ABSTRACT
Ehrlichia chaffeensis secretes tandem repeat protein (TRP) effectors that are involved in a diverse array of host cell interactions, some of which directly activate cell signaling pathways and reprogram host gene transcription to promote survival in the mononuclear phagocyte. However, the molecular details of these effector-host interactions and roles in pathobiology are incompletely understood. In this study, we determined that the E. chaffeensis effector TRP120 is posttranslationally modified by ubiquitin (Ub) and that ubiquitination occurs through intrinsic and host-mediated HECT ligase activity. A functional HECT E3 ligase domain with a conserved catalytic site was identified in the C-terminal region of TRP120, and TRP120 autoubiquitination occurred in vitro in the presence of host UbcH5b/c E2 enzymes. TRP120 ubiquitination sites were mapped using a high-density microfluidic peptide array and confirmed by ectopic expression of TRP120 lysine mutants in cells. Moreover, we determined that the HECT E3 ubiquitin ligase, Nedd4L, interacts with TRP120 during infection and also mediates TRP120 ubiquitination. Nedd4L knockdown resulted in the reduction of TRP120-Ub, decreased ehrlichial infection, and reduced recruitment of a known TRP120-interacting host protein, PCGF5, to ehrlichial inclusions. TRP120-mediated PCGF5 polyubiquitination was associated with a reduction in PCGF5 levels. Inhibition of ubiquitination with small molecules also significantly decreased ehrlichial infection, indicating that the Ub pathway is critical for ehrlichial intracellular replication and survival. The current study identified a novel E. chaffeensis ubiquitin ligase and revealed an important role for the ubiquitin pathway in effector-host interactions and pathogen-mediated host protein stability in order to promote intracellular survival.
KEYWORDS: Ehrlichia, tandem repeat protein, effector, ubiquitin, SUMO, HECT ligase
INTRODUCTION
Ehrlichia chaffeensis is an obligately intracellular bacterium that resides and replicates within cytoplasmic vacuoles of mononuclear phagocytes and causes human monocytotropic ehrlichiosis (HME), an emerging, life-threatening, tick-borne zoonosis (1, 2). E. chaffeensis has evolved molecular strategies to successfully survive in the mononuclear phagocyte mediated by tandem repeat protein (TRP) effectors, which are secreted by the type 1 secretion system (T1SS) (3). Insight into the functional roles of TRPs has been revealed in studies that have defined TRP interactions with a diverse network of host proteins, suggesting TRP moonlighting capability. Through these interactions, host cellular processes targeted by ehrlichial TRPs include transcriptional and translational regulation, posttranslational modification, signaling and immune responses, intracellular trafficking and cytoskeletal organization, and apoptosis (4, 5). Most recently, E. chaffeensis via the TRP120 effector was shown to activate Wnt and Notch signaling pathways to promote intracellular survival (6, 7).
E. chaffeensis TRP120 is expressed by infectious dense-cored ehrlichiae in both arthropod and mammalian cells and is a major target of the human antibody response (8). TRP120 translocates across the vacuolar membrane through an unknown mechanism and localizes to different host subcellular compartments, including the nucleus (3, 9). Notably, TRP120 is known to interact with several host cell proteins involved in posttranslational modification, including enzymes required for ligation and conjugation of ubiquitin (Ub) and ubiquitin-like modifiers (10). Moreover, we have recently demonstrated that recombinant TRP120 is modified by SUMO at a canonical consensus SUMO conjugation motif located in the C-terminal domain (11), and this TRP120 SUMOylation site was further confirmed in vitro using a high-density microfluidic peptide array (12). Functionally, TRP120 SUMOylation facilitates interactions with host cell targets, such as polycomb group proteins (PCGF5), actin and myosin cytoskeleton components, and vesicular trafficking protein GGA1. Inhibition of host SUMO pathway with a small-molecule inhibitor significantly decreased interaction between TRP120 and PCGF5, as well as decreasing PCGF5 recruitment to the ehrlichial cytoplasmic inclusion. More importantly, inhibition of the SUMO pathway also decreased E. chaffeensis intracellular survival (11).
Ubiquitination and SUMOylation are two important posttranslational modifications that play pivotal roles in regulating cell signaling, protein trafficking, protein stability, and transcription (13, 14). Ubiquitination involves the covalent attachment of either a single Ub molecule (monoubiqutination) or a chain of covalently linked Ub molecules (polyubiquitination) to a lysine residue(s) on the substrate protein. Poly-Ub chains are formed through the linkage of Ub at one of seven lysine residues. Although mixed-linkage chains exist, the most commonly studied Ub chains are homotypic, with differently linked Ub chains targeting proteins to different fates. Alternatively, ubiqutinated lysine residues can be modified by Ub-like molecules (such as SUMO). In contrast with SUMOylation, ubiquitination is the most widely recognized posttranslational modification and is associated with proteasomal protein degradation. Although TRP120 is SUMOylated, it is unknown whether TRP120 is ubiquitinated and if the functional consequences of TRP120 ubiquitination are different from those of SUMOylation.
Bacterial effectors have recently been shown to interfere with the host ubiquitination system. Several bacterial effectors target specific steps in the ubiquitination pathway, such as the Shigella effectors OspI and OspG, which interact with host E2 enzymes and interfere with their function (15–17), or the NEL family effectors, which mimic E3 ligases to promote Ub-mediated degradation of specific targets (18). Bacteria can also utilize ubiquitination to regulate the activity and localization of their effectors. The Salmonella type III secretion system effector SopB (an inositol polyphosphate phosphatase that modulates several host functions) typically acts at the cell surface to modulate actin-mediated bacterial entry; however, upon ubiquitination, SopB relocates to the Salmonella-containing vacuole (SCV), where it is required to recruit Rab5 (19–21). SopA, a T3SS effector of Salmonella enterica serovar Typhimurium, has E3 ubiquitin ligase activity, suggesting a direct regulatory role for SopA in host ubiquitination pathways (22, 23).
In this study, we demonstrated that E. chaffeensis TRP120 has a functional C-terminally located HECT E3 ligase domain. TRP120 is ubiquitinated by intrinsic and host HECT ligase activity in vitro and during infection, and TRP120 ubiquitination enhances interactions with defined TRP120-interacting host proteins. TRP120-Ub influences host protein recruitment to the ehrlichial vacuole, and our results demonstrate that TRP120 ubiquitinates host proteins, resulting in decreased protein levels during infection.
RESULTS
TRP120 has functional C-terminal HECT E3 ligase and autoubiquitinates.
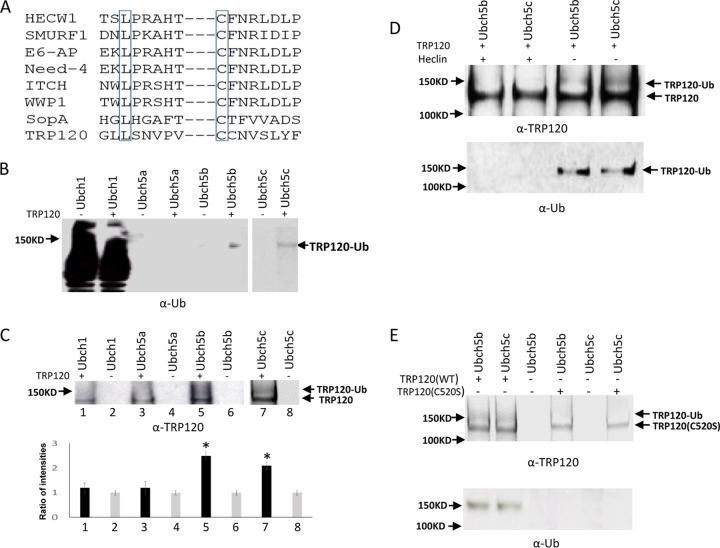
The C terminus of TRP120 was aligned with those of several known HECT E3 ligases, and we determined that TRP120 contains conserved leucine and cysteine residues in the C terminus at positions 514 and 520. The conserved leucine is thought to be involved in substrate recognition. The cysteine residue at 520 is consistent with the catalytic cysteine site in eukaryote HECT E3s (Fig. 1A). Next, we investigated the E2 and E3 ligases responsible for TRP120 ubiquitination in vitro by using recombinant protein in the presence of E1 and different E2 enzymes. Immunoblot analysis with anti-TRP120 and anti-Ub demonstrated the formation of higher-molecular-mass species of TRP120 in the presence of UbcH5b/c, whereas these bands were not significantly detected in the presence of UbcH1 and UbcH5a after quantification analysis (Fig. 1B and C). Ubiquitinated TRP120 was not detected when incubated with UbcH5b/c in the presence of HECT E3 ligase inhibitor (Fig. 1D). In addition, we did not observe any ubiquitinated TRP120 or E3 ubiquitin ligase activity for the TRP120C520S mutant (Fig. 1E). Collectively, these findings demonstrate that TRP120 has intrinsic HECT ligase activity and autoubiquitinates in the presence of UbcH5b/c. The study also confirmed that Cys520 is the active site of TRP120 E3 ubiquitin ligase activity.
FIG 1.
TRP120 effector autoubiquitinates and contains a C-terminal HECT E3 ligase domain. (A) Alignment of TRP120 HECT E3 catalytic domain with other HECT-like E3 ubiquitin ligases. Conserved cysteine and leucine residues are boxed. (B) TRP120 in vitro ubiquitination assay was performed in separate reactions using recombinant protein in the presence of Ubch1, Ubch5a, and Ubch5b (left) and Ubch5c (right), with immunoblot analysis of assay products with anti-Ub. (C) Immunoblot analysis of assay products with anti-TRP120 and densitometric analysis using ImageJ software shows elevated ubiquitinated TRP120 in the presence of UbcH5b/c compared with control (bottom; *, Student's t test; P < 0.05). (D) TRP120 in vitro ubiquitination assay using recombinant protein and UbcH5b/c with or without heclin HECT E3 inhibitor, with immunoblot analysis of assay products with anti-Ub and anti-TRP120. (E) TRP120 in vitro ubiquitination assay using TRP120C520S recombinant mutant protein in the presence of E1 and UbcH5b/c, with immunoblot analysis of assay products with anti-Ub and anti-TRP120.
HECT E3 ligase (Nedd4L) ubiquitinates TRP120.
The conserved catalytic cysteine residue in the C termini of known HECT E3 ligases has an important role in mediating E2 specificity and catalysis (24). The C terminus of TRP120 is similar to that of SopA effector; no significant similarity of sequence was observed with several known HECT E3 ligases except the conserved catalytic cysteine residues in the C terminus at position 520 (23). Nedd4L belongs to the Nedd4 family of E3 HECT domain ubiquitin ligases (25). The crystal structure of a complex between the HECT domain of Nedd4L and the E2 UbcH5b has been determined and shows that Ub is readily transferred from the cysteine of UbcH5b to the catalytic cysteine of Nedd4L (24). Because UbcH5b is required for TRP120 autoubiquitination, we considered the possibility that Nedd4L may also ubiquitinate TRP120 in the presence of UbcH5b. To test this hypothesis, we first examined Nedd4L protein levels and the interaction between TRP120 and Nedd4L in E. chaffeensis-infected host cells. Immunoblot analysis demonstrated that Nedd4L protein levels were significantly increased during E. chaffeensis infection at 48 and 72 h (Fig. 2A). Immunofluorescence microscopy also revealed that Nedd4L colocalized with TRP120-expressing E. chaffeensis morulae (Fig. 2B). To further demonstrate the interaction between TRP120 and Nedd4L, coimmunoprecipitation was performed using E. chaffeensis-infected whole-cell THP-1 lysates and anti-TRP120 and Nedd4L antibodies. As shown in Fig. 2C, Nedd4L coimmunoprecipitated with TRP120, suggesting that these proteins interact in infected cells. We then examined whether Nedd4L could ubiquitinate TRP120 in the presence of UbcH5b. We used an in vitro ubiquitination assay to show the generation of a higher-molecular-mass TRP120 isoform (∼150 kDa) in the presence of recombinant Nedd4L using both anti-TRP120 and anti-ubiquitin antibodies. Quantitative densitometric analysis was performed to examine TRP120 ubiquitination level and showed an ∼4-fold increase in the presence of Nedd4L. This indicated that Nedd4L catalyzes TRP120 ubiquitination in vitro (Fig. 3A). In addition, to examine E. chaffeensis TRP120 ubiquitination during infection, uninfected and E. chaffeensis-infected THP-1 cell lysates were subjected to ubiquitin enrichment. We used TRP120 antibody to screen ubiquitinated proteins that had been enriched from uninfected and E. chaffeensis-infected lysates. Ubiquitin affinity beads recovered ubiquitinated TRP120 from infected-cell lysates as a minor proportion of the total TRP120, migrating primarily as a single band with an approximate mobility of 150 kDa (Fig. 3B). Collectively, these results demonstrated that both native and recombinant TRP120 is ubiquitinated.
FIG 2.
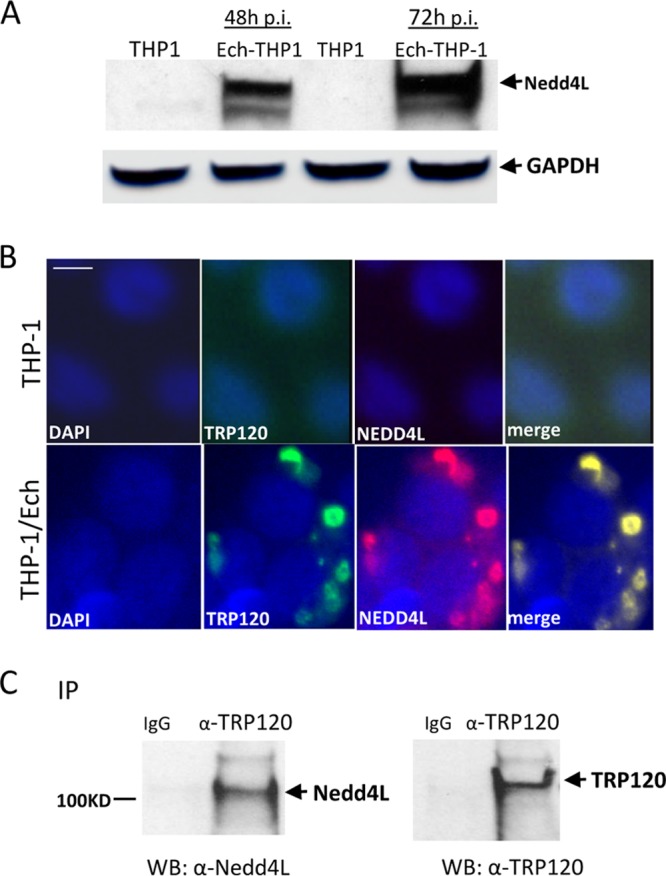
Human HECT E3 ligase, Nedd4L, colocalizes with E. chaffeensis inclusions and interacts with TRP120. (A) Western immunoblot detection of Nedd4L protein levels during E. chaffeensis infection at 48 and 72 h p.i. (B) Immunofluorescence microscopy visualization shows that Nedd4L colocalized with TRP120-expressing E. chaffeensis morulae. Uninfected THP-1 cells were probed with DAPI (DNA; blue), anti-TRP120 (green), and anti-Nedd4L (red) (top). E. chaffeensis-infected THP-1 cells were probed (72 h p.i.) with DAPI (DNA; blue), anti-TRP120 (green), and anti-Nedd4L (red) (bottom). Bar, 10 μm. (C) Western immunoblot (WB) detection of TRP120 and Nedd4L from immunoprecipitated (IP) samples prepared from E. chaffeensis-infected cells using TRP120-specific antibody.
FIG 3.
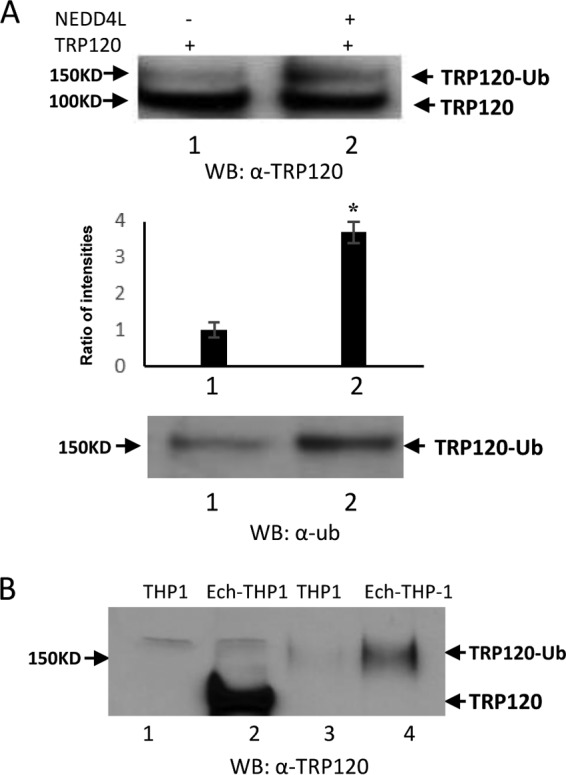
Both native TRP120 and recombinant TRP120 are ubiquitinated and detected by Western immunoblotting. (A) In vitro TRP120 ubiquitination in the presence of Nedd4L using anti-TRP120 (top) and anti-Ub (bottom). The densitometric analysis shows elevated ubiquitinated TRP120 in the presence of Nedd4L compared with control (middle; Student's t test; *, P < 0.05). (B) Uninfected and infected THP-1 cell lysates were subjected to Ub enrichment and Western blot detection of TRP120 ubiquitination before (lanes 1 and 2) and after (lanes 3 and 4) Ub enrichment.
TRP120 ubiquitination site mapping.
Recently, we developed a powerful in vitro Ub/SUMO assay using a novel, high-density peptide array incorporated within a microfluidic device that allows rapid identification of ubiquitination and SUMOylation sites on target proteins (12). To further identify the ubiquitination sites on TRP120, a microarray on a microfluidic chip containing TRP120 peptides (n = 15 [Table 1]) was incubated with the E1, Ub, ATP, Mg2+, and freshly prepared cell lysate. We analyzed TRP120 peptides that were spotted on the peptide microarray which exhibited fluorescent signals higher than the negative-control mutant peptides. A representative image of the TRP120 K432 ubiquitination site showing signal intensity compared to the control (lysine to alanine) peptide is shown in Fig. 4A. Peptides representing 5 lysine residues in TRP120 (K33, K120, K130, K396, and K432) had a fluorescent signal that was significantly higher than those of the mutant peptides (P < 0.05), demonstrating ubiquitination of these lysines (Fig. 4B). To confirm that K432 was one of the ubiquitination sites in vitro and vivo, we investigated TRP120 ubiquitination in vitro by using TRP120 wild-type (WT) and TRP120K432R recombinant proteins in the presence of UbcH5b/c E2 enzymes. Immunoblot analysis with anti-TRP120 and anti-Ub indicated that TRP120K432R mutant was less ubiquitinated than WT TRP120 (Fig. 4C). In addition, green fluorescent protein (GFP)-tagged WT and K432R TRP120 was transiently expressed in HeLa cells, and cells were lysed 24 h posttransfection in the presence of isopeptidase inhibitors (N-ethylmaleimide and iodoacetamide). The cell lysates were subjected to Ub pulldown and analyzed by immunoblotting with anti-TRP120. GFP-tagged WT TRP120 probed with anti-TRP120 exhibited a higher-molecular-mass protein band than that of unmodified WT GFP-TRP120. Mutation of K432 diminished the TRP120-Ub level ∼2.5-fold compared with the level of TRP120 (WT) in cells (Fig. 4D). These results demonstrate that ubiquitination occurs at K432 during in vitro and in cells, but all 5 lysine residues are potential sites of ubiquitination.
TABLE 1.
TRP120 peptides corresponding to lysinea
| Modified K | Peptide sequence |
|
|---|---|---|
| Wild type | Control | |
| 33 | ILGFGNKNIVQP | ILGFGNANIVQP |
| 71 | AESEVSKVEQEK | AESEVSAVEQEK |
| 76 | SKVEQEKTNPEV | SKVEQEATNPEV |
| 84 | NPEVLIKDLQDV | NPEVLIADLQDV |
| 120 | HQGETEKESGIT | HQGETEAESGIT |
| 130 | ITESHQKEDEIV | ITESHQAEDEIV |
| 151 | AESEVSKVEQEE | AESEVSAVEQEE |
| 396 | SKVEQEKTNPEI | SKVEQEATNPEI |
| 418 | IPVVVEKDEMFA | IPVVVEADEMFA |
| 432 | FNPIVIKEEDKV | FNPIVIAEEDKV |
| 436 | VIKEEDKVCETC | VIKEEDAVCETC |
| 449 | QEFEIVKDSQTV | QEFEIVADSQTV |
| 455 | KDSQTVKGSEDI | KDSQTVAGSEDI |
| 488 | MLCPMSKPGQYV | MLCPMSAPGQYV |
| 507 | YGFQDVKDLLGG | YGFQDVADLLGG |
Amino acid residues in the peptides carrying a lysine (K; indicated in bold). For each peptide, a corresponding negative control sequence was included with alanine (A) substituted for lysine (indicated in bold).
FIG 4.
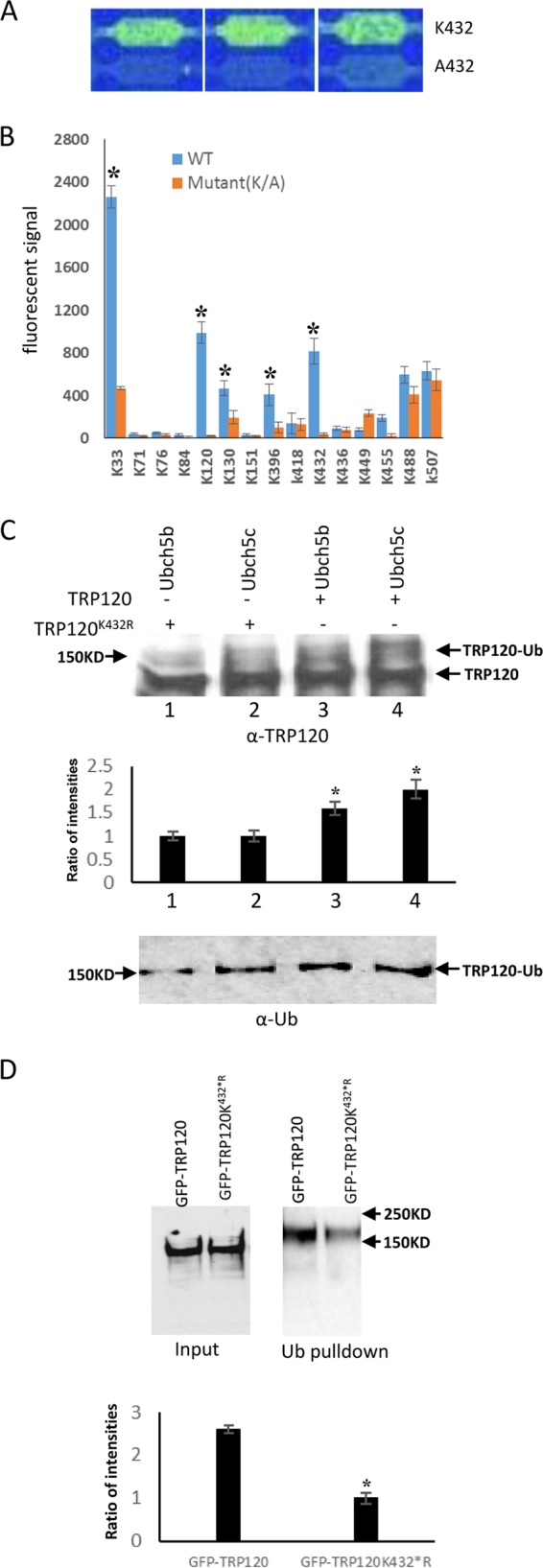
TRP120 ubiquitination site mapping. (A) Representative images of three nonadjacent replicates of the peptide chip showing ubiquitination signals at K432 of TRP120 compared to the mutant control (A432). (B) Histogram plot of the ubiquitination signals at different TRP120 lysine residues compared to the corresponding mutant controls for 15 peptides. Data represent the means ± SD of triplicate determinations (*, P < 0.05). (C) Western immunoblot detection of in vitro TRP120 and TRP120K432R ubiquitination in the presence of UbcH5b/c using anti-TRP120 (top) and anti-Ub (bottom). The densitometric analysis shows decreased ubiquitinated TRP120 in the presence of TRP120 (K432R) compared with TRP120 (WT) (middle; *, Student's t test; P < 0.05). (D) GFP-tagged TRP120 and TRP120 K432R were transfected into HeLa cells. Transfected HeLa cells were lysed, subjected to ubiquitin pulldown, and analyzed using anti-TRP120 antibody. Quantitative densitometric analysis was performed to examine TRP120 ubiquitination level and shows around a 2.5-fold decrease in the presence of TRP120 K432R mutant (Student's t test; *, P < 0.05).
TRP120 ubiquitination mediates host protein recruitment to the ehrlichial vacuole.
In vitro and cellular analysis of TRP120 ubiquitination demonstrated that this effector is ubiquitinated by intrinsic and extrinsic ligase activity. In order to demonstrate that TRP120-Ub is functionally important during E. chaffeensis infection, THP-1 cells were treated with Nedd4L small interfering RNA (siRNA) and control (scrambled) siRNA and then infected with E. chaffeensis. We first examined changes in PCGF5 abundance compared to control siRNA using uninfected THP-1 cell and observed no reduction in PCGF5 abundance after Nedd4L siRNA treatment. Colocalization of TRP120 and that of a defined interacting protein, PCGF5, were compared using immunofluorescence microscopy. Nedd4L knockdown cells showed significantly less PCGF5 colocalization than control cells, as determined by fluorescence intensity profiles (Fig. 5A and B). PCGF5 levels were not affected by control or Nedd4L siRNA (Fig. 5A). We examined changes in infection status in Nedd4L knockdown and control cells and observed no difference in bacterial loads at 24 h postinfection (p.i.) compared with control siRNA. However, at 48 h p.i., significant decreases in bacterial load were observed by real-time quantitative PCR (qPCR) analysis of dsb copy number (Fig. 5C). These studies demonstrate that during E. chaffeensis infection, TRP120 ubiquitination is important for PCGF5 interaction and recruitment to ehrlichial inclusions and that ubiquitination of TRP120 by Nedd4L promotes intracellular survival.
FIG 5.
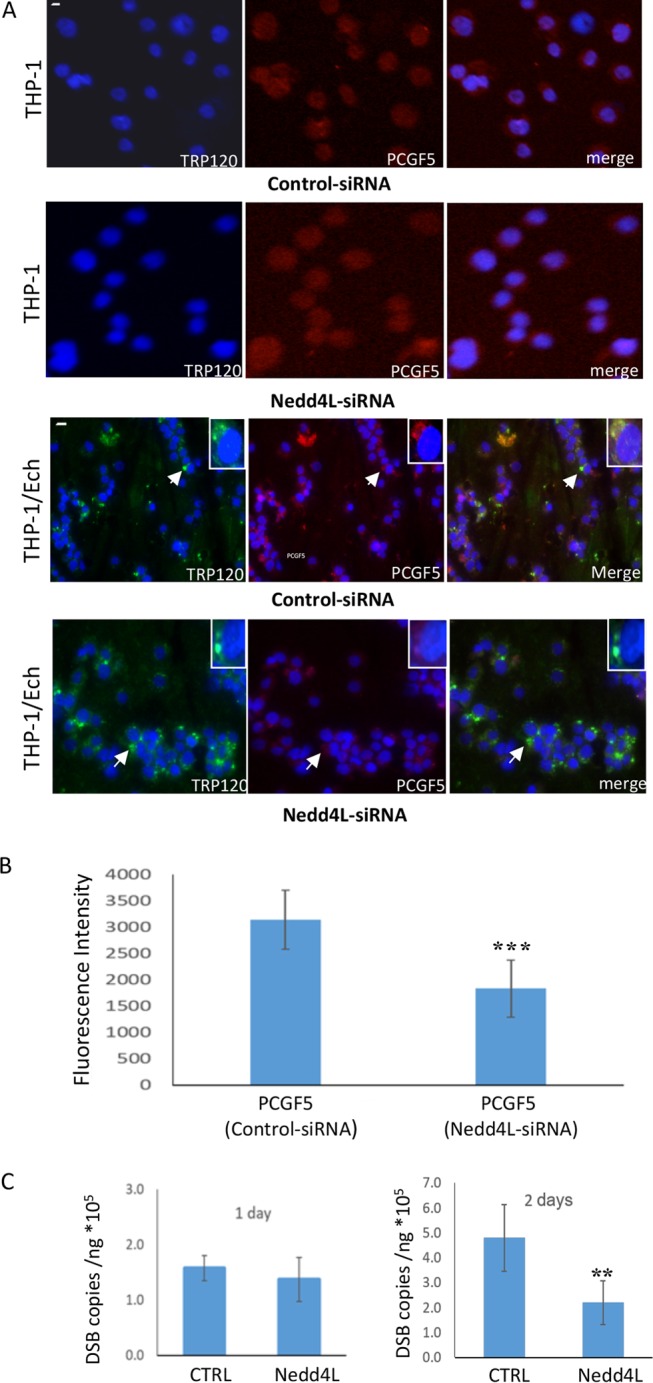
TRP120 ubiquitination facilitates host protein recruitment to the ehrlichial vacuole. (A) Immunofluorescence microscopy shows that Nedd4L siRNA does not reduce PCGF5 abundance in uninfected cells compared with control siRNA and that PCGF5 recruitment to the ehrlichial vacuole decreased with a Nedd4L siRNA knockdown compared with control siRNA (bar, 10 μm) in infected cells. Arrows indicate areas magnified (insets) to show the colocalization of PCGF5 with TRP120. Decreased colocalization of PCGF5 (red) with TRP120 (green) was observed following Nedd4L siRNA knockdown at 48 h p.i. (B) Histogram plot of PCGF5 fluorescence at the ehrlichial inclusion in NEDD4L knockdown cells compared to corresponding control siRNA. Data represent the means ± SD of 20 cell determinations (Student's t test; ***, P < 0.001). (C) Bacterial loads were determined by qPCR analysis of the dsb gene after siRNA silencing of Nedd4L in THP-1 cells and compared to control siRNA during E. chaffeensis infection (24 and 48 h) (Student's t test; **, P < 0.01).
TRP120 targets PCGF5 for ubiquitination.
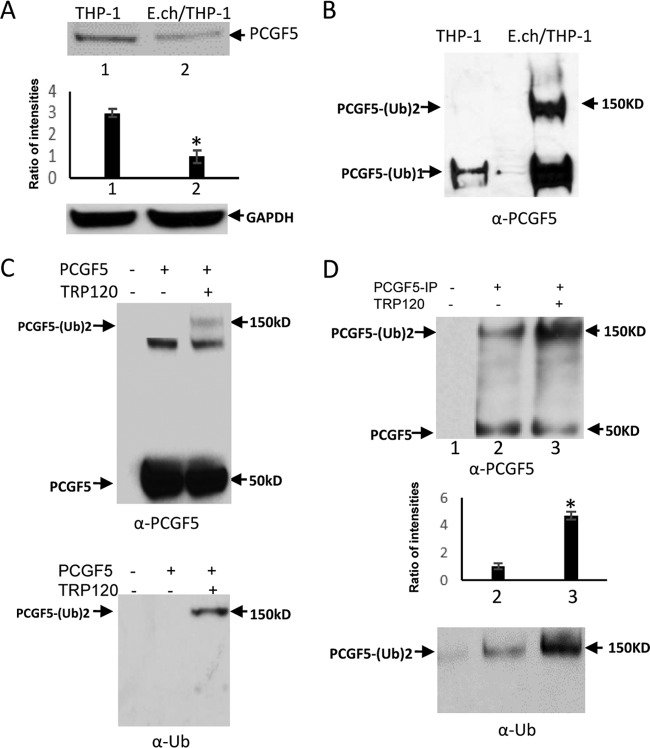
We previously demonstrated that PCGF5, a component of polycomb repressive complex 1 (PRC1) strongly interacts with TRP120 during infection and these these interactions occur at the PCGF5 RING domain (10). We first examined the change in PCGF5 protein level during infection. Indeed, a significant decrease (∼3-fold) in PCGF5 protein occurred during infection (Fig. 6A). Next, we examined PCGF5 ubiquitination levels during infection. E. chaffeensis-infected cell lysates were subjected to ubiquitin enrichment, and a novel PCGF5 band (∼150 kDa) was identified in infected cells but not in uninfected cells (Fig. 6B), demonstrating that PCGF5 is ubiquitinated during infection in cells. In mammalian cells, PCGF5 ubiquitination is highly UbcH5c (E2) dependent (26) and TRP120 autoubiquitinates through intrinsic HECT ligase activity in the presence of UbcH5b/c. To elucidate a mechanism of TRP120-mediated PCGF5 degradation through the ubiquitin-proteasome pathway, we examined the ubiquitination of PCGF5 in vitro using recombinant protein with or without recombinant TRP120. Immunoblot analysis of assay products with anti-PCGF5 and anti-Ub demonstrated the presence of a higher-molecular-mass species of PCGF5 (150 kDa) in the presence of TRP120, whereas this band was not detected in the absence of TRP120 (Fig. 6C). Next, we performed an in vitro ubiquitination assay with native PCGF5 purified by immunoprecipitation; the higher-molecular-mass species of PCGF5 (150 kDa) with or without TRP120 were observed by immunoblotting using anti-PCGF5 and anti-Ub antibody. Levels of native PCGF5-Ub were greatly increased (∼5-fold) in the presence of TRP120 (Fig. 6D). These studies indicate that TRP120 directly targets PCGF5 for ubiquitination and may result in PCGF5 degradation by the ubiquitin-proteasome pathway during infection.
FIG 6.
In vivo and in vitro immunodetection of ubiquitination of PCGF5 by TRP120. (A) Immunoblot analysis of PCGF5 protein level in uninfected and E. chaffeensis-infected THP-1 cells with anti-PCGF5 and densitometric analysis shows decreased PCGF5 during infection (bottom; Student's t test; *, P < 0.05). (B) Cell lysates were subjected to ubiquitin enrichment, and PCGF5 ubiquitination detection with anti-PCGF5 showed multiubiquitination of native PCGF5. (C) In vitro ubiquitination assay of recombinant PCGF5 with or without recombinant TRP120 protein. PCGF5 ubiquitination detection with anti-PCGF5 (top) and anti-Ub (bottom) shows higher-molecular-mass (150 kDa) wild-type PCGF5 in the presence of TRP120, indicative of polyubiquitination events. (D) In vitro ubiquitination assay of PCGF5 in the presence of purified native PCGF5 with or without recombinant TRP120 protein. Immunoblot analysis performed with anti-PCGF5 (top) and anti-Ub (bottom) shows higher-molecular-mass (150 kDa) wild-type PCGF5 in the presence of TRP120, indicative of polyubiquitination events. The densitometric analysis to examine PCGF5 ubiquitination level and shows around a 5-fold increase in the presence of TRP120 (middle; Student's t test; *, P < 0.05).
Heclin inhibits TRP120 ubiquitination and E. chaffeensis infection.
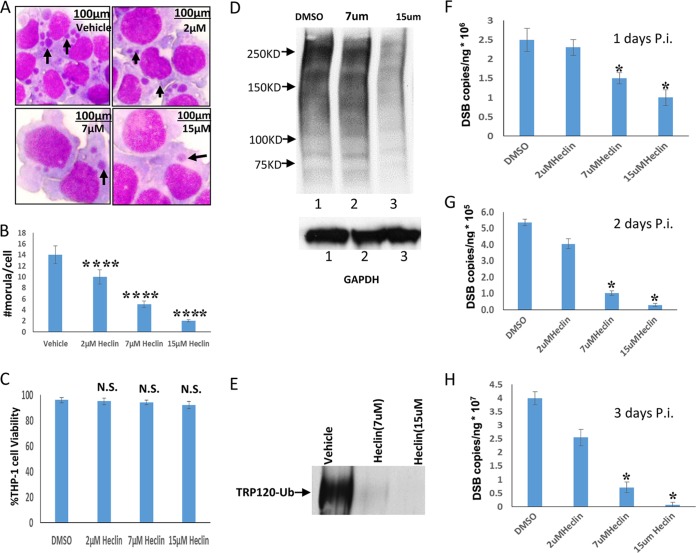
Heclin selectively inhibits several HECT E3 ubiquitin ligases, including Nedd4L. To study the role of HECT E3 ligase activity and specifically of Nedd4L, we examined the effects of heclin on E. chaffeensis infection and TRP120 ubiquitination. THP-1 cells were pretreated with the inhibitor heclin, which has a significant impact on Nedd4L activity (27). Significantly fewer ehrlichial inclusions per cell were observed for heclin-treated cells than for vehicle-treated cells at 48 h p.i. (Fig. 7A), indicating that heclin has a significant impact on ehrlichial morula count (Fig. 7B) without toxicity to host cells (Fig. 7C). Immunoblot analysis showed that global host ubiquitination and TRP120 ubiquitination decreased with treatment at heclin concentrations of 7 and 15 μM compared to that of vehicle-treated cells at 48 h p.i. (Fig. 7D and E). Samples were collected at 24-, 48-, and 72-h intervals p.i. to examine the impact of heclin treatment on E. chaffeensis infection. Heclin treatment (2 μM) did not significantly affect bacterial replication at these time points; however, significant decreases in ehrlichial load were observed relative to those in vehicle-treated cells at 24, 48, and 72 h p.i. for the 7 μM and 15 μM treatments (Fig. 7 F, G, and H). These findings demonstrate that heclin inhibits ehrlichial replication at a concentration consistent with a mechanism involving inhibition of the host and TRP120 HECT ubiquitin ligase activity.
FIG 7.
HECT ligase inhibitor inhibits host cell ubiquitination and decreases E. chaffeensis survival. (A) Bright-field images (magnification, ×100) of Diff-Quik-stained samples collected at 48 h p.i. demonstrate a decreased number of ehrlichial inclusions per cell following treatment with increasing concentrations of heclin. (B) Ehrlichial inclusion (morula) count per cell (THP-1) was determined using Diff-Quik-stained samples at 20 cells per condition. Heclin-treated samples were compared to vehicle-treated infection cells to test significance (Student's test; ****, P < 0.0001). (C) Trypan blue staining was performed to assess host cell viability graphed as a percentage of total cell count for THP-1 cells exposed to 72-h treatments with a vehicle (DMSO) or heclin (Student's test; N.S., not significant). (D) Immunoblot analysis was performed with Ub antibody (or GAPDH antibody for a loading control) and THP-1 whole-cell lysate, with or without heclin, at 48 h infection. (E) THP-1 cells were treated with DMSO (vehicle) or heclin for 3 h and then infected with E. chaffeensis. Cell lysates were subjected to ubiquitin enrichment and immunoblot detection of TRP120 ubiquitination levels. (F, G, and H) THP-1 cells were treated with DMSO (vehicle) or heclin for 3 h and then infected with E. chaffeensis. Bacterial loads were determined by qPCR analysis of the dsb gene and compared to those in vehicle-treated infected cells (24, 48, or 72 h p.i.). (Student's t test; *, P < 0.05).
DISCUSSION
E. chaffeensis invades and survives in mononuclear phagocytes by modulating host cell processes and evading innate defenses, but the mechanisms are not fully defined. However, there is an increasing appreciation for the role of Ehrlichia TRP effectors in the modulation of host innate immune response during infection via direct interactions with host proteins and activation of cellular pathways (4, 6, 7). Previously, we have reported that TRP120 interacts with a functionally diverse array of host proteins, including components of the Ub PTM pathways (10). More recently, we reported that TRP120 is conjugated with SUMO2/3 isoforms in a C-terminal canonical SUMO motif, and that this modification mediates interactions with some of its target proteins (11). In this study, we demonstrated that TRP120 ubiquitination occurs through intrinsic and host HECT ligase activity during E. chaffeensis infection. A proposed model of this modification and the functional outcomes is presented in Fig. 8. TRP120 is a HECT-like E3 ligase, and Cys520 is critical for its E3 ubiquitin ligase activity and autoubiquitination in the presence of UbcH5b/c. Interestingly, NEDD4L ubiquitinates TRP120 and influences TRP120 interactions and levels during infection. Disruption of ubiquitination through the use of HECT ligase inhibitors or siRNA targeting Nedd4L significantly decreased levels of PCGF5 recruitment to the ehrlichial inclusions and had a significant impact on E. chaffeensis infection. This indicates that HECT mediated ubiquitination is critical for E. chaffeensis intracellular survival. Additionally, we have shown that TRP120 directly targets PCGF5 for ubiquitination in vitro and may result in PCGF5 degradation by the ubiquitin-proteasome pathway during infection.
FIG 8.
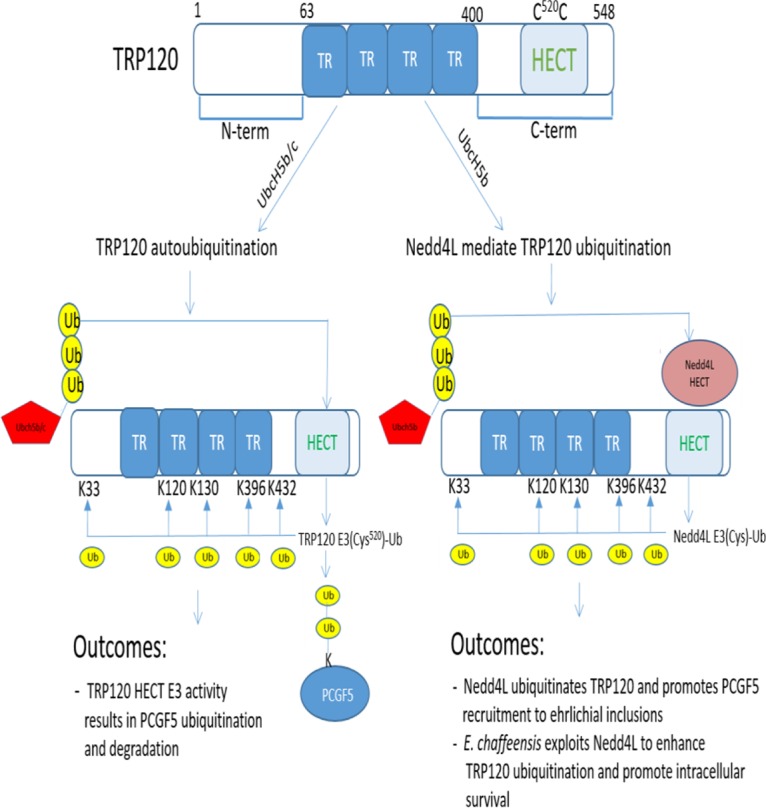
Depiction of TRP120 ubiquitination and the functional outcomes during E. chaffeensis infection of the mononuclear phagocyte. TRP120 is an E. chaffeensis type I effector which contains tandem repeats (TRs). TRP120 has E3 ubiquitin-protein ligase activity in the presence of UbcH5b/c, and Cys520 is the active site of TRP120 E3 ubiquitin ligase activity. TRP120 ubiquitination sites were mapped to K33, K120, K130, and K432. TRP120 mediates PCGF5 polyubiquitination, which is associated with a reduction in PCGF5 levels. A HECT E3 ubiquitin ligase, Nedd4L, mediates TRP120 ubiquitination and regulates recruitment of known TRP120-interacting host protein, PCGF5, to the ehrlichial inclusion during infection. E. chaffeensis exploits Nedd4L to enhance TRP120 ubiquitination and promote intracellular survival.
E2 enzymes are central components in Ub pathways. Our studies reveal that TRP120 preferentially utilizes UbcH5b/c to catalyze autoubiquitination. Interestingly, SopA, a Salmonella type III effector, mimics the mammalian HECT E3 ubiquitin ligase and also utilizes UbcH5 to autoubiquitinate (22, 23). When TRP120 and SopA are aligned, the region from 358 to 377 of SopA and the region from 446 to 465 of TRP120 exhibit 30% identity. Notably, these homologous regions do not encompass the C-terminal 40 amino acids of SopA and TRP120, which include the catalytic cysteine. It is not clear if these regions contain UbcH5 binding domain, because structural information with regard to TRP120/UbcH5 interaction is unknown. In addition, the precise physiological roles of UbcH5 are not well defined. However, in mammalian cells, there is evidence that UbcH5b/c plays a significant role in regulating the NF-κB pathway and inflammatory responses (28–30).
In the ubiquitin system, the interaction between the E2 enzymes and the target protein is mediated by E3 enzymes (31, 32). We previously reported that TRP120 interacts with a number of Ub E3 ligases, including KLHL12 and FBW7 (F-box and WD repeat domain-containing 7, part of SCF, which is an E3 ligase complex) in addition to Ub isoforms UBB and UBC (10). This suggested that TRP120 is a target of the Ub pathway, consistent with our current findings. Nedd4L is a member of the Nedd4 family of HECT ubiquitin ligases, which are implicated in several cellular processes (25, 33). Nedd4L was not previously identified as a TRP120-interacting host protein by yeast two-hybrid screening (10). However, in this study, we found that Nedd4L levels steadily increased during infection and demonstrated that Nedd4L strongly interacts with TRP120 and facilitates TRP120 ubiquitination in vitro. Indeed, Nedd4L localizes primarily in the cytoplasm, where it targets proteins and receptors upon Ca2+-mediated activation (34). Our recent studies showed that Ehrlichia and TRPs stimulate intracellular Ca2+ release during E. chaffeensis entry and activates the noncanonical Wnt/Ca2+ pathway (6). This suggests that Nedd4L may act as a critical regulator for TRP120 ubiquitination during infection. On the other hand, we report here that TRP120 is a E3 ubiquitin ligase, the treatment of cells with HECT E3 inhibitor significantly decreased the ubiquitination levels of TRP120, suggesting a direct regulatory role for TRP120 in host ubiquitination pathways. Although both TRP120 itself and Nedd4L play pivotal roles in defining TRP120 ubiquitination specificities, Nedd4L functions more as an E3 to catalyze the transfer of ubiquitin to target substrates in the presence of Ubch5b (24), while TRP120 has enzymatic activity in the presence of both UbcH5b and UbcH5c. Thus, TRP120 has a distinct functional mimicry of the mammalian E3 ubiquitin ligase by targeting self- and host protein ubiquitination.
We previously demonstrated that E. chaffeensis TRP120 strongly interacts with PCGF5, a component of the polycomb group (PcG) multiprotein PRC1-like complex, which maintains the transcriptionally repressive state of many genes, including hox genes (3, 10). Coimmunoprecipitation of TRP120 with PCGF5 showed that TRP120 SUMOylation at Lys 432 regulates PCGF5 recruitment to the ehrlichial vacuole (11). In this study, the role of Lys 432 of TRP120 was further expanded, as it is also targeted by ubiquitination. We demonstrated that Nedd4L knockdown in infected cells also causes a reduction in PCGF5 recruitment. TRP120 ubiquitination decreased after Nedd4L knockdown, which also significantly decreased levels of PCGF5 colocalization with ehrlichial inclusions, suggesting that TRP120 ubiquitination at K432 also facilitates the interaction with PCGF5 (10).
Previous studies demonstrated that E. chaffeensis T1S effectors TRP47 and TRP120 robustly interact with PCGF5, a polycomb-repressive protein. TRP120-PCGF5 interaction occurs through the TRP120 TR domain, while the amino- and carboxy-terminal domains did not interact (10). The significantly lower level of PCGF5 colocalization observed with Nedd4L knockdown cells suggests that the ubiquitinated TRP120 conformation engages PCGF5. In addition, E. chaffeensis-infected cells exhibited a significant reduction in the expression of PCGF5 and increased levels of ubiquitinated PCGF5 compared to uninfected cells. This was observed despite a decrease in RING1B, a host E3 previously reported to ubiquitinate PCGF5. The fact that the TRP120-PCGF5 interaction occurs through the TRP120 TR domain while maintaining the HECT domain of TRP120 in an active state indicates that PCGF5 is a substrate of TRP120 ligase activity. Consistent with this conclusion, we demonstrated that TRP120 enhances PCGF5 ubiquitination in vitro and that proteasomal degradation of PCGF5 occurs in vivo. We have demonstrated that like most E3 Ub ligases, TRP120 can also autoubiquitinate; however, how TRP120 autoubiquitination influences its ubiquitin ligase activity is undefined.
Lysine residues act as acceptors not only for SUMO modification but also for all other ubiquitin-like proteins. They are also sites of methylation and acetylation. It is possible that SUMO conjugation could block other lysine-dependent modifications, such as ubiquitination. Therefore, the cooperation between ubiquitin and SUMO raises an intriguing question as to whether the two modifiers can coexist on TRP120, or whether SUMOylation and ubiquitination affect the same target protein independently. Colocalization of enzymes involved in SUMO conjugation and deconjugation within the cells indicates that SUMO modification and deconjugation may be part of a highly dynamic process in which substrates undergo rapid modification by SUMO followed by equally rapid deconjugation (35, 36). Supported for this concept is provided by our recent observation that only a small proportion of the total TRP120 is SUMOylated in vivo during infection. Conversely, we detected high levels of ubiquitinated TRP120 in vivo during infection, indicating that TRP120 SUMO conjugation may not block its ubiquitination. In addition, both SUMOylation and ubiquitination of TRP120 can occur independently of E3 ligases. In fact, a number of Ub E3 ligases, but not SUMO E3 ligases, have been shown to interact with TRP120 (10). Notably, in this study we found that Nedd4L protein levels were significantly increased during E. chaffeensis infection and that TRP120 ubiquitination was greatly facilitated with Nedd4L in vitro, suggesting that TRP120 ubiquitination may play a major role in facilitating host protein interactions, including PCGF recruitment and degradation.
We previously demonstrated that TRP120 SUMOylation mediates interactions with PCGF5. Inhibition of the host SUMO pathway decreases PCGF5 recruitment to the ehrlichial vacuole and ehrlichial intracellular survival (11). Although there is a growing family of ubiquitin-like proteins, including SUMO, that modify cellular targets in a pathway (37), it was not surprising that inhibition of host ubiquitination significantly decreased E. chaffeensis infection and survival. Heclin, a selective HECT E3 ubiquitin ligase inhibitor, inhibits Nedd4L with a 50% inhibitory concentration (IC50) of 6.3 μM (27). The median lethal concentration for heclin on HEK293 growing cells was 45 μM, which is about seven times higher than the IC50. In the studies described here, the heclin IC50 for bacterial load at 72 h p.i. was less than 10 μM, suggesting that the inhibitory effect on E. chaffeensis replication occurs through inhibition of pathogen and host HECT E3 ligases. The current studies suggest that HECT-mediated ubiquitination plays an important role for E. chaffeensis infection and replication and may be a novel target for antiehrlichial therapeutics.
This study reveals a novel functional Ub ligase domain on the ehrlichial effector TRP120. Our findings illustrate the evolving knowledge regarding multiple functional roles of TRP120 in ehrlichial pathobiology. However, we have uncovered an important role of TRP120 intrinsic and host HECT ligase activity in host-pathogen interactions and ehrlichial survival. In addition, TRP120 is likely not the only example of a bacterial substrate of this pathway; other bacterial effectors may also possess ligase function or be targets of host PTM pathways. Thus, further studies will be required to elucidate the mechanism between TRP120 ubiquitination and pathogen-host interactions and host protein degradation, to identify additional substrates targeted by TRP120 for host Ub/proteasome pathway (UPP) modulation, and to determine the role of pathogen-host ubiquitination in promoting replication and survival of Ehrlichia.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
Human monocytic leukemia cells (THP-1) were propagated in RPMI medium 1640 with l-glutamine and 25 mM HEPES buffer (Invitrogen) supplemented with 1 mM sodium pyruvate (Sigma, St. Louis, MO), 2.5 g/liter of d-(+)-glucose (Sigma), and 10% fetal bovine serum (HyClone). E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described (38).
Cell lysis and protein extraction.
Whole-cell lysates were extracted from E. chaffeensis-infected THP-1 cells using a whole-cell extraction kit (Abcam, Cambridge, MA). Extractions were performed according to the manufacturer's protocol at 24, 48, or 72 h postinfection. The concentration was determined by Lowry method (DC assay; Bio-Rad).
Antibodies and siRNAs.
Rabbit and mouse anti-TRP120 antibodies have been described previously (39). Other antibodies used in this study were anti-Ub Alexa Fluor 647 (Santa Cruz, CA), anti-FK2 (Enzo), anti-Nedd4L (Proteintech, Rosemont, IL), anti-PCGF5 (Abcam), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; Proteintech), All antibodies used for immunofluorescence were tested by the vendor to ensure the specificity and confirmed by Western immunoblotting, immunofluorescence microscopy, or both. Nedd4L (siRNAs) and a control siRNA were obtained from GE Dharmacon (Lafayette, CO).
Recombinant TRP120, PCGF5, and Nedd4L.
Generation of the TRP120 expression construct (pBAD/Thio-TRP120) has been described previously (9). TRP120 point mutants were generated with pAcGFP1 and pBAD/Thio constructs using a QuikChange II site-directed mutagenesis (Agilent, Santa Clara, CA) with sense and antisense primers (TRP120 K432R sense primer, 5′-AAATGTTTGCACCTTCATTTAATCCAATCGTTATACGGGAGGAAGATAAAGTTTG-3′; TRP120 K432R antisense primer, 5′-CAAACTTTATCTTCCTCCCGTATAACGATTGGATTAAATGAAGGTGCAAACATTT-3′; TRP120 C520S sense primer, 5′-CATAAAATAAAGGCTAACATTACAACTCACAGGAACATTACTTAATAAACCAC-3′; and TRP120 C520S antisense primer, 5′-GTGGTTTATTAAGTAATGTTCCTGTGAGTTGTAATGTTAGCCTTTATTTTATG-3′).
Expression and purification of recombinant TRP120 and mutants were performed as previously described (9, 11). PCGF5 and Nedd4L recombinant protein were obtained from a commercial source (Abnova, Taipei, Taiwan).
Western blot analyses.
Protein samples for Western immunoblot analysis were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and blocked in Tris-buffered saline containing 5% nonfat dry milk. Primary antibodies were rabbit anti-TRP120 (1:5,000), rabbit anti-Nedd4L (1:1,000), rabbit anti-PCGF5 (1:1,000), and rabbit anti-GAPDH (1:4,000). Secondary antibodies were horseradish peroxidase-labeled goat anti-rabbit IgG and goat anti-mouse IgG (Kirkegaard & Perry, Gaithersburg, MD). Densitometric quantification of the immunoblot bands was performed using ImageJ densitometry software.
Microfluidic peptide array technology for analysis of TRP120 ubiquitination.
The workflow for the TRP120 ubiquitination studies by peptide microarray, including peptide design, peptide chip synthesis, ubiquitination and reaction, chip image, data processing, and analysis, was previously described (12). E. chaffeensis TRP sequences can be found on the Integrated Microbial Genomes and Microbiomes website (40). A peptide array was designed using an in-house-developed PERL program (41). Peptides (12-mer) representing TRP regions with lysine residues were synthesized by flanking a central lysine residue with 6 N-terminal amino acids and 5 C-terminal amino acids. For each peptide, a corresponding negative control was included with alanine (A) substituted in place of lysine (K). The significant fluorescent signals of ubiquitination were determined by comparison with the control peptide (A peptide) using anti-Ub Alexa Fluor 647 antibody (Santa Cruz Biotechnology; 1:300). The sequences for peptides used in this study, including the position of each lysine, are shown in Table 1.
RNA interference.
THP-1 cells (2 × 105/well on a 24-well plate) were transfected with human siRNA (5 pmol) using Lipofectamine 3000 (Invitrogen). A control siRNA consisting of a scrambled sequence was used as a negative control. At 1 day posttransfection, the cells were synchronously infected by cell-free E. chaffeensis at a multiplicity of infection (MOI) of ∼50, and they were collected at 1, 2, and 3 days postinoculation (dpi) and observed using immunofluorescence microscopy to determine PCGF5 colocalization.
Immunofluorescence microscopy and coimmunoprecipitation.
Uninfected or E. chaffeensis-infected THP-1 cells were cytocentrifuged onto glass slides, fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, permeabilized, and blocked with 0.3% Triton X-100 and 2% bovine serum albumin in PBS for 1 h. Cells were then incubated with rabbit anti-Nedd4L (1:100) and mouse anti-TRP120 (1:1,000) for 1 h, washed, and stained with fluorescein isothiocyanate (FITC)-conjugated or Alexa Fluor 594-IgG (H+L) and Alexa Fluor 488-IgG (H+L) secondary antibodies (1:100; Molecular Probes) for 30 min. THP-1 cells were grown to 50% confluence in 24-well plates. After Nedd4L siRNA transfection (24 h), cells were synchronously infected by cell-free E. chaffeensis at an MOI of ∼50. Infected cells were collected at 1 and 2 dpi and processed as described above. After siRNA treatment, uninfected and infected cells were incubated with anti-TRP120 (1:1,000) and anti-PCGF5 (1:100) primary antibodies for 1 h, washed, and treated with Alexa Fluor 594-IgG (H+L) and Alexa Fluor 488-IgG (H+L) secondary antibodies (1:100; Molecular Probes) for 30 min. All slides were washed then mounted with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Images were obtained using an Olympus BX61 epifluorescence microscope and were analyzed using Slidebook software (version 5.0; Intelligent Imaging Innovations, Denver, CO). Coimmunoprecipitation was performed on highly infected (95%) THP-1 cells 3 dpi using the EZ-Magna ChIP A/G immunoprecipitation kit (EMD Millipore, Billerica, MA) with anti-E. chaffeensis TRP120 and IgG control antibodies according to the manufacturer's instructions.
In vitro TRP120 and PCGF5 ubiquitination assay.
All in vitro ubiquitination assays were performed using an in vitro ubiquitination kit (Enzo Life Sciences, Farmingdale, NY). TRP120 autoubiquitination reactions were performed using wild-type TRP120 and TRP120K432R and TRP120C520S mutants. Briefly, TRP120 (200 nM) was added to reaction mixtures containing an E1, one of several E2s, and a buffer containing Ub, ATP, inorganic pyrophophatase, and Mg2+ (ubiquitination buffer). The ubiquitination of TRP120, TRP120K432R, and TRP120C520S was performed, respectively, in the presence of UbcH5b/c E2s with or without the addition of heclin HECT E3 inhibitor (15 μM). The ubiquitination of TRP120 (400 nM) was performed in the presence of Nedd4L (40 nM) and UbcH5b. PCGF5 ubiquitination was performed with recombinant TRP120, PCGF5, and purified native PCGF5 protein. TRP120 (10 nM) and PCGF5 (100 nM), or purified native PCGF5 protein, were added to ubiquitination buffer in the presence of E1 and UbcH5c. Native PCGF5 protein was isolated from THP-1 cells using a specific anti-PCGF5 antibody that was later incubated with bead-conjugated protein G. The pulled-down PCGF5 was eluted with elution buffer (0.1 M glycine, pH 3), and the eluted solution was immediately neutralized with neutralization buffer (1 M Tris, pH 8.5). Eluted PCGF5 fractions were subjected to in vitro ubiquitination reactions in a 50-μl total volume according to the manufacturer's protocol with or without TRP120. All of the ubiquitination reactions were performed at 37°C for 3 h, and the reactions were stopped by addition of Laemmli buffer. Samples were boiled for 5 min, resolved by 4 to 20% SDS-PAGE, and processed for Western blotting using anti-TRP120, anti-PCGF5, and anti-Ub antibodies.
In vivo TRP120 and PCGF5 ubiquitination assay.
E. chaffeensis-infected cell lysates (400 μg) were subjected to ubiquitin pulldown (Enzo Life Sciences) using a ubiquitin enrichment kit (Pierce) by following the manufacturer's protocol, and eluates were analyzed by Western blotting using anti-TRP120 and anti-PCGF5 antibodies. GFP-tagged (pAcGFP1) TRP120 and TRP120K432R were prepared as previously described (11). HeLa cells were seeded in 6-well plates and transfected with 4.0 to 8.0 μg of endotoxin-free plasmid purified with a plasmid maxikit (Qiagen, Valencia, CA) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Transfected HeLa were lysed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 200 mM iodoacetamide (Sigma) with protease inhibitor cocktail (Roche Diagnostics). Lysates (400 μg) were subjected to ubiquitin pulldown, and eluates were analyzed by Western blotting using anti-TRP120 antibody.
Small-molecule treatment and real-time qPCR.
THP-1 cells were pretreated with 1% dimethyl sulfoxide (DMSO) or different concentrations of heclin (Tocris Bioscience, Avonmouth, Bristol, UK) for 3 h and then infected with E. chaffeensis. Samples were collected at 24, 48, and 72 h postinfection. Diff-Quik-stained slides were collected and analyzed with an Olympus BX61 epifluorescence microscope using a color camera. Lysates were prepared using a whole-cell extract kit (Abcam) at room temperature and analyzed by Western blotting. Total DNA was extracted from E. chaffeensis-infected THP-1 cells with a DNeasy blood and tissue kit (Qiagen), and bacterial load was measured by real-time quantitative PCR (qPCR). Amplification of the integral ehrlichial gene dsb was performed using Brilliant II SybrGreen master mix (Agilent, Santa Clara, CA), 200 nM forward primer (5′-GCTGCTCCACCAATAAATGTATCCCT-3′), and 200 nM reverse primer (5′-GTTTCATTAGCCAAGAATTCCGACACT-3′). The absolute E. chaffeensis dsb copy number in the cells was determined against the standard curve as previously described (42).
Statistical analysis.
Statistical analyses were performed with a two-tailed unpaired t test. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants AI106859 and AI115449 and the Clayton Foundation for Research.
REFERENCES
- 1.Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat Rev Microbiol 8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- 2.McBride JW, Walker DH. 2011. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev Mol Med 13:e3. doi: 10.1017/S1462399410001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakeel A, den Dulk-Ras A, Hooykaas PJ, McBride JW. 2011. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front Cell Infect Microbiol 1:22. doi: 10.3389/fcimb.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lina TT, Farris T, Luo T, Mitra S, Zhu B, McBride JW. 2016. Hacker within! Ehrlichia chaffeensis effector driven phagocyte reprogramming strategy. Front Cell Infect Microbiol 6:58. doi: 10.3389/fcimb.2016.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunphy PS, Luo T, McBride JW. 2013. Ehrlichia moonlighting effectors and interkingdom interactions with the mononuclear phagocyte. Microbes Infect 15:1005–1016. doi: 10.1016/j.micinf.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo T, Dunphy PS, Lina TT, McBride JW. 2016. Ehrlichia chaffeensis exploits canonical and noncanonical host Wnt signaling pathways to stimulate phagocytosis and promote intracellular survival. Infect Immun 84:686–700. doi: 10.1128/IAI.01289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lina TT, Dunphy PS, Luo T, McBride JW. 2016. Ehrlichia chaffeensis TRP120 activates canonical Notch signaling to downregulate TLR2/4 expression and promote intracellular survival. mBio 7:e00672-16. doi: 10.1128/mBio.00672-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Popov VL, Yu X, Walker DH. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb Pathog 28:71–80. doi: 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 9.Zhu B, Kuriakose JA, Luo T, Ballesteros E, Gupta S, Fofanov Y, McBride JW. 2011. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect Immun 79:4370–4381. doi: 10.1128/IAI.05422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. 2011. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect Immun 79:4382–4391. doi: 10.1128/IAI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunphy PS, Luo T, McBride JW. 2014. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun 82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu B, Farris T, Milligan S, Chen HS, Zhu RJ, Hong AL, Zhou XC, Gao XL, McBride JW. 2016. Rapid identification of ubiquitination and SUMOylation target sites by microfluidic peptide array. Biochem Biophys Rep 5:430–438. doi: 10.1016/j.bbrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, Otsubo R, Morikawa H, Nishide A, Takagi K, Sasakawa C, Mizushima T. 2014. Bacterial effectors and their functions in the ubiquitin-proteasome system: insight from the modes of substrate recognition. Cells 3:848–864. doi: 10.3390/cells3030848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishide A, Kim M, Takagi K, Himeno A, Sanada T, Sasakawa C, Mizushima T. 2013. Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI. J Mol Biol 425:2623–2631. doi: 10.1016/j.jmb.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 16.Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, Oyama A, Ashida H, Kobayashi T, Koyama T, Nagai S, Shibata Y, Gohda J, Inoue J, Mizushima T, Sasakawa C. 2012. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483:623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- 17.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A 102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piscatelli H, Kotkar SA, McBee ME, Muthupalani S, Schauer DB, Mandrell RE, Leong JM, Zhou D. 2011. The EHEC type III effector NleL is an E3 ubiquitin ligase that modulates pedestal formation. PLoS One 6:e19331. doi: 10.1371/journal.pone.0019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. 2009. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol 11:1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel JC, Hueffer K, Lam TT, Galan JE. 2009. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin DY, Diao J, Zhou D, Chen J. 2011. Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J Biol Chem 286:441–449. doi: 10.1074/jbc.M110.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diao J, Zhang Y, Huibregtse JM, Zhou D, Chen J. 2008. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol 15:65–70. doi: 10.1038/nsmb1346. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. 2006. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol 62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 24.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. 2009. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell 36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Kumar S. 2010. Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ 17:68–77. doi: 10.1038/cdd.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taherbhoy AM, Huang OW, Cochran AG. 2015. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat Commun 6:7621. doi: 10.1038/ncomms8621. [DOI] [PubMed] [Google Scholar]
- 27.Mund T, Lewis MJ, Maslen S, Pelham HR. 2014. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci U S A 111:16736–16741. doi: 10.1073/pnas.1412152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchi K, Meier P. 2009. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol Cell 36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 29.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D. 2010. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickart CM, Eddins MJ. 2004. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 33.Bernassola F, Karin M, Ciechanover A, Melino G. 2008. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Escobedo A, Gomes T, Aragon E, Martin-Malpartida P, Ruiz L, Macias MJ. 2014. Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure 22:1446–1457. doi: 10.1016/j.str.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. 2004. A mechanism for inhibiting the SUMO pathway. Mol Cell 16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Saitoh H, Matunis MJ. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol 22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beyer AR, Truchan HK, May LJ, Walker NJ, Borjesson DL, Carlyon JA. 2015. The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol 17:504–519. doi: 10.1111/cmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuriakose JA, Miyashiro S, Luo T, Zhu B, McBride JW. 2011. Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS One 6:e24136. doi: 10.1371/journal.pone.0024136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo T, Zhang X, McBride JW. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin Vaccine Immunol 16:982–990. doi: 10.1128/CVI.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wall LC, Orwant TJ. 2000. Programming Perl. O'Reilly, Sebastopol, CA. [Google Scholar]
- 42.Luo T, McBride JW. 2012. Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infect Immun 80:2297–2306. doi: 10.1128/IAI.00154-12. [DOI] [PMC free article] [PubMed] [Google Scholar]