ABSTRACT
During bacterial infection, the pathogenic agent and the host battle for iron, due to its importance for fundamental cellular processes. However, iron redistribution and sequestration during infection can culminate in anemia. Although hepcidin has been recognized as the key regulator of iron metabolism, in some infections its levels remain unaffected, suggesting the involvement of other players in iron metabolism deregulation. In this work, we use a mouse model to elucidate the main cellular and molecular mechanisms that lead to iron redistribution during infection with two different pathogens: Listeria monocytogenes and Salmonella enterica serovar Typhimurium. Both infections clearly impacted iron metabolism, causing iron redistribution, decreasing serum iron levels, decreasing the saturation of transferrin, and increasing iron accumulation in the liver. Both infections were accompanied by the release of proinflammatory cytokines. However, when analyzing iron-related gene expression in the liver, we observed that hepcidin was induced by S. Typhimurium but not by L. monocytogenes. In the latter model, the downregulation of hepatic ferroportin mRNA and protein levels suggested that ferroportin plays a major role in iron redistribution. On the other hand, S. Typhimurium infection induced the expression of hepcidin mRNA, and we show here, for the first time in vivo, that this induction is Toll-like receptor 4 (TLR4) dependent. In this work, we compare several aspects of iron metabolism alterations induced by two different pathogens and suggest that hepcidin-(in)dependent mechanisms contribute to iron redistribution upon infection.
KEYWORDS: iron metabolism, innate immunity, infection, host-pathogen interactions, Listeria monocytogenes, Salmonella Typhimurium
INTRODUCTION
Due to its capacity to act as an ideal redox catalyst for processes such as DNA synthesis, mitochondrial respiration, and oxygen transport, iron is an essential element for almost all living organisms (1–4). Not surprisingly, iron plays a crucial role in host-pathogen interactions: pathogenic agents developed sophisticated strategies to acquire iron from their environment, while the host innate immune response includes several mechanisms to reduce iron availability for microbes (5–7). The hepatocyte-derived peptide hormone hepcidin has been considered a central player in this process. Hepcidin causes a decrease in plasma iron levels by blocking the activity of ferroportin (FPN), the only known iron exporter, responsible for iron release from macrophages, hepatocytes, and enterocytes (4). The expression of hepcidin is known to be induced by the cytokine interleukin-6 (IL-6) (8), released after the detection of different microbes or microbial products such as lipopolysaccharide (LPS) (9, 10). Continuously high levels of hepcidin lead to decreases of serum iron levels, presumably reducing bacterial growth and contributing to the avoidance of sepsis. Consequently, high levels of hepcidin are thought to be responsible for “anemia of chronic disease,” due to the reduced availability of iron for erythropoiesis (11). However, a direct link between hepcidin, hypoferremia, and anemia of infection has very rarely been demonstrated in vivo.
Curiously, previous work from our laboratory showed that Mycobacterium avium infection leads to moderate anemia, without a significant alteration of hepcidin levels (12). In the present work, we aimed to extend the study of the in vivo role of hepcidin during infection, using two different intracellular pathogens: Listeria monocytogenes and Salmonella enterica serovar Typhimurium (Gram-positive and Gram-negative bacteria, respectively).
L. monocytogenes and S. Typhimurium are intracellular foodborne pathogens, which can cross the intestinal barrier and cause fatal septicemia in susceptible hosts (13, 14). The pathogenicity of these bacteria is known to be highly dependent on iron availability (15–17). In this work, we infected mice with L. monocytogenes or S. Typhimurium and analyzed the impact on host iron metabolism. Our data show that different pathogens impact host iron metabolism by different mechanisms, including hepcidin-independent pathways.
RESULTS
Infections with L. monocytogenes and S. Typhimurium cause alterations in host iron metabolism and distribution.
In this study, we evaluated the alterations in host iron metabolism resulting from infection with two species of facultative intracellular bacteria: the Gram-positive organism L. monocytogenes and the Gram-negative organism S. Typhimurium. After intravenous injection of 104 CFU, both bacteria colonized the liver and spleen and grew exponentially until 72 h postinfection (Fig. 1). The bacterial loads in the liver were similar for the two bacteria, while in the spleen, the bacterial loads of Salmonella reached higher levels than those of Listeria. Of note, while the growth of L. monocytogenes was controlled after 3 days of infection, S. Typhimurium continued to grow and caused the death of the mice by day 4 postinfection.
FIG 1.
Bacterial loads in the liver and spleen of animals infected with L. monocytogenes or S. Typhimurium. C57BL/6 mice were infected intravenously with 104 CFU of L. monocytogenes (A and C) or S. Typhimurium (B and D). Groups of at least 3 animals were sacrificed at different time points, livers (A and B) and spleens (C and D) were aseptically harvested, and the bacterial load was evaluated. The graphs represent mean log10 CFU per organ and standard deviations.
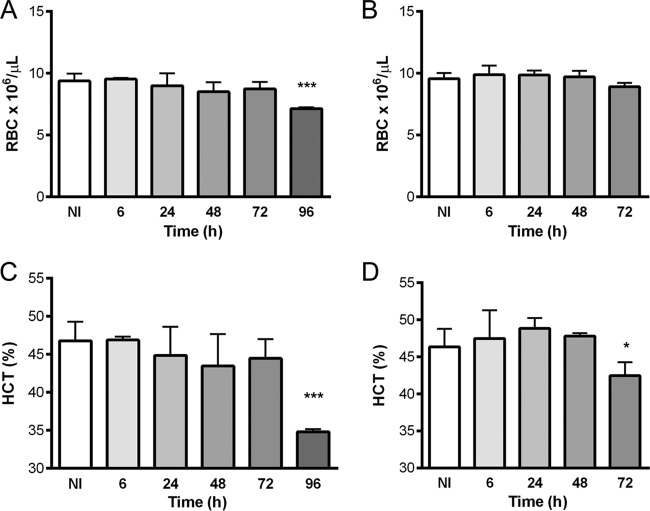
At different time points after infection, blood was collected to evaluate hematological (Fig. 2) and iron (Fig. 3) parameters. A significant reduction in the number of red blood cells (RBCs) was observed in mice infected with L. monocytogenes (Fig. 2A) (from 9.4 × 106 ± 0.6 × 106 to 7.1 × 106 ± 0.1 × 106 RBCs/μl), and a clear reduction of the hematocrit value was observed in mice infected with both pathogens at late time points, namely, 96 h after L. monocytogenes infection (from 46.8% ± 2.5% to 34.9% ± 0.3%) (Fig. 2C) and 72 h after infection with S. Typhimurium (from 46.4% ± 2.4% to 42.4% ± 1.8%) (Fig. 2D).
FIG 2.
Hematological parameters in C57BL/6 mice infected with Listeria monocytogenes (A and C) or Salmonella Typhimurium (B and D). After sacrifice at different time points, blood was collected, and the numbers of red blood cells (A and B) and the hematocrit (HCT) values (C and D) were determined by a certified laboratory. The graphs represent means plus standard deviations for at least 4 animals. Statistical analysis was performed by applying one-way ANOVA followed by a Bonferroni multiple-comparison post hoc test. *, P < 0.05; ***, P < 0.001 (compared with noninfected [NI] mice).
FIG 3.
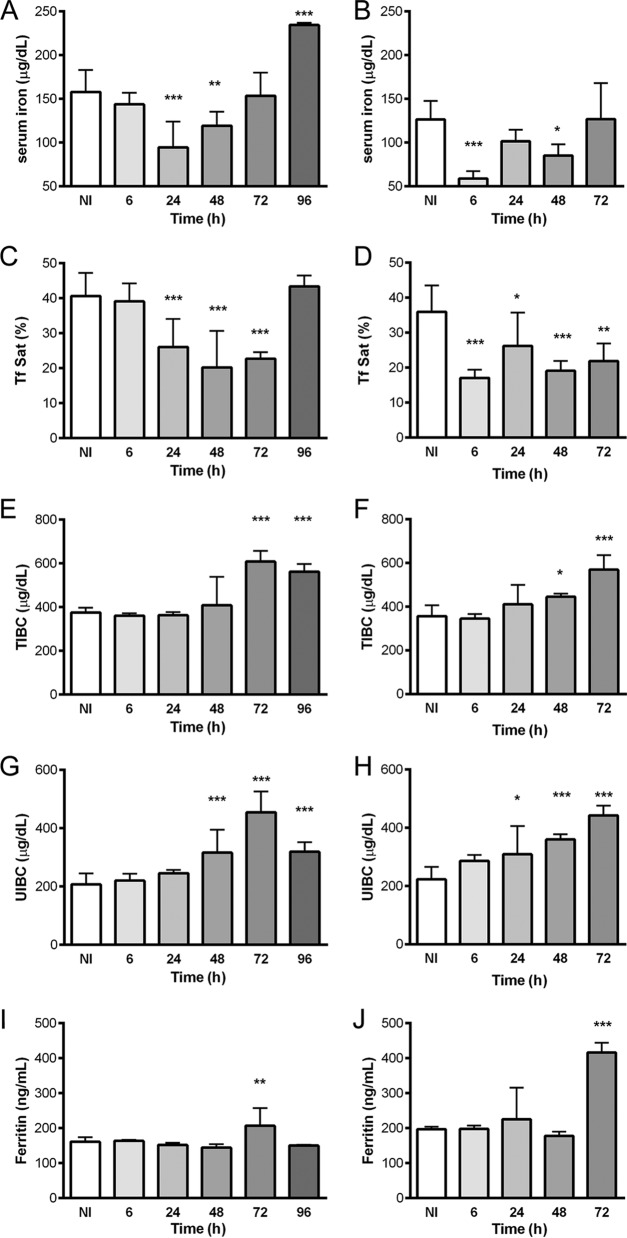
Serum iron parameters in C57BL/6 mice infected with Listeria monocytogenes (A, C, E, G, and I) or Salmonella Typhimurium (B, D, F, H, and J). After sacrifice at different time points, blood was collected, and serum was obtained by centrifugation. The serum iron levels (A and B), transferrin saturation (Tf Sat) (C and D), total iron binding capacity (E and F), unsaturated iron binding capacity (G and H), and ferritin levels (I and J) were measured by a certified laboratory. Statistical analysis was performed by applying one-way ANOVA followed by a Bonferroni multiple-comparison post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with noninfected [NI] mice).
Both infections also impacted serum iron parameters (Fig. 3), causing decreases in levels of serum iron and transferrin saturation (Fig. 3A to D). However, kinetic differences were evident. While in L. monocytogenes-infected mice, a significant decrease (of approximately 40%) in serum iron levels was observed only 24 h after infection, which completely recovered at 72 h (Fig. 3A), mice infected with S. Typhimurium exhibited a decrease in serum iron levels of approximately 65% as early as 6 h after infection (Fig. 3B). In accordance, the percentage of saturated transferrin decreased more quickly and severely in S. Typhimurium-infected mice than in L. monocytogenes-infected mice (Fig. 3D and C, respectively). Interestingly, the total iron binding capacity (TIBC) and the unsaturated iron binding capacity (UIBC) increased during both infections at later time points (Fig. 3E to H), with a slight decrease in the case of Listeria at 96 h postinfection. Moreover, the levels of serum ferritin were significantly increased at 72 h in both models (Fig. 3I and J).
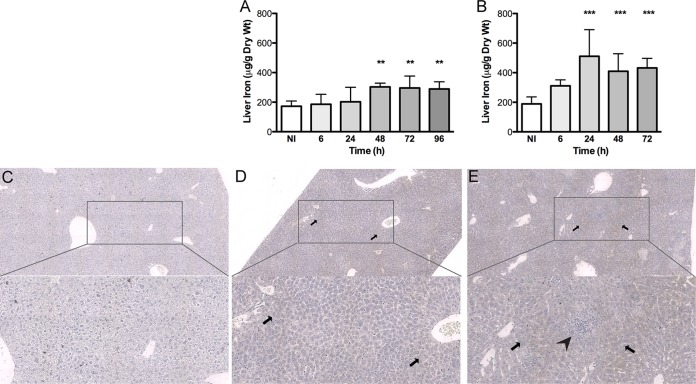
Given the observed decrease in serum iron levels, we decided to investigate the iron content of the liver, a central organ for both iron metabolism and iron storage. We observed a clear increase in tissue iron levels in mice infected with both types of bacteria (Fig. 4A and B), although the increase in tissue iron levels reached statistical significance earlier and iron loads were higher in S. Typhimurium-infected mice than in mice infected with L. monocytogenes. Iron levels in the liver of infected mice remained above control values until the end of the experimental protocol. In order to evaluate iron distribution in the liver, we performed Perl's-DAB (diaminobenzidine) staining in liver sections obtained from mice infected for 72 h. In control noninfected mice, iron was hardly seen (Fig. 4C), in contrast to infected animals (Fig. 4D and E). In Listeria-infected mice, no significant tissue damage was evident, and iron was found predominantly in the parenchyma perivascular regions (Fig. 4D). In the case of Salmonella-infected mice, centers of tissue necrosis could be seen, and iron seemed more evenly distributed (Fig. 4E). In both cases, iron was found exclusively inside hepatocytes (Fig. 4D and E).
FIG 4.
Iron quantification and distribution in the livers of C57BL/6 mice infected with L. monocytogenes or S. Typhimurium. (A and B) The amount of total tissue iron was quantified by using the bathophenanthroline method. (A) L. monocytogenes-infected mice; (B) S. Typhimurium-infected mice. Data represent means and standard deviations for at least 4 animals and are expressed as micrograms of iron per gram of dry liver. Statistical analysis was performed by applying one-way ANOVA followed by a Bonferroni multiple-comparison post hoc test. **, P < 0.01; ***, P < 0.001 (compared with noninfected [NI] mice). (C to E) The iron distribution in the liver was analyzed after Perl's-DAB staining at 72 h postinfection. Representative images of at least three animals per group, including images of noninfected animals (C), L. monocytogenes-infected animals (D), and S. Typhimurium-infected animals (E), were obtained with the D-Sight plus f2.0 analyzer at ×4 and ×10 magnifications. Iron deposition is shown in brown and indicated by the arrows. The arrowhead indicates an area of tissue necrosis.
Taken together, these results indicate that, despite differences in kinetics, both L. monocytogenes and S. Typhimurium have an impact on host iron metabolism, causing decreases in levels of serum iron and transferrin saturation, increases in hepatic iron levels, and decreases in erythrocyte counts.
Both L. monocytogenes and S. Typhimurium infections induce the release of proinflammatory cytokines.
In the case of L. monocytogenes infection, the kinetics of serum proinflammatory cytokines follows that of organ bacterial loads, with the highest levels of tumor necrosis factor (TNF) and IL-6 being found at 72 h postinfection and with levels decreasing thereafter (Fig. 5A and C). Interferon gamma (IFN-γ) is an exception, with the highest level being found at 24 h postinfection (Fig. 5E). In the case of S. Typhimurium, high levels of the three cytokines were found at 6 h postinfection (Fig. 5B, D, and F) (i.e., much earlier than in L. monocytogenes-infected mice). After transient decreases at 24 h postinfection, the levels of the three cytokines increased and tended to be higher in S. Typhimurium-infected than in L. monocytogenes-infected mice. Importantly, at 72 h postinfection, IL-6 levels (Fig. 5C and D) in the sera of S. Typhimurium-infected animals were significantly higher than those in the sera of L. monocytogenes-infected mice (2,907 ± 1,019 pg/ml versus 1,347 ± 532.9 pg/ml; P = 0.0204 by using a t test). Moreover, when we evaluated the liver mRNA expression levels of proinflammatory cytokines, we found 4-fold-higher expression levels of IL-1β and 20-fold-higher expression levels of TNF-α at 6 h postinfection in S. Typhimurium-infected than in L. monocytogenes-infected mice (not shown).
FIG 5.
Proinflammatory profile of mice infected with L. monocytogenes or S. Typhimurium. At the indicated time points after infection, blood of C57BL/6 mice infected with L. monocytogenes (A, C, and E) or S. Typhimurium (B, D, and F) was collected, and serum was obtained to measure TNF-α (A and B), IL-6 (C and D), and IFN-γ (E and F) levels by using a cytometric bead array mouse inflammation kit. Graphs represent means plus standard deviations of data from at least 4 animals. Statistical analysis was performed by applying one-way ANOVA followed by a Bonferroni multiple-comparison post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with noninfected mice).
L. monocytogenes and S. Typhimurium infections have distinct impacts on the liver expression of iron-related genes.
Given the alterations in serum iron levels and the expression levels of proinflammatory cytokines, namely, IL-6, we evaluated the expression of iron-related genes, including hepcidin, in the livers of infected mice. As a positive control for hepcidin induction, a group of C57BL/6 mice was injected with lipopolysaccharide (LPS) and sacrificed 8 h later. These mice exhibited on average a 26-fold increase in hepcidin expression levels relative to those in control mice.
As shown in Table 1, S. Typhimurium infection caused a significant downregulation of ferroportin and an upregulation of hemeoxygenase-1, besides a 6- to 11-fold increase in the expression levels of hepcidin, which remained significantly increased throughout the course of infection. Surprisingly, in the case of L. monocytogenes infection, hepcidin levels remained unaltered (Table 1), despite the clear alterations in the iron distribution described above. In order to try to identify additional players that could be causing iron redistribution in L. monocytogenes-infected animals, we measured the mRNA expression levels of other iron-related genes in the liver. As shown in Table 1, the expression levels of H- and L-ferritins, as well as that of transferrin, were not significantly altered by L. monocytogenes infection. Heme oxygenase 1 expression increased only at later time points, while transferrin receptor was downregulated at early time points. Interestingly, the expression of ferroportin was significantly affected by infection by L. monocytogenes. After an initial increase 6 h after infection, the expression level of ferroportin significantly decreased at 24 h and 48 h, recovering to basal levels at the last time points studied. As ferroportin may be regulated posttranscriptionally, we also evaluated the protein levels of this iron exporter in liver lysates by Western blotting. We confirmed that the levels of the ferroportin protein decreased during infection with L. monocytogenes (Fig. 6), which provides a possible explanation for the hepcidin-independent iron redistribution in the host.
TABLE 1.
Iron-related gene expression induced by infectiona
| Time (h) | Mean fold change in expression level of gene ± SD |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hepcidin |
Ferroportin 1 |
Heme oxygenase 1 |
Transferrin receptor |
Transferrin |
H-ferritin |
L-ferritin |
||||||||
| L. monocytogenes | S. Typhimurium | L. monocytogenes | S. Typhimurium | L. monocytogenes | S. Typhimurium | L. monocytogenes | S. Typhimurium | L. monocytogenes | S. Typhimurium | L. monocytogenes | S. Typhimurium | L. monocytogenes | S. Typhimurium | |
| 6 | 1.85 ± 0.65 | 6.05 ± 1.69** | 3.99 ± 2.03* | 0.06 ± 0.05*** | 0.73 ± 0.18 | 3.27 ± 1.45** | 0.28 ± 0.07 | 0.84 ± 0.07 | 1.63 ± 0.17** | 0.26 ± 0.05 | 0.94 ± 0.06 | 1.09 ± 0.44 | 1.20 ± 0.15 | 0.11 ± 0.05 |
| 24 | 1.00 ± 0.30 | 6.91 ± 1.16*** | 0.33 ± 0.33* | 1.19 ± 0.72 | 1.82 ± 0.73 | 3.84 ± 1.41*** | 0.27 ± 0.11* | 0.90 ± 0.44 | 1.29 ± 0.63 | 1.54 ± 0.74 | 0.68 ± 0.18 | 2.11 ± 1.57 | 0.70 ± 0.34 | 1.23 ± 1.04 |
| 48 | 1.12 ± 0.53 | 10.75 ± 0.78** | 0.36 ± 0.09* | 0.27 ± 0.11** | 2.99 ± 2.02* | 13.73 ± 4.52*** | 1.75 ± 0.68 | 0.63 ± 0.26 | 5.31 ± 3.36 | 2.06 ± 0.99 | 0.86 ± 0.23 | 1.04 ± 0.22 | 1.26 ± 0.67 | 1.14 ± 0.10 |
| 72 | 2.48 ± 1.16* | 11.19 ± 7.8** | 0.80 ± 0.34 | 0.26 ± 0.09** | 7.60 ± 3.33*** | 7.23 ± 0.98*** | 1.02 ± 0.2 | 0.49 ± 0.21* | 3.08 ± 2.95 | 2.19 ± 0.82* | 0.46 ± 0.20 | 3.93 ± 2.51* | 0.70 ± 0.16 | 0.69 ± 0.22* |
| 96 | 1.62 ± 0.87 | 0.62 ± 0.20 | 9.06 ± 8.54* | 0.67 ± 0.23 | 1.27 ± 0.9 | 0.65 ± 0.52 | 1.10 ± 0.46 | - | ||||||
C57BL/6 mice were infected with L. monocytogenes or S. Typhimurium or injected with an equivalent amount of saline. At the indicated time points after infection, mice were sacrificed, and mRNA levels of different genes in liver extracts were quantified by real-time PCR. Data shown are the means ± standard deviations of the fold increases in expression levels in the infected group versus the noninfected group, with at least 4 animals per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with the respective noninfected group).
FIG 6.

Listeria monocytogenes infection causes a decrease of FPN protein contents in liver lysates. Shown are data from Western blot analyses of liver lysates. Beta-actin was used as the housekeeping protein. The graph represents data from densitometry analysis of the FPN protein under each condition, normalized to beta-actin densitometry values and expressed as a percentage relative to values for noninfected mice. Data represent means plus standard deviations of data from at least 4 animals. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with noninfected mice). S, standard molecular mass markers.
Hepcidin upregulation during S. Typhimurium infection is mediated by TLR4 and proinflammatory cytokines.
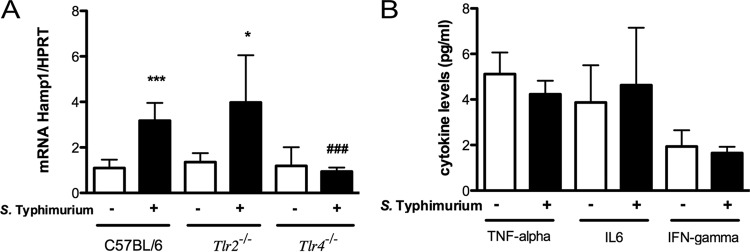
The increase in hepcidin expression levels during infection has been associated with the release of proinflammatory cytokines, namely, IL-6, by infected macrophages. The exuberant proinflammatory response observed in S. Typhimurium-infected mice is compatible with the stimulation of Toll-like receptors (TLRs) by this Gram-negative bacterium. To investigate whether hepcidin induction during S. Typhimurium infection was dependent on TLR activation, we used TLR2 and TLR4 knockout mice infected with this bacterium to evaluate hepcidin induction. While both wild-type and Tlr2−/− mice exhibited a 4-fold increase in hepcidin expression levels in the liver 24 h after infection with S. Typhimurium, no significant induction of this gene was observed in infected Tlr4−/− mice (Fig. 7A). Accordingly, no significant increases in levels of proinflammatory cytokines were seen in Tlr4−/− mice infected with S. Typhimurium (Fig. 7B). The liver bacterial loads were the same in wild-type and TLR4 knockout mice (data not shown). Overall, these data strongly suggest that the Gram-negative bacterium S. Typhimurium is recognized by TLR4 in the host, inducing the production of proinflammatory cytokines, which in turn activates the expression of hepcidin, impacting host iron metabolism.
FIG 7.
Inflammatory cytokines and hepcidin expression induced by S. Typhimurium in TLR-deficient mice. C57BL/6, Tlr2−/−, and Tlr4−/− mice were infected intravenously. (A) Twenty-four hours after infection, mice were sacrificed, and hepcidin mRNA levels in liver extracts were quantified by real-time PCR. (B) At the same time point after infection, blood was collected, and serum was obtained to measure TNF-α, IL-6, and IFN-γ levels by using a cytometric bead array mouse inflammation kit (B). The graphs represent means plus standard deviations of data from at least 4 animals per group. Statistical analysis was performed by using Student's t test. *, P < 0.05; ***, P < 0.001 (compared with the respective noninfected group). ###, P < 0.001 (compared with infected C57BL/6 mice).
These observations indicate that L. monocytogenes and S. Typhimurium impact host iron metabolism by different molecular mechanisms.
DISCUSSION
Iron, being an essential micronutrient, plays a central role in host-pathogen interactions. During evolution, pathogens developed different strategies to acquire iron from the host, contributing to their proliferation and virulence. Conversely, iron withholding is an important part of innate resistance to infection.
In this work, we used the mouse model of infection to show that two important pathogens, L. monocytogenes and S. Typhimurium, cause hypoferremia and mild anemia through different molecular pathways.
It has been known for a long time that hypoferremia and anemia accompany infectious diseases, especially those that evolve to a chronic state. With the discovery of hepcidin about 16 years ago, the mechanisms of infection-associated anemia were thought to be completely understood. Hepcidin is produced mostly in the liver in response to inflammatory stimuli and binds to ferroportin, blocking cellular iron efflux and consequently decreasing serum iron levels (9, 18). However, although several studies have confirmed the induction of hepcidin production and hypoferremia after the injection of microbial products such as LPS, fewer studies have analyzed the alterations in iron metabolism and hepcidin production in the context of in vivo infection with different pathogens.
In this work, we compared the impacts of infection with L. monocytogenes and S. Typhimurium in vivo on host iron metabolism using the mouse model. Our data clearly show that both infections affect host iron distribution and induce moderate anemia, as the hematocrit values decreased after a few days (Fig. 2C and D). Moreover, the levels of serum iron and saturated transferrin are also decreased during infection, indicating that the host responded by withholding iron from pathogens to prevent their growth (Fig. 3A to D). In time, this strategy resulted in iron retention in the liver (Fig. 4).
When we investigated whether hepcidin was induced by these infections, we found an increase in hepcidin mRNA levels only in the liver of S. Typhimurium-infected mice and not in L. monocytogenes-infected mice (Table 1). Previous works showed increased levels of hepcidin during mouse in vivo infections with Salmonella as well as with Pseudomonas aeruginosa, group A Streptococcus, Vibrio vulnificus, Candida albicans, or influenza A virus (9, 19, 20). Additionally, Salmonella was shown to induce the production of hepcidin by myeloid cells in vitro in a TLR4-dependent manner (20). Here, we show for the first time that this association between TLR4 activation and hepcidin production by Salmonella is relevant in vivo (Fig. 7). Of note, this hepcidin induction was observed in total liver lysates, highly suggesting that hepatocytes are its major source in vivo. This observation is also in agreement with data from previous studies showing the TLR4-dependent induction of hepcidin by LPS (21–23). As expected from this Gram-negative bacterium, along with the increased hepcidin mRNA levels, we observed high levels of proinflammatory cytokines, including IL-6, in the serum of S. Typhimurium-infected mice. The regulatory process that mediates hepcidin synthesis is known to rely on IL-6. IL-6 receptor activation is followed by JAK2-mediated phosphorylation of the signal transducer and activator of transcription STAT3, which stimulates hepcidin (18, 23–25). Recently, Estrogen-related receptor γ was added as an intermediate player in this signaling cascade (26, 27).
Notwithstanding these considerations, the predominant role of hepcidin in the development of hypoferremia of infection was recently challenged by several studies showing that TLR agonists can induce hypoferremia without a concomitant increase in hepcidin expression levels (28, 29). Given that our experiments showed that L. monocytogenes did not induce alterations in hepcidin levels, although promoting iron redistribution, we decided to evaluate the expression levels of other players involved in the iron metabolism of the host. A significant induction of the Hmox1 gene was observed for both infections although with differences in intensity and kinetics. We previously reported that Hmox1 is highly induced during mycobacterial infection and plays an important role in host protection against this type of pathogen (30). The induction of Hmox1 has also been reported for other infection models, namely, sepsis. The exact mechanisms involved in Hmox1 induction during infection are not known but may include an increased release of heme resulting from tissue damage and also the combined effects of oxidative stress and inflammatory signals (31). Particularly important for the understanding of iron metabolism deregulation was the observation that both L. monocytogenes and S. Typhimurium infections were associated with a decrease in ferroportin expression. Moreover, we saw that the decrease in ferroportin expression in the L. monocytogenes model could also be seen at the protein level. This alteration per se may explain the liver iron retention and the decrease in serum iron levels observed in infected mice. Our data are in apparent contrast with previously reported observations of an increase in ferroportin expression associated with Salmonella infection (32, 33). These discrepancies may be explained by two main reasons. One reason is the infection models used, since we infected mice by the intravenous route, and the authors of those studies used oral infections, with a much longer time course. Another important point is that the previous studies focused on the spleen and/or specifically on macrophages. In the present work, we focused our study on the liver, and the differences in mRNA levels that we saw are probably attributed to hepatocytes. Interestingly, while the manuscript was in preparation, a new study showing a marked hepcidin-independent downregulation of ferroportin, in both the liver and cultured macrophages, in response to Salmonella infection was reported (34). The exact mechanisms of ferroportin transcriptional regulation in different cell types are far from clear and deserve further evaluation in future studies, as different TLR agonists were shown to cause ferroportin downregulation by hepcidin-independent mechanisms (29, 35).
A deeper understanding of the mechanisms underlying the battle for iron between the pathogen and the host during infection will have important consequences for the treatment of infectious diseases. New treatment strategies can be envisioned, which can exploit iron-withholding mechanisms as an antimicrobial weapon. On the other hand, the specific inhibition of pathways leading to anemia will allow the prevention of this severe comorbidity. Our work contributes to this knowledge, showing that different pathogens impact host iron metabolism by different mechanisms, both hepcidin dependent and hepcidin independent.
MATERIALS AND METHODS
Chemicals.
All chemicals used in this work are of the highest analytical grade and were purchased from Sigma-Aldrich Co. (St. Louis, MO), unless specified otherwise.
Animals.
C57BL/6 and C57BL/6.TLR2 knockout (Tlr2−/−) mice were bred and housed at the Instituto de Investigação e Inovação em Saúde (i3S)/Instituto de Biologia Molecular e Celular (IBMC) animal facility. TLR4 knockout (Tlr4−/−) mice (B6.B10ScN-Tlr4lps-del/JthJ) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). The animals were kept inside individually ventilated cages bearing high-efficiency particulate air (HEPA) filters and were fed with sterilized food and water ad libitum. For experimental treatments, 12- to 16-week-old males were used. All animal experiments were carried out in compliance with the animal ethics guidelines of the institute and national and European regulations for the care and handling of laboratory animals. A.C.M., J.V.N., T.S., P.N.R., and M.S.G. are accredited by the Federation of European Laboratory Animal Science Associations (FELASA) for animal experimentation at category C. P.O. is accredited by FELASA for animal experimentation at the category B level.
Bacteria.
Listeria monocytogenes EGDe was generously provided by Didier Cabanes (molecular microbiology group of i3S/IBMC). L. monocytogenes was precultured in brain heart infusion (BHI) broth medium (BD Biosciences, San Jose, CA, USA) overnight at 37°C, after which the bacterial suspension was diluted in BHI medium and incubated to mid-log phase. Bacteria were then harvested by centrifugation and washed with ice-cold phosphate-buffered saline (PBS). Salmonella enterica serovar Typhimurium strain ATCC 14028 was kindly provided by Luísa Peixe (Faculdade de Farmácia da Universidade do Porto [FFUP]). S. Typhimurium was grown to mid-log phase in tryptic soy broth (TSB) medium (Conda, Torrejón de Ardoz, Madrid, Spain) for 6 h at 37°C, after which bacteria were harvested by centrifugation and washed with PBS. Aliquots in PBS were kept at −80°C until use.
Mouse infection and sacrifice.
Mice were intravenously infected in the lateral tail vein with 1 × 104 CFU of each type. Control animals were injected with the same volume of PBS by the same route. At different time points, animals were anesthetized with isoflurane (B. Braun Medical, Portugal) and sacrificed by cervical dislocation. At each time point, livers were harvested for different analyses, including bacterial load quantification, gene expression evaluation, nonheme iron determination, histological alterations, and examination of the iron distribution in tissue. Liver samples collected for gene expression and iron analyses were quickly frozen in liquid nitrogen and stored at −80°C until use.
Hematological and serum iron parameters.
Blood samples were collected by retro-orbital puncture under anesthesia, and 150 μl was transferred to EDTA tubes (Vacutainer; BD, NJ, USA) to evaluate the erythron. Serum was obtained by high-speed centrifugation of the remaining blood. Blood and serum parameters were determined by a certified laboratory in a blind manner (CoreLab, Centro Hospitalar do Porto, Porto, Portugal).
Quantification of bacterial loads in organs.
Tissues were aseptically collected, weighed, and homogenized in sterile PBS. Serial dilutions were performed, and dilutions were plated in duplicate onto BHI agar medium and Salmonella Shigella agar medium for L. monocytogenes and S. Typhimurium, respectively. After incubation at 37°C for 24 h, the colonies were counted. The number of CFU per liver was calculated, taking into consideration the dilution at which colonies were counted, the total organ weight, and the volume of the organ homogenate used for plating.
Liver iron content.
Liver iron levels were determined by the bathophenanthroline method (36). Briefly, 100 to 180 mg of fresh liver tissue was placed into iron-free Teflon vessels (ACV-Advanced Composite Vessel; CEM Corporation, Matthews, NC, USA) and dried in a microwave oven (MDS 2000; CEM Corporation), after which the tissue was weighed. Digestion was performed for 20 h at 65°C with a mixture of 10% trichloroacetic acid–30% hydrochloric acid. After cooling to room temperature, samples were mixed with a chromogen reagent solution (5 volumes of deionized water, 5 volumes of saturated sodium acetate, and 1 volume of 0.1% bathophenanthroline sulfonate–1% thioglycolic acid). The absorbance was read at 535 nm. The extinction coefficient for bathophenanthroline is 22.14 mM−1 cm−1 and was used to calculate the amount of iron that reacted with this compound.
Perl's-DAB staining.
Liver samples were fixed in a 10% neutral buffered formalin solution and included in paraffin blocks prior to Perl's blue staining. Ferric iron was detected by Perl's-enhanced DAB staining. Briefly, paraffin sections were deparaffinized and processed through downgraded alcohols, rehydrated, and incubated in a 2% potassium ferrocyanide trihydrate–2% HCl solution for 30 min. Sections were rinsed five times in water and incubated in a mixture containing 10 mg DAB–40 ml 0.01 M Tris HCl (pH 7.4)–160 μl 30% H2O2 for 1 min. Sections were rinsed twice with water, dehydrated (reverse of downgraded alcohols and deparaffinization), and coverslipped in Entellan. Sections were imaged in D-Sight Plus f2.0 and analyzed with D-Sight viewer (both from A. Menarini Diagnostics).
Gene expression.
Liver samples were collected, frozen in liquid nitrogen, and stored at −80°C prior to analysis. According to the manufacturer's instructions, RNA extraction from mouse liver was performed with the PureLink RNA minikit (Life Technologies, Carlsbad, CA, USA), and for real-time PCR quantification, total RNA was transcribed into cDNA by using an NZY First-Strand cDNA synthesis kit (NZYTech, Lisbon, Portugal). For the amplification of each gene of interest, a corresponding specific pair of primers (STAB Vida, Lisbon, Portugal) was used. All reactions were performed in a 20-μl total volume with SYBR green PCR master mix (Bio-Rad Laboratories, Hercules, CA, USA) and carried out with an iQ5 instrument (Bio-Rad Laboratories, Hercules, CA, USA). Baseline thresholds were obtained by using the Bio-Rad iQ5 program and the threshold cycles (CTs) were calculated by the 2CT method, where CT values for the genes of interest were normalized to the level of the hypoxanthine guanine phosphoribosyl transferase housekeeping gene (hprt1). Data are shown as n-fold differences relative to values for noninfected samples, calculated with the 2−ΔΔCT method. Primers used were as follows: hprt forward primer 5′-GGTGGAGATGATCTCTCAAC-3′, hprt reverse primer 5′-TCATTATAGTCAAGGGCATATCC-3′, hamp1 forward primer 5′-CCTATCTCCATCAACAGATG-3′, hamp1 reverse primer 5′-AACAGATACCACACTGGGAA-3′, transferrin forward primer 5′-ACCTGGAACAACCTGAAAGG-3′, transferrin reverse primer 5′-GGCCAATACACAGGTCACAG-3′, h-ferritin forward primer 5′-GCTGAATGCAATGGAGTGTGCA-3′, h-ferritin reverse primer 5′-GGCACCCATCTTGCGTAAGTTG-3′, l-ferritin forward primer 5′-ACCTACCTCTCTCTGGGCTT-3′, l-ferritin reverse primer 5′-TGGCTTCTGCACATCCTGGA-3′, fpn forward primer 5′-TTGGTGACTGGGTGGATAAGAATGC-3′, fpn reverse primer 5′-CGCAGAGGATGACGGACACATTC-3′, transferrin receptor forward primer 5′-GCAGCATTGGTCAAAACATGG-3′, transferrin receptor reverse primer 5′-GCTTTGGGCATTTGCAACCC-3′, Hmox-1 forward primer 5′-GCCACCAAGGAGGTACACAT-3′, and Hmox-1 reverse primer 5′-GCTTGTTGCCCTCTATCTCC-3′.
Cytokine profile analysis.
Analysis of cytokine levels was performed by using the BD cytometric bead array (CBA) mouse inflammation kit (catalog no. 552364; BD Biosciences, San Jose, CA, USA), which allows the simultaneous measurement of levels of TNF-α, IL-6, and IFN-γ, among others. Serum samples were mixed with PE detection reagent. The mixture was incubated for 2 h at room temperature (RT) in the dark. The samples were washed, the supernatant was discarded, and the beads were resuspended in washing buffer. Samples were then analyzed by using the BD FACSCanto II Bioanalyzer and FCAP Array software (BD Biosciences, San Jose, CA, USA) according to the manufacturer's instructions.
Western blotting.
Liver extracts were prepared in Laemmli buffer (Bio-Rad, Hercules, CA, USA). Equivalent amounts of protein (30 μg) were separated by electrophoresis in 10% SDS-polyacrylamide gels and electrophoretically transferred onto a polyvinylidene difluoride (PVDF) membrane for 90 min at 100 V. After the membrane was blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline–Tween (TBST) (50 mM Tris-HCl [pH 8], 154 mM NaCl, 0.1% Tween 20) for 2 h at RT, membranes were incubated with the following primary antibodies according to the manufacturers' instructions: rabbit anti-FPN (ferroportin/SLC40A1 antibody, catalog no. NPB1-21502; Novus, Littleton, CO, USA) (1:300) for 1 h at RT and rabbit anti-beta-actin (catalog no. 8227; Abcam, Cambridge, UK) (1:5,000) overnight at 4°C. Membranes were washed and incubated with the secondary anti-rabbit antibody (1:10,000) in 1% BSA in TBST. Membranes were then incubated with the Luminata Crescendo Western horseradish peroxidase (HRP) substrate, imaged with the ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA), and analyzed with ImageLab software (Bio-Rad, Hercules, CA, USA).
Statistical analysis.
Data analysis was performed by using the GraphPad Prism 5.0 program for Macintosh (GraphPad Software, Inc., La Jolla, CA, USA), and data were expressed as means plus standard deviations of the numbers of samples indicated in the legends of the figures. Multiple comparisons were performed by using one-way analysis of variance (ANOVA) followed by a Bonferroni multiple-comparison post hoc test. Comparisons between two groups were performed by using Student's t test. Significance was accepted when the P value was <0.05.
ACKNOWLEDGMENTS
This work is a result of the project “Norte-01-0145-FEDER-000012-Structured program on bioengineered therapies for infectious diseases and tissue regeneration,” supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER). It was also supported by project “NORTE-07-0124-FEDER-000002-Host-Pathogen Interactions” cofunded by Programa Operacional Regional do Norte (ON.2—O Novo Norte), under the Quadro de Referência Estratégico Nacional (QREN), through the Fundo Europeu de Desenvolvimento Regional (FEDER) and by FCT (Fundação para a Ciência e Tecnologia). A.C.M., J.V.N., and T.S. were recipients of SFRH/BPD/101405/2014, SFRH/BPD/86380/2012, and SFRH/BD/77564/2011 fellowships, respectively, attributed by the FCT.
The funders had no role in study design, data collection and interpretation, or the decision to submit work for publication.
Listeria monocytogenes EGDe was generously provided by Didier Cabanes (i3S/IBMC). We thank Rita Pombinho (i3S/IBMC) for the help in the manipulation of Listeria monocytogenes. Salmonella Typhimurium 14028 was kindly given by Luísa Peixe (Faculdade de Farmácia da Universidade do Porto). We thank Graça Henriques from CoreLab, Centro Hospitalar do Porto, Porto, Portugal; Gonçalo Mesquita for technical assistance; and the following i3S scientific services: Cell Culture and Genotyping (CCGen), Histology and Electron Microscopy Services (HEMS), and the animal facility.
M.S.G. and P.N.R. designed the experiments; A.C.M., J.V.N., T.S., and P.O. performed the experiments; A.C.M., M.S.G., and P.N.R. wrote the manuscript; and all authors revised the manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Wang J, Pantopoulos K. 2011. Regulation of cellular iron metabolism. Biochem J 434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. 2010. Two to tango: regulation of mammalian iron metabolism. Cell 142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Park CH, Valore EV, Waring AJ, Ganz T. 2001. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 4.Ward DM, Kaplan J. 2012. Ferroportin-mediated iron transport: expression and regulation. Biochim Biophys Acta 1823:1426–1433. doi: 10.1016/j.bbamcr.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nairz M, Haschka D, Demetz E, Weiss G. 2014. Iron at the interface of immunity and infection. Front Pharmacol 5:152. doi: 10.3389/fphar.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. 2004. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 7.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 8.Wrighting DM, Andrews NC. 2006. Interleukin-6 induces hepcidin expression through STAT3. Blood 108:3204–3209. doi: 10.1182/blood-2006-06-027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arezes J, Jung G, Gabayan V, Valore E, Ruchala P, Gulig PA, Ganz T, Nemeth E, Bulut Y. 2015. Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe 17:47–57. doi: 10.1016/j.chom.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns M, Muthupalani S, Ge Z, Wang TC, Bakthavatchalu V, Cunningham C, Ennis K, Georgieff M, Fox JG. 2015. Helicobacter pylori infection induces anemia, depletes serum iron storage, and alters local iron-related and adult brain gene expression in male INS-GAS mice. PLoS One 10:e0142630. doi: 10.1371/journal.pone.0142630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michels K, Nemeth E, Ganz T, Mehrad B. 2015. Hepcidin and host defense against infectious diseases. PLoS Pathog 11:e1004998. doi: 10.1371/journal.ppat.1004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues PN, Gomes SS, Neves JV, Gomes-Pereira S, Correia-Neves M, Nunes-Alves C, Stolte J, Sanchez M, Appelberg R, Muckenthaler MU, Gomes MS. 2011. Mycobacteria-induced anaemia revisited: a molecular approach reveals the involvement of NRAMP1 and lipocalin-2, but not of hepcidin. Immunobiology 216:1127–1134. doi: 10.1016/j.imbio.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Allerberger F, Wagner M. 2010. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 14.Ooi ST, Lorber B. 2005. Gastroenteritis due to Listeria monocytogenes. Clin Infect Dis 40:1327–1332. doi: 10.1086/429324. [DOI] [PubMed] [Google Scholar]
- 15.Kortman GA, Mulder ML, Richters TJ, Shanmugam NK, Trebicka E, Boekhorst J, Timmerman HM, Roelofs R, Wiegerinck ET, Laarakkers CM, Swinkels DW, Bolhuis A, Cherayil BJ, Tjalsma H. 2015. Low dietary iron intake restrains the intestinal inflammatory response and pathology of enteric infection by food-borne bacterial pathogens. Eur J Immunol 45:2553–2567. doi: 10.1002/eji.201545642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma S, Srikanth CV. 2015. Understanding the complexities of Salmonella-host crosstalk as revealed by in vivo model organisms. IUBMB Life 67:482–497. doi: 10.1002/iub.1393. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez R, Jung CL, Gabayan V, Deng JC, Ganz T, Nemeth E, Bulut Y. 2014. Hepcidin induction by pathogens and pathogen-derived molecules is strongly dependent on interleukin-6. Infect Immun 82:745–752. doi: 10.1128/IAI.00983-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armitage AE, Eddowes LA, Gileadi U, Cole S, Spottiswoode N, Selvakumar TA, Ho LP, Townsend AR, Drakesmith H. 2011. Hepcidin regulation by innate immune and infectious stimuli. Blood 118:4129–4139. doi: 10.1182/blood-2011-04-351957. [DOI] [PubMed] [Google Scholar]
- 20.Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V. 2006. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood 107:3727–3732. doi: 10.1182/blood-2005-06-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. 2001. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Du F, Qian ZM, Ge XH, Zhu L, Yung WH, Yang L, Ke Y. 2008. Lipopolysaccharide induces a significant increase in expression of iron regulatory hormone hepcidin in the cortex and substantia nigra in rat brain. Endocrinology 149:3920–3925. doi: 10.1210/en.2007-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanamori Y, Sugiyama M, Hashimoto O, Murakami M, Matsui T, Funaba M. 2016. Regulation of hepcidin expression by inflammation-induced activin B. Sci Rep 6:38702. doi: 10.1038/srep38702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganz T. 2011. Hepcidin and iron regulation, 10 years later. Blood 117:4425–4433. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meynard D, Babitt JL, Lin HY. 2014. The liver: conductor of systemic iron balance. Blood 123:168–176. doi: 10.1182/blood-2013-06-427757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DK, Jeong JH, Lee JM, Kim KS, Park SH, Kim YD, Koh M, Shin M, Jung YS, Kim HS, Lee TH, Oh BC, Kim JI, Park HT, Jeong WI, Lee CH, Park SB, Min JJ, Jung SI, Choi SY, Choy HE, Choi HS. 2014. Inverse agonist of estrogen-related receptor gamma controls Salmonella Typhimurium infection by modulating host iron homeostasis. Nat Med 20:419–424. doi: 10.1038/nm.3483. [DOI] [PubMed] [Google Scholar]
- 27.Misra J, Kim DK, Choi HS. 2017. ERRgamma: a junior orphan with a senior role in metabolism. Trends Endocrinol Metab 28:261–272. doi: 10.1016/j.tem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Deschemin JC, Vaulont S. 2013. Role of hepcidin in the setting of hypoferremia during acute inflammation. PLoS One 8:e61050. doi: 10.1371/journal.pone.0061050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guida C, Altamura S, Klein FA, Galy B, Boutros M, Ulmer AJ, Hentze MW, Muckenthaler MU. 2015. A novel inflammatory pathway mediating rapid hepcidin-independent hypoferremia. Blood 125:2265–2275. doi: 10.1182/blood-2014-08-595256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva-Gomes S, Appelberg R, Larsen R, Soares MP, Gomes MS. 2013. Heme catabolism by heme oxygenase-1 confers host resistance to Mycobacterium infection. Infect Immun 81:2536–2545. doi: 10.1128/IAI.00251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wegiel B, Hauser CJ, Otterbein LE. 2015. Heme as a danger molecule in pathogen recognition. Free Radic Biol Med 89:651–661. doi: 10.1016/j.freeradbiomed.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Nairz M, Schleicher U, Schroll A, Sonnweber T, Theurl I, Ludwiczek S, Talasz H, Brandacher G, Moser PL, Muckenthaler MU, Fang FC, Bogdan C, Weiss G. 2013. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J Exp Med 210:855–873. doi: 10.1084/jem.20121946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown DE, Nick HJ, McCoy MW, Moreland SM, Stepanek AM, Benik R, O'Connell KE, Pilonieta MC, Nagy TA, Detweiler CS. 2015. Increased ferroportin-1 expression and rapid splenic iron loss occur with anemia caused by Salmonella enterica serovar Typhimurium infection in mice. Infect Immun 83:2290–2299. doi: 10.1128/IAI.02863-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willemetz A, Beatty S, Richer E, Rubio A, Auriac A, Milkereit RJ, Thibaudeau O, Vaulont S, Malo D, Canonne-Hergaux F. 2017. Iron- and hepcidin-independent downregulation of the iron exporter ferroportin in macrophages during Salmonella infection. Front Immunol 8:498. doi: 10.3389/fimmu.2017.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu XB, Nguyen NB, Marquess KD, Yang F, Haile DJ. 2005. Regulation of hepcidin and ferroportin expression by lipopolysaccharide in splenic macrophages. Blood Cells Mol Dis 35:47–56. doi: 10.1016/j.bcmd.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Torrance JD, Bothwell TH. 1980. Tissue iron stores, p 104–109. In Cook JD. (ed), Methods in hematology, vol 1 Churchill Livingstone Press, New York, NY. [Google Scholar]