterologous infections is of high importance for controlling influenza in pig herds. The identification of protective cross-reacting antigens thus remains an open challenge.
Some previous studies have shown that infection with SwIVs of European ‘avian-like’ H1N1 lineage can induce complete protection against the influenza A virus of the 2009 pandemic H1N1 lineage [3, 4]. Such cross-protection was conferred in the absence of cross-reactive antibodies capable of inhibiting hemagglutination or virus neutralization, suggesting that other factors, like cross-reacting T-cell responses, might have been involved.
Reverse vaccinology tries to overcome problems related to the empirical identification of antigens. Previously, a combination of different bioinformatics prediction methods and in vitro testing has been used to identify T-cell epitopes in pigs [5]. In addition, recent approaches that use immunoinformatic tools have been used to identify MHC class I and class II T-cell epitopes that are highly conserved in SwIVs circulating in the US swine population [6]. In the present study, reverse vaccinology technologies were extended to identify cross-reactive T-cell epitopes to two different H1N1 influenza A virus (IV) strains during the infection of pigs expressing SLA-1*0702 allele. Strain A/swine/Spain/SF11131/2007 (SpH1N1) is an ‘avian-like’ H1N1 SwIV [7], whereas strain A/swine/Denmark/101310-1/2011 (pdmH1N1) is an H1N1 strain that circulated in pigs, but belongs to the 2009 pandemic lineage.
Firstly, T-cell epitopes residing in SpH1N1 and pdmH1N1 were predicted for SLA-1*0702 binding using a previously described strategy that combines two methods for in silico prediction [8]. Briefly, the neural network NetMHCpan v. 2.8 prediction tool (www.cbs.dtu.dk) and a positional scanning combinatorial peptide library (PSCPL) for the SLA-1*0702 binding motif were used to predict T-cell epitopes. The experimental strategy of PSCPL has been described previously for both HLA [9] and SLA [10] proteins.
Secondly, the peptide sequences that were consistently predicted by these two methods in combination were further analysed for actual MHC binding affinities (Kd less than 1000 nM) to recombinant SLA-1*0702 (Table 1) by an in vitro immunosorbent assay [11]. Following these criteria, 11 sequences were predicted to be T-cell epitopes for the selected SLA allele and seven of them were conserved between both SwIV strains (Table 1). These sequences happened to be located within HA, NA and M1 proteins. Finally, tetramers were generated, as previously described [10, 12], using the 11 peptides identified in vitro and a specifically designed peptide as a negative control. Theoretically, the detection of T-cell populations by recognizing specific peptides presented on SLA-1*0702 would indicate the recognition of viral T-cell epitopes.
Table 1. SwIV peptides in silico predicted to bind with SLA-1*0702 with their respective in vitro binding results.
Strong binding peptides with a Kd less than 1000 nM were used to generate tetramers.
| Peptide sequence | Viral protein of origin (no.) | Position | Virus | Kd (nM) | Tetramer name | |
|---|---|---|---|---|---|---|
| pdmH1N1 | SpH1N1 | |||||
| NADTLCIGY | HA(1) | 16–24 | + | 1 543 | ||
| SLSTASSWSY | HA(2) | 86–95 | + | 35 | 57 | |
| TLYQNNHTY | HA(3) | 207–215 | + | 6 | 66 | |
| YVSVGSSKY | HA(4) | 215–223 | + | 987 | 93 | |
| SVKNGTYDY | HA(5) | 295–303 | + | 20 000 | ||
| GMIDGWYGY | HA(6) | 300–308 | + | 20 000 | ||
| EIGNGCFEFY | HA(7) | 476–485 | + | + | 132 | 63 |
| CPVSGWAIY | NA(1) | 92–100 | + | + | 6 | 55 |
| CPIGEVPSPY | NA(2) | 171–180 | + | + | 37 | 60 |
| GPSNGQASY | NA(3) | 245–253 | + | + | 25 | 59 |
| SVELNAPNY | NA(4) | 266–274 | + | 20 000 | ||
| EMNAPNYHY | NA(5) | 268–278 | + | 231 | 61 | |
| NMDRAVKLY | M1(1) | 92–100 | + | + | 293 | 62 |
| ALASCMGLIY | M1(2) | 123–132 | + | + | 127 | 56 |
| LASCMGLIY | M1(3) | 124–132 | + | + | 9 | 64 |
| VSYAAAAAY | Negative control | 831 | ||||
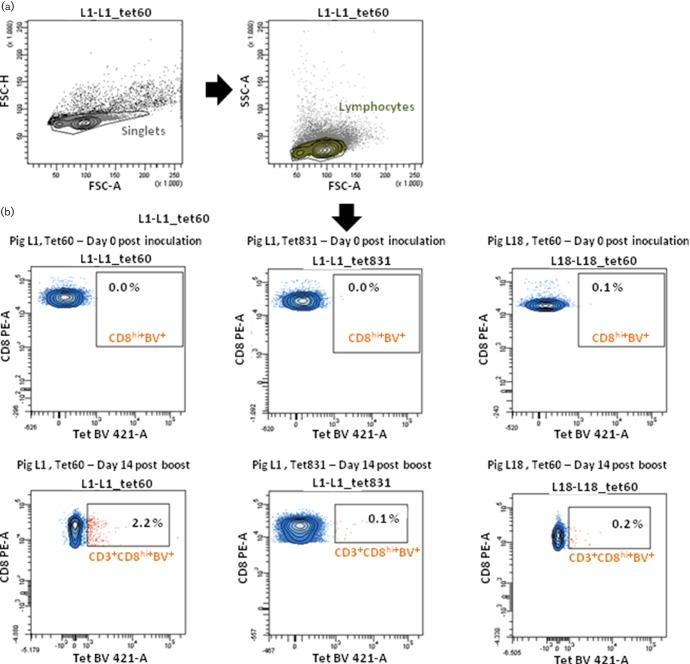
Four pigs bearing the allele SLA-1*0702 and a mismatched pig (not bearing the selected SLA allele) were selected following genomic SLA-I amplification by high-resolution sequence-specific primers [13] and infected with SpH1N1. All animals but one (L12) were intranasally infected with 3 ml, 9.04 logTCID50 of SpH1N strain. L12, the SLA-I-matched pig, was intratracheally infected with the same dose. At 139 days post-primary infection (d.p.i.), animals were intranasally infected with 6 ml, 5.58 logTCID50 of the Danish pdmH1N1strain. The study was carried out in accordance with Danish legislation on animal experimentation and EU regulations on the use of laboratory animals for research (protocol number 2012‐15‐293–00682). All animals were euthanized 14 days post-second infection (boost; d.p.b.), corresponding to 153 d.p.i. Serum samples were collected to monitor antibody levels against both SwIVs by hemagglutination inhibition assay [14] and influenza A Ab ELISA (IDEXX). PBMCs were isolated from animals at different days post-infection. Biotinylated tetramers (Table 1) were used to stain the PBMCs and were visualized with a streptavidin-BV421 fluorophore. PE-conjugated MAb against porcine CD8α (clone 76-2-11, BD Pharmingen) and FITC-conjugated MAb against porcine CD3ε (clone PPT3, Southern Biolegend) were additionally used to characterize cells that stained positive with the tetramers. CD3+CD8high cells contain a CD8+αβ T-cell subpopulation which recognises class-I epitopes [5, 15]. Tetramers showing frequencies two times higher compared to background (negative control or tetramer 831) and two times higher compared to the mismatched pig staining were regarded as positive.
None of the pigs developed clinical signs due to infection. Humoral SpH1N1- and pdmH1N1-specific responses were generated against each virus after infection (data not shown). Tetramer-positive T-cell subpopulations were below detection levels at 4, 7 and 9 d.p.b. (data not shown). At 14 d.p.b. specific T-cell responses were detected within the CD3+CD8high subpopulations (Tet+CD3+CD8high) as compared with the negative controls (Fig. 1, Table 2). The frequency of Tet+CD3+CD8high ranged from 0.4 to 2.6 %, depending on individual peptides and animals (Table 2). Nine peptides were identified as T-cell epitopes by tetramer staining; three located in the M1 protein (Tet62, Tet56 and Tet64), three in NA (Tet55, Tet60 and Tet59) and three in HA (Tet93, Tet57 and Tet66). These results highlighted the high efficiency of the discovery of class I restricted T-cell epitopes using this pipeline. The NA171-180 (CPIGEVPSPY) peptide was considered to be immunodominant, as it was consistently detected in all pigs with the highest frequency within CD3+CD8high cells. Importantly, most of the other T-cell epitopes were only found in animal L6, in which NA171–180 was not dominant.
Fig. 1.
Tetramer staining of PBMCs of infected pigs. Example of gating strategy and staining of cells from pig L1 and pig L18 (mismatched pig) at days 0 and 153 post-infection (14 d.p.b.). (a) Singlet and lymphocyte analysis. (b) CD3+CD8high cells are shown. Tetramer 60 (Tet 60) is shown to specifically stain PBMCs from pig L1 at 14 d.p.b. The same tetramer and negative control tetramer (Tet 381) do not stain PBMCs at 0 d.p.i. or PBMCs from the mismatched pig (L18) at any time point.
Table 2. Frequency of Tet+CD3+CD8+ cells at 14 d.p.b. Numbers in bold show values two times higher than the background (negative control or tetramer 831).
Those values that were also two times higher than the respective peptide in the mismatched pig (L18) are underlined.
| Pig | Frequency of tetramers | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 831 | 93 | 55 | 56 | 57 | 59 | 60 | 61 | 62 | 63 | 64 | 66 | |
| L1 | 0.1 | 0.1 | 0.1 | 0.7 | 1.9 | 0.2 | 2.2 | 0.0 | 0.0 | 0.1 | 0.7 | 0.0 |
| L6 | 0.1 | 0.4 | 1.7 | 1.2 | 1.2 | 2.1 | 1.0 | 0.4 | 0.7 | 0.1 | 0.1 | 1.0 |
| L11 | 0.7 | 0.1 | 0.2 | 0.0 | 1.2 | 1.2 | 2.6 | 0.6 | 1.3 | 0.1 | 0.8 | 0.7 |
| L12 | 0.3 | 0.1 | 0.2 | 0.1 | 0.4 | 0.2 | 0.7 | 0.1 | 0.4 | 0.6 | 0.3 | 0.5 |
| L18 | 0.1 | 0.1 | 0.4 | 0.5 | 0.8 | 0.7 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.3 |
Several factors can account for the different immunodominance hierarchy among animals [16]. For example, the pigs in this study were not inbred; therefore, different levels of SLA-1*0702 might have been expressed on pig cells, depending on whether these animals were homozygous or heterozygous for this allele.
All T-cell epitopes identified in NA and M1 proteins were conserved between both IVs. By contrast, the T-cell epitopes identified in HA were not conserved. However, it is noteworthy that SpH1N1-HA-specific T-cells (Tets 66 and 93) were detected after pdmH1N1 infection, indicating that a population of T-cells was primed after the second infection, which cross-reacted. A previous study in pigs suggested that there is flexibility in T-cell receptor recognition of class I epitopes and thus they can recognize non-homologous peptides [5]. It could also be speculated that HA-SpH1N1-specific T-cells, generated during the first infection, were re-activated by pdmH1N1 in a non-specific or bystander way, as has been described for other systems [17].
A recent study identified a number of SLA-I epitopes by using another immunoinformatic tool in pigs with a different SLA-I [6]. Interestingly, seven of the eleven selected epitopes in this study (HA1, HA2, HA5, HA6, NA5, M1-1 and M1-3) were identical to or had just one amino acid change compared to those reported by Gutiérrez et al. [6] using outbred pigs expressing SLA-I alleles other than SLA-1*0702. The combination of these data suggests that these epitopes potentially have cross-reactive properties and may be relevant in a broad range of pigs. Out of these seven sequences, HA2 (Tet 57), NA5 (Tet 61), M1-1 (Tet 62) and M1-3 (Tet 64) were selected for the detection of epitope-specific T-cells (CD3+CD8+). All of them depicted specific T-cell subpopulations in some of the pigs, although they were not dominant. Remarkably, the majority of the predicted epitopes in this study and in Gutiérrez et al. [6] had tyrosine in position 9, suggesting that an amino acid with a hydrophobic side chain might be the preferable option for SLA-I antigen presentation in pigs.
In conclusion, the data in this work further extend the accuracy and versatility of our pipeline for SLA class I T-cell epitope identification and confirm the epitope predictions generated using other technologies. Several cross-reacting T-cell epitopes were identified and specific subpopulations were detected following heterologous SwIV infection. The functionality of these epitopes and their involvement in cross-protection will require further assessment in the future with the aim of applying these technologies to reverse vaccinology for SwIV in pigs.
Funding information
The research leading to these results received funding from the European Community’s Seventh Framework Programme (FP7, 2007–2013), Research Infrastructures Action, under grant agreement no. FP7-228393 (NADIR project), and from the Spanish Government's MINECO AGL2013-22200-C02-01 and BBSRC BBS/E/I/00002014 grants.
Acknowledgements
We thank the staff in the animal facility for their support with the infection experiments. We also thank Dr Bryan Charleston at the Pirbright Institute for his support with this work and the manuscript. We thank Dr E. Reid for critically reviewing and editing the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, et al. Molecular epidemiology and evolution of influenza viruses circulating within European swine between 2009 and 2013. J Virol. 2015;89:9920–9931. doi: 10.1128/JVI.00840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crisci E, Mussá T, Fraile L, Montoya M. Review: influenza virus in pigs. Mol Immunol. 2013;55:200–211. doi: 10.1016/j.molimm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Busquets N, Segalés J, Córdoba L, Mussá T, Crisci E, et al. Experimental infection with H1N1 European swine influenza virus protects pigs from an infection with the 2009 pandemic H1N1 human influenza virus. Vet Res. 2010;41:74. doi: 10.1051/vetres/2010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu Y, de Hert K, van Reeth K. Cross-protection against European swine influenza viruses in the context of infection immunity against the 2009 pandemic H1N1 virus: studies in the pig model of influenza. Vet Res. 2015;46:105. doi: 10.1186/s13567-015-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen LE, Breum SØ, Riber U, Larsen LE, Jungersen G. Identification of swine influenza virus epitopes and analysis of multiple specificities expressed by cytotoxic T cell subsets. Virol J. 2014;11:163. doi: 10.1186/1743-422X-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutiérrez AH, Loving C, Moise L, Terry FE, Brockmeier SL, et al. In vivo validation of predicted and conserved T cell epitopes in a swine influenza model. PLoS One. 2016;11:e0159237. doi: 10.1371/journal.pone.0159237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baratelli M, Córdoba L, Pérez LJ, Maldonado J, Fraile L, et al. Genetic characterization of influenza A viruses circulating in pigs and isolated in North-East Spain during the period 2006–2007. Res Vet Sci. 2014;96:380–388. doi: 10.1016/j.rvsc.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen LE, Rasmussen M, Harndahl M, Nielsen M, Buus S, et al. A combined prediction strategy increases identification of peptides bound with high affinity and stability to porcine MHC class I molecules SLA-1*04:01, SLA-2*04:01, and SLA-3*04:01. Immunogenetics. 2016;68:157–165. doi: 10.1007/s00251-015-0883-9. [DOI] [PubMed] [Google Scholar]
- 9.Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, et al. Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur J Immunol. 1996;26:1911–1918. doi: 10.1002/eji.1830260836. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen LE, Harndahl M, Rasmussen M, Lamberth K, Golde WT, et al. Porcine major histocompatibility complex (MHC) class I molecules and analysis of their peptide-binding specificities. Immunogenetics. 2011;63:821–834. doi: 10.1007/s00251-011-0555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sylvester-Hvid C, Kristensen N, Blicher T, Ferré H, Lauemøller SL, et al. Establishment of a quantitative ELISA capable of determining peptide–MHC class I interaction. Tissue Antigens. 2002;59:251–258. doi: 10.1034/j.1399-0039.2002.590402.x. [DOI] [PubMed] [Google Scholar]
- 12.Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, et al. One-pot, mix-and-read peptide-MHC tetramers. PLoS One. 2008;3:e1678. doi: 10.1371/journal.pone.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen LE, Jungersen G, Sorensen MR, Ho CS, Vadekær DF. Swine Leukocyte Antigen (SLA) class I allele typing of Danish swine herds and identification of commonly occurring haplotypes using sequence specific low and high resolution primers. Vet Immunol Immunopathol. 2014;162:108–116. doi: 10.1016/j.vetimm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Van Reeth K, Brown IH, Dürrwald R, Foni E, Labarque G, et al. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002–2003. Influenza Other Respir Viruses. 2008;2:99–105. doi: 10.1111/j.1750-2659.2008.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerner W, Talker SC, Koinig HC, Sedlak C, Mair KH, et al. Phenotypic and functional differentiation of porcine αβ T cells: current knowledge and available tools. Mol Immunol. 2015;66:3–13. doi: 10.1016/j.molimm.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Chu T, Tyznik AJ, Roepke S, Berkley AM, Woodward-Davis A, et al. Bystander-activated memory CD8 T cells control early pathogen load in an innate-like, NKG2D-dependent manner. Cell Rep. 2013;3:701–708. doi: 10.1016/j.celrep.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]