Abstract
Neuroinflammatory processes are a central feature of Alzheimer’s disease (AD) in which microglia are over-activated, resulting in the increased production of pro-inflammatory cytokines. Moreover, deficiencies in the anti-inflammatory system may also contribute to neuroinflammation. Recently, advanced methods for the analysis of genetic polymorphisms have further supported the relationship between neuroinflammatory factors and AD risk because a series of polymorphisms in inflammation-related genes have been shown to be associated with AD. In this review, we summarize the polymorphisms of both pro- and anti-inflammatory cytokines related to AD, primarily interleukin-1 (IL-1), IL-6, tumor necrosis factor alpha, IL-4, IL-10, and transforming growth factor beta, as well as their functional activity in AD pathology. Exploration of the relationship between inflammatory cytokine polymorphisms and AD risk may facilitate our understanding of AD pathogenesis and contribute to improved treatment strategies.
Keywords: Alzheimer’s disease, Neuroinflammation, Cytokine, Genetic polymorphism
Introduction
Alzheimer’s disease (AD) is a common neurodegenerative disease characterized by progressive declines in cognitive and functional abilities. Amyloid-beta (Aβ) plaques and neurofibrillary tangles (NFTs) are its main pathological hallmarks. Aβ aggregates seem to initiate the pathogenesis of AD, while NFTs may be more involved in its progression [1]. However, the exact mechanisms by which AD occurs and develops remain ill-defined, and effective methods to cure the disease or halt the associated cognitive decline remain undiscovered. Nevertheless, neuroinflammation and oxidative stress have received increasing attention as accumulating evidence suggests their involvement in its development [2, 3]. Activation of microglia, the brain-specific macrophages, has been reported in both AD patients and animal models [4], accompanied by an activated complement system [5] and increased levels of chemokines and cytokines [6, 7]. In addition, the protective effects of non-steroidal anti-inflammatory drugs against AD risk [8] further support the neuroinflammation hypothesis of AD.
Moreover, AD is a multifactorial disease with a hereditary component, and advanced technologies for the analysis of genetic polymorphisms have identified numerous genetic loci that affect AD. Recent genome-wide association studies (GWASs) strongly support the interaction of inflammation and AD because several inflammation-related genes, such as CR1 (complement receptor 1), MS4A (membrane-spanning 4-domains subfamily A), TREM2 (triggering receptor expressed on myeloid cells 2), and CD33 [9], have been identified as AD risk modifiers in Caucasians. The association has been further supported in other populations. For instance, variants of immune-related genes, including complement factor H [10], MS4A [11], CR1 [12], and CD33 [13] are associated with AD susceptibility in Han Chinese. Moreover, based on the findings that the levels of cytokines are altered in AD patients and that cytokines are key components of neuroinflammation, numerous case-control studies have explored the association between AD risk and genetic polymorphisms of inflammatory cytokines, primarily interleukin-1 (IL-1), IL-4, IL-6, IL-10, tumor necrosis factor alpha (TNFα), and transforming growth factor beta (TGFβ). However, the results have been inconsistent. Thus, a series of meta-analyses was conducted, and conclusions with high reliability were obtained. However, because genetic polymorphism analyses are increasingly used in different populations, the data in this field are continually updated.
Accordingly, in the current review, we comprehensively summarize the key genetic polymorphisms of inflammatory cytokines associated with AD, including the latest evidence (summarized in Table 1). In addition, progress in the exploration of the functional activity of these polymorphisms and the mechanisms by which they modify AD susceptibility are illustrated.
Table 1.
Genetic polymorphisms of cytokines associated with AD.
| Gene | Variant | rs number | Functional consequence | Association with AD | Ethnicity | Key ref. |
|---|---|---|---|---|---|---|
| IL1A | −889 C/T | rs1800587 | 5′ UTR variant | Increased risk | Caucasian | [14, 15] |
| Increased microglial activity | Caucasian | [16] | ||||
| IL1B | −511C/T | rs16944 | Upstream variant | Decreased risk | Non-European | [17] |
| Possible increased risk | Caucasian | [18] | ||||
| Increased Aβ42 in CSF | German | [19] | ||||
| Decreased plasma homocysteine | Italian | [20] | ||||
| +3953 C/T | rs1143634 | Synonymous codon | Increased risk | Caucasian | [18] | |
| Earlier onset | Italian | [21] | ||||
| Delayed onset Reduced NP Reduced NFT |
American Caucasian | [22] | ||||
| −31 T/C | rs1143627 | Upstream variant | Decreased risk | Italian | [23] | |
| IL-6 | −174 G/C | rs1800795 | Intron variant | Reduced risk | Asian, Caucasian | [23–25] |
| Increased IL-6 in blood and brain | Italian | [26] | ||||
| −572 C/G | rs1800796 | Intron variant | Reduced risk | Han Chinese | [27, 28] | |
| TNFA | −850 C/T | rs1799724 | Upstream variant | Increased risk | Australian Caucasian, European | [29] |
| −308 G/A | rs1800629 | Upstream variant | Increased risk | East Asian | [30] | |
| Earlier onset | Italian | [31] | ||||
| VEGF | −2578 C/A | rs699947 | Upstream variant | Increased risk in APOE ε4 (-) | Caucasian, Asian | [32] |
| −1154 G/A | rs1570360 | Upstream variant | Decreased risk in APOE ε4 (-) | Caucasian, Asian | [32] | |
| IL-18 | −607 C/A | rs1946518 | Upstream variant | Decreased risk | Han Chinese, Italian Caucasian | [33–35] |
| Decreased IL-18 production | Italian Caucasian | [35] | ||||
| −137 G/C | rs187238 | Upstream variant | Decreased risk | Han Chinese | [34] | |
| Faster cognitive decline | Italian Caucasia | [33] | ||||
| IL-33 | rs7044343 | rs7044343 | Intron variant | Decreased risk Decreased CAA |
Caucasian | [36] |
| rs1157505 | rs1157505 | Intron variant | ||||
| rs11792633 | rs11792633 | Intron variant | ||||
| IL-12A | rs568408 | rs568408 | Intron variant | Decreased risk | Han Chinese | [37] |
| IL-12B | rs3212227 | rs3212227 | 3′ UTR variant | Decreased risk | Han Chinese | [37] |
| IL-4 | −590 C/T | rs2243250 | Upstream variant | Decreased risk | Caucasian, Han Chinese | [38, 39] |
| −1098 T/G | rs2243248 | Upstream variant | Decreased risk | Caucasian | [38] | |
| Increased risk | Han Chinese | [39] | ||||
| IL-10 | −1082G/A | rs1800896 | Intron variant | Increased risk | European | [40, 41] |
| Decreased risk | Brazilian | [42] | ||||
| TGFB1 | +10 T/C | rs1800470 | Missense mutation | Increased risk | Caucasian | [43, 44] |
| Decreased CAA Increased neocortical plaques |
Japanese-American, Japanese | [45, 46] | ||||
| −509 C/T | rs1800469 | Upstream variant | Increased risk | Caucasian | [47] |
CAA cerebral amyloid angiopathy; NFT neurofibrillary tangle; NP neuritic amyloid plaque
Pro-inflammatory Cytokines
IL-1
Increased expression of IL-1 has been reported in AD brains [48]. Subsequent investigations suggest that the overexpressed IL-1 restrains the function of cholinergic systems [49] and favors the formation of Aβ plaques and accumulation of NFTs [50, 51], supporting the important role of IL-1 in AD development. This concept has been further supported by gene association analysis, as several variants located in the genes of IL-1A and IL-1B have been found to influence AD risk.
The relationship between AD risk and the IL1A −889 C/T polymorphism has been tested in numerous experiments that include different ethnic groups from different regions, and the results vary because a polymorphism in the 5’ regulatory region was shown to either increase the AD risk [52, 53] or failed to influence it [54, 55]. Two recent meta-analyses have consistently found significant associations in Caucasians but not in Asians [14, 15]. Hayes and colleagues showed that microglial activation is greater in the brains of AD patients with the T allele, particularly those with the apolipoprotein E (APOE) ε4 allele [16]. Moreover, the functional significance of this polymorphism may be attributable to its promotional effect on IL1A gene transcription and IL-1α protein synthesis, as has been shown in both a human pancreatic tumor cell line and an astrocyte cell line [56, 57]. Overall, the T allele of IL1A −889 C/T diminishes AD progression via the promotion of microglial activation and an increase of IL1A gene transcription, causing overexpression of pro-inflammatory cytokines, which can facilitate neuroinflammatory processes and AD pathology. However, the causes of the differing results in Caucasians and Asians remain unclear. Furthermore and paradoxically, patients who are homozygous for the C allele exhibit an accelerated rate of cognitive decline [58]. Therefore, further work, especially in vivo investigations, are needed to explore the mechanisms by which this polymorphism influences AD-related phenotypes.
Several polymorphisms of the IL1B gene have been suggested to increase AD risk [59, 60], while negative associations are also frequently reported. Meta-analysis by Di Bona et al. [18] revealed that the IL1B +3953 TT genotype is associated with an increased AD risk and that the IL1B −511 TT genotype might be a risk factor for AD only in Caucasians, supporting an association between IL1B genetic polymorphisms and AD. However, according to the latest meta-analysis, the IL1B −511 single nucleotide polymorphism (SNP) has no significant relationship with overall AD risk while the CC genotype is a risk factor for AD in non-Europeans [17]. The inconsistency concerning IL1B −511 partially results from the inclusion criteria that involve subgroup analysis, because a significant effect could be obtained only when the investigators analyzed the four studies of Caucasians with the highest statistical power [18]. Thus, studies with larger sample sizes are needed to confirm or refute these conclusions. The significance of the IL1B −511 SNP is under investigation, and the findings that monocytes from healthy carriers of the T allele and lymphocytes from patients with epilepsy carrying the T allele have a slight but non-significant elevated capacity to produce IL-1β in vitro [61, 62], indicating that the SNP in the promoter region may have a promotional effect on IL-1β production. In addition, IL1B −511 may influence IL-1β in coordination with other genetic loci, such as IL1B +3953, as suggested by Santtila et al [62]. Furthermore, the findings that the IL1B −511 T allele is related to both increased Aβ42 in the cerebrospinal fluid (CSF) and decreased homocysteine in the plasma suggest that this SNP is associated with AD risk factors other than neuroinflammation [19, 20]. Regarding IL1B +3953, AD patients carrying the T allele have a significantly earlier onset, particularly those who carry the IL1A −889 TT genotype [21]. However, the T allele is also associated with a delayed age at AD onset in patients without the APOE ε4 allele and is accompanied by reduced amyloid plaques and NFTs [22]. These findings suggest that gene-gene interactions are involved in AD progression, and analyses of haplotypes should be considered in further studies. Similarly, Payão et al. suggested that the haplotypes of IL1B −511C, IL1B −31T, and an IL1 receptor antagonist (VNTR2) have protective effects against AD [63]. Moreover, the IL1B −31 TT genotype seems to be a risk factor for AD in Caucasians [23], but fails to influence the risk in the Chinese population [64]; therefore, additional studies with larger populations are needed to confirm or deny this association.
IL-6
Associations between AD and IL-6 have also been identified, as AD patients show increased IL-6 expression in both the periphery [65] and the CNS [66]. An in vitro study suggested that overexpressed IL-6 influences the cdk5/p53 pathway to induce the phosphorylation of tau, increasing the AD risk [67]. Regarding genetic polymorphisms of the IL-6 gene, the protective role of IL-6 −174 in AD development has been highlighted [68, 69]. However, inconsistent results have also been reported [70]. Several meta-analyses have suggested that the CC genotype is likely a protective factor for AD [24, 71]. Moreover, the latest study involving 1,246 individuals in Italy revealed that the frequency of the GG genotype in AD patients is significantly higher than that in controls [23]. This finding further supports a protective effect of the −174 C allele on AD risk. Based on findings that the G allele is associated with both increased IL-6 secretion in vitro and increased levels of plasma IL-6 [72], it has been assumed that the SNP in the IL-6 gene promoter region may modify the risk of AD by influencing IL-6 protein release in the brain, playing an important role in the neuroinflammatory cascade and the AD process. However, the association between this genotype and IL-6 plasma level has been queried, and negative results have been reported [73]. Furthermore, a study by Licastro and colleagues provided results that further confuse the association because they reported increased levels of IL-6 in the blood and brain of AD patients with the CC genotype. Similarly, the same study showed that the C allele also increased AD risk [26]. Very recently, this SNP was shown to influence the CSF level of clusterin protein, which is significantly associated with AD risk and the tau/Aβ ratio in CSF [74]. These contradictory results may arise from the different genetic backgrounds, but they also strongly imply multifaceted roles of IL-6 −174 in AD. The G allele of the IL-6 −572 C/G polymorphism also has protective effects against AD risk in Han Chinese populations [27, 28], whereas contrary results have been reported in other populations [65, 75, 76]. Moreover, protective effects of the IL-6 −174G and IL-6 −597A haplotypes have been found [76], further suggesting gene-gene interactions and the complicated effects of genetic polymorphisms on AD.
TNFA
The role of TNF-α seems to be multifunctional. In animal models of AD, increased TNF-α is a key element in inflammatory cascade and increases the Aβ and tau pathology [77]. Moreover, short-term anti-TNF-α treatment improves cognition in AD patients [78], probably by relieving the Aβ pathology [79]. However, neuroprotective roles of TNF-α have also been reported, as long-term and non-specific inhibition of TNF-α signaling worsens the AD-related pathology in the brains of AD transgenic mice [80]. Correspondingly, TNF-α mediates the microglial phagocytosis of Aβ [80]. Two types of TNF-α receptors (TNF-RI and TNF-RII) have been identified, and they activate different downstream pathways to mediate distinct biological effects. In AD brains, TNF-RI is over-activated, while TNF-RII is inhibited [81]. Furthermore, activation of TNF-RI promotes the deposition of Aβ and Aβ-induced neuronal death [82, 83], while TNF-RII seems to counteract the TNF-RI-mediated injury [84]. The interactions between the two receptors, at least partially, lead to the diverse effects of TNF-α on AD. Concerning genetic polymorphisms, several SNPs have been reported to influence the risk of AD. A meta-analysis conducted in 2009 by De Bona et al. [29] supported the −850 T allele as a risk factor for AD susceptibility, particularly in individuals who carry the APOE ε4 allele. Although no further studies on this SNP and AD risk have been published, the conclusion is credible because no evidence of heterogeneity between studies was found. However, as noted by Bona et al., larger samples are needed to confirm the association because only five medium-sized studies have been performed thus far. Correlations of AD risk with other TNFA SNPs (−238, −308, −863, and −1031) have also been included in meta-analyses, and negative results have been reported [29]; however, significant between-study heterogeneities may have influenced the reliability of these conclusions. Subsequently, several studies concerning the association between the TNFA −308 G/A polymorphism and AD risk involving different ethnic groups have been performed. Negative results were obtained in the majority of these investigations [23, 38, 85], strongly suggesting that the TNFA −308 polymorphism alone does not significantly influence AD susceptibility. As reported by Vural et al. [86], the AA haplotype TNF –308 and IL-10 –1082 and the AC haplotype TNFA –308 and IL-6 –174 significantly increase the risk of AD, suggesting that TNFA –308 exerts its function via gene-gene interaction. However, inconsistent results have also been reported [87–89], and the latest meta-analysis by Lee et al. [30] implied that the TNFA −308 polymorphism may influence the AD risk in certain ethnic groups. Moreover, Lio et al. [31] reported an association of TNFA −308 A with the earlier onset of AD among Italians. Thus, TNF −308 G/A might be a disease-modifying polymorphism in AD patients of certain ethnic groups or in coordination with other polymorphisms; however, this interaction needs further examination. Little progress has been made on the interactions between AD risk and the other three SNPs of the TNF gene noted above, with the exception of a negative result reported for the TNFA −863 polymorphism in an Azeri Turk population [88]. Therefore, the TNFA −308 and −850 genetic polymorphisms may be associated with AD susceptibility, and they have been hypothesized to act by influencing TNFA gene transcription. Some in vitro experiments have provided support for this hypothesis [90], but no direct evidence is available [91].
Other Pro-inflammatory Cytokines
Changes in the expression of vascular endothelial growth factor (VEGF) have been reported in AD patients; a cross-sectional study showed that it significantly increased in the early stage of AD while decreasing as the disease progressed [92]. In AD brains, VEGF immunoreactivity is mainly located in reactive astrocytes [93], and oxidative stress can regulate its expression [94]. Two functional SNPs (−2578C/A and −1154G/A) located in the promoter region of VEGF have been suggested to influence AD risk, but the results in this area are ambiguous. Previous meta-analysis has suggested that the AA genotype of −2578C/A is a risk factor for AD development in Europeans [95], but further studies with larger sample sizes seem to have negated this association [32, 96]. However, in the subgroup of APOE ε4 non-carriers, −2578C/A increases AD risk while −1154G/A plays a protective role [32]. These results seem puzzling, as both of these SNPs have been shown to reduce VEGF expression via controlling the activity and responsiveness of the gene promoter [97]. Therefore, it will be of interest to investigate the functional role of VEGF in AD. Initially defined as an endothelium-specific vascular permeability factor, VEGF has neuroprotective effects, as it promotes adult neurogenesis and associated memory and learning [98]. Meanwhile, it may also be neurotoxic, as a high dose has been demonstrated to reduce neuronal survival and promote neuronal apoptosis in vitro [94]. Therefore, the inconsistency concerning the associations of AD risk with −2578C/A and −1154G/A may be attributable to the complex roles of VEGF in AD.
IL-18 is a member of the IL-1 family. In addition to its pro-inflammatory activity, IL-18 is involved in aging and neurodegeneration [99]. Elevated expression of IL-18 has been detected in AD brains [100], and peripheral cells from AD patients show increased production of IL-18 upon stimulation [35]. Gene association analysis further supports the involvement of IL-18 in AD development. Two SNPs in the promoter region of its gene have been reported to influence AD, −137 G/C and −607 C/A. The C allele of IL-18 −137 G/C has a negative correlation with AD risk in Han Chinese [34], but the CC genotype is strongly associated with accelerated cognitive decline in Caucasians [33]. Meanwhile, the C allele of IL-18 −607 and the CC genotype are associated with increased AD risk in Han Chinese and Italian Caucasian populations, respectively [33, 34]. Functional analyses have revealed that the IL-18 −607 CC genotype results in significantly elevated IL-18 production by blood mononuclear cells in AD patients [35], implying that this polymorphism exacerbates AD processes via the promotion of IL-18 secretion. However, negative interactions have also been found in Italian Caucasians by Segat et al. [101], implying that this interaction needs to be confirmed in further studies with larger numbers of AD patients.
IL-33 also belongs to the IL-1 family. Decreased expression of IL-33 has been reported in the brains of AD patients [36]. In a cellular model, overexpressed IL-33 was reported to promote the production of Aβ [36]. A study by Chapuis et al. initially identified the association between IL-33 polymorphisms and AD risk in Caucasian populations. They found protective effects of rare alleles of three intronic SNPs (rs7044343, rs1157505, and rs11792633) and the associated haplotypes [36]. In addition, functional analyses have revealed interactions between these protective allele and decreased cerebral amyloid angiopathy in the brains of non-APOE ε4 individuals with AD [36]. Subsequently, consistent results for rs11792633 were obtained in a Han Chinese population; however, rs7044343 failed to affect AD risk, and the rs1157505 polymorphism was not observed [102]. These data suggest that IL-33 polymorphism modifies AD risk by influencing Aβ metabolism, but this needs further confirmation.
IL-12, another member of the IL-1 family, has recently been implicated in the AD process. Elevated levels of the p40 subunit of IL-12 have been detected in AD brains [103]. In AD animal models, inhibition of p40 alleviates the cognitive impairments and AD-related pathology [103, 104]. Rs568408 and rs3212227, which are located in IL-12A and IL-12B, respectively, have recently been reported to influence AD risk in the Han Chinese population [37]. Based on previous research, these SNPs may influence the levels of IL-12 mRNA and protein, which may account for their functional activity [105, 106]. However, associations of these two SNPs with AD risk require further validation in other ethnic groups.
Anti-Inflammatory Cytokines
IL-4
IL-4 is an anti-inflammatory cytokine and plays a role in neutralizing the neuroinflammatory process in AD brains. A previous study found that peripheral mononuclear cells from AD patients showed reduced IL-4 production upon stimulation [107]. In vitro, IL-4 induces the microglial clearance of Aβ via promoting the expression of CD36 and Aβ-degrading enzymes (neprilysin and insulin-degrading enzyme) [108]. Moreover, in vivo treatment with IL-4 reduces the accumulation of Aβ and alleviates the cognitive impairments in AD animal models [109]. Two SNPs in the promoter region of IL-4 (−590 C/T and −1098 T/G) have been reported to influence AD risk in a study of Han Chinese and another study of Caucasians [38, 39]. The −590 T allele had protective effects against AD in both studies, but the results varied for the −1098 T/G allele. In Caucasians, the −1098 T allele appears to be a risk factor [38], but the −1098 G allele may increase the susceptibility to AD in Han Chinese [39]. This discrepancy may be due to ethnic characteristics, but it should be noted that the study by Ribizzi et al. was conducted with only 39 individuals (19 patients and 20 controls); therefore, further studies with larger samples are needed to confirm or deny this interaction. The significance of the SNPs might be related to the control of IL-4 gene transcription or IL-4 protein expression [110, 111]; this would result in an imbalance between pro- and anti-inflammatory cytokines and ultimately accelerate the AD process. The +33 T/C polymorphism of IL-4 has been identified and was assumed to affect AD risk, but negative results have been obtained in all three relevant studies conducted with Asians [39, 112, 113].
IL-10
IL-10 plays a neuroprotective role in the CNS, as it is anti-inflammatory, anti-apoptotic, and promotes cellular survival [114], but its implications for AD are complex. IL-10 expression mediated by adeno-associated virus (AAV) in the brains of AD animal models was reported to enhance neurogenesis and cognition [115]. Unexpectedly, further investigation opposed these findings, as AAV-mediated IL-10 expression increased the Aβ burden and worsened cognitive impairment in AD animal models [116]. Exploring the interactions between AD-related phenotypes and polymorphisms of IL-10 will help understand its roles in AD. The −1082G/A polymorphism has been extensively investigated, and the GG genotype has been suggested to decrease [86, 117], increase [38], or have no influence on AD risk [118]. According to the two recent meta-analyses, IL-10 −1082G may be associated with reduced AD risk, particularly among Europeans [40, 41]. However, negative results were reported in two studies conducted in Asia [112, 119]. Moreover, contradictory conclusions have been drawn by Moraes et al., who very recently found a significant association between the IL-10 −1082 AA genotype and decreased AD risk in a large sample in Brazil [42]. These data query the relationship between the IL-10 −1082 genetic polymorphism and AD risk and strongly imply differences between ethnic groups. Moreover, in a recent investigation by Medway et al., association of the IL-10 −1082 genetic polymorphism with AD was only found in women [120], suggesting a sex-specific effect of this SNP. It has been reported that this SNP alters IL-10 gene transcription and plasma IL-10 concentrations [119, 121], which may account for the functional activity. Moreover, interactions between the other two SNPs in the promoter regions of IL-10 (IL-10 −819 and IL-10 −592) and AD risk have also been investigated, and strong or even complete linkage disequilibrium (−592C and −819C) has been reported in most studies [117, 119, 122]. The GG genotype has been reported to promote [119], decrease [117], or have no effect [122] on AD risk, and the meta-analysis by Bona et al. [41] suggested that neither individual SNPs nor haplotypes were associated with AD risk. The differences between individual experiments may have resulted from ethnic characteristics, and further clarification of this association requires additional studies involving larger numbers of participants from various regions.
TGFB1
TGF-β signaling has been reported to be insufficient in the brains of AD patients [123]. This deficiency has been shown to cause more Aβ deposition and neurodegeneration [123], while treatment with TGF-β prevents Aβ-induced neurotoxicity [124]. All these results suggest a neuroprotective effect of TGF-β in the AD process. Both the +10 T/C and −509 C/T polymorphisms have been suggested to influence TGFB1 gene transcription and TGF-β1 protein expression [47, 125], and a series of studies has explored their interactions with AD risk. The +10 T/C polymorphism was associated with increased AD risk in studies by Arosio et al. and Caraci et al. [43, 44], and the −509 C/T polymorphism exhibited the same association in a study by Luedecking et al [47]. Moreover, patients with mild cognitive impairment carrying the +10 C allele had an increased risk of developing AD over a 4-year follow-up conducted by Arosio et al. [43]. Furthermore, the +10 CC genotype has been shown to be associated with increased neocortical plaques in Japanese-Americans [46], but the T allele of the +10 T/C polymorphism exhibited a positive correlation with the severity of cerebral amyloid angiopathy in another Japanese sample [45]. These data indicate the involvement of TGFB1 polymorphisms in AD development and AD-related pathology. However, inconsistent results have been reported in the majority of studies conducted in various populations [126], and a negative association has been further supported by a recent meta-analysis [126]. No positive results have been obtained for the other SNPs in TGFB1 [43, 44, 46, 73, 127–129], so it remains controversial whether TGFB1 polymorphisms influence AD risk.
Conclusions and Perspectives
In the current review, we have summarized the key findings on genetic polymorphisms of the inflammatory cytokines associated with AD (Table 1). These discoveries highlight the importance of cytokines in AD pathology and support the neuroinflammatory hypothesis of AD. Moreover, the majority of loci identified thus far are located in the promoter regions of cytokine genes, suggesting that the polymorphic loci function primarily by influencing the expression of cytokines in AD. This may partially account for the microglial priming in AD because the polymorphism-induced increases in gene transcriptional activity could lead to excessive pro-inflammatory cytokine release by microglia upon stimulation [130, 131].
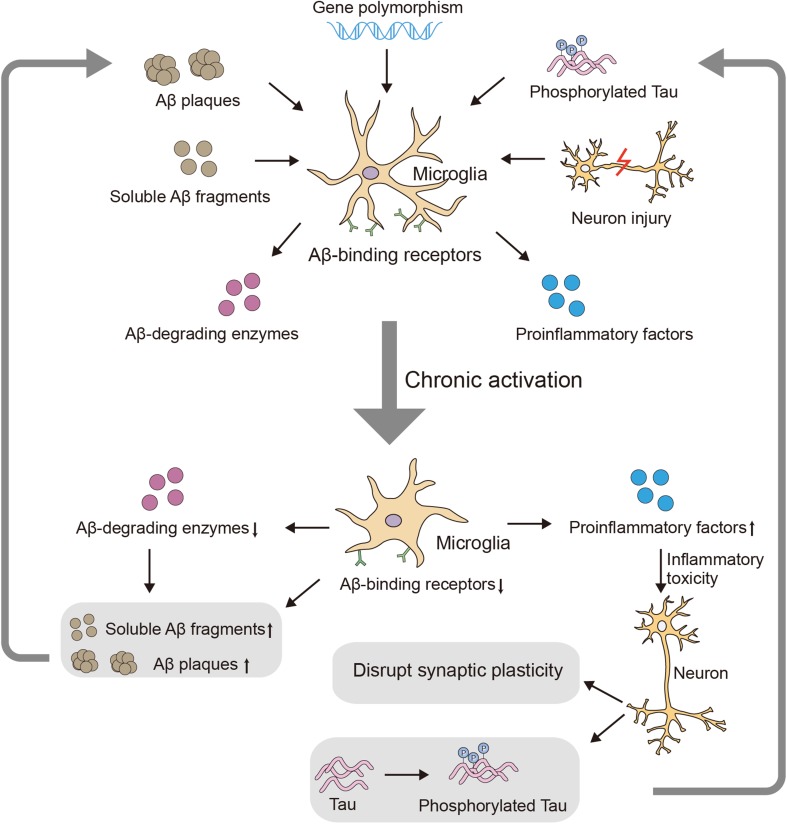
Accumulating evidence has demonstrated the involvement of microglia in AD [132], and cytokines are key components that mediate microglial function (Fig. 1). Pro-inflammatory cytokines acutely benefit the CNS and play major roles in immune attack when pathogens invade the brain. However, due to their non-specific nature, these cytokines can simultaneously damage surrounding healthy tissue [133] via “by-stander” injury. Thus, the chronic increase in pro-inflammatory cytokines in the AD brain [134, 135] could induce sustained inflammatory-toxicity, causing neuronal dysfunction and ultimately the deterioration observed in AD progression. Furthermore, as noted above, cytokines can also influence AD-related pathology, including Aβ and tau [67, 79, 136, 137]. Moreover, indoleamine 2, 3-dioxygenase, which is activated by cytokines, may be an additional candidate mechanism of cytokine-induced AD pathology and could lead to increased levels of the neurotoxic factor, quinolinic acid [138], and promote tau hyperphosphorylation [139]. In summary, chronic over-expression of pro-inflammatory cytokines exacerbates AD, and the influence of anti-inflammatory cytokines in the process of AD also merits attention. The functional significance of anti-inflammatory cytokines is to down-regulate the pro-inflammatory process and initiate tissue reconstruction [140]. Therefore, it can be hypothesized that anti-inflammatory cytokines have protective effects against AD by neutralizing the harmful effects of pro-inflammatory cytokines, and their deficiency may also promote the risk of AD. In addition, TGF-β is protective against Aβ-induced cytotoxicity both in vivo and in vitro [124]. Deficiency of TGF-β1 promotes not only Aβ accumulation but also NFT formation [141], directly highlighting the importance of TGF-β in AD pathology. However, no direct evidence has been obtained concerning the beneficial effects of other anti-inflammatory cytokines in AD, and further work is needed. In conclusion, dysfunction of the cytokine system, which is an important component of neuroinflammation, may be involved in AD pathology.
Fig. 1.
Involvement of microglia in AD development. AD-related pathology induces chronic activation of microglia, exacerbating neuroinflammation and inhibiting the clearance of Aβ, in turn leading to AD-related deterioration.
However, it must be noted that a recent GWAS did not highlight the SNPs discussed here in AD development (apart from IL-6 572 C/G and TNFA −308 G/A, all the SNPs were included in the GWAS analysis, but none reached genome-wide significance) [142]. It may be that the effect size of each individual SNP was too small. In addition, the influence of the same SNP on AD risk varies among populations. The population-based differences arise, at least partially, from natural selection during evolution, causing the minor allele frequency to shift over time. Furthermore, other factors such as diet, infection (cytokines play crucial roles in immune defense against pathogens), and the environment can also contribute to ethnic diversity. Therefore, further research on the interactions between AD and cytokine polymorphisms is needed in additional independent samples that include different ethnic groups, and validation of the functional activity of these loci deserve high priority. Moreover, several other cytokines, such as IL-17, have been shown to have interactions with AD [143], but no studies seem to have investigated the association of polymorphisms of these cytokines with AD. More importantly, gene-gene interactions merit more attention, and studies of such interactions could provide a more comprehensive understanding of heredity in AD. Therefore, the genetic polymorphisms of inflammatory cytokines are promising targets for exploring the etiology of AD, and extensive work remains to be done.
Acknowledgements
This review was partly supported by the National Natural Science Foundation of China (91332104 and 81201080); the Program for New Century Excellent Talents in University, China (NCET-13-0117); the Key Program for Clinical Medicine and Science and Technology of Jiangsu Province Clinical Medical Research Center, China (BL2013025); the Natural Science Foundation of Jiangsu Province, China (BK2012337); the National High-Tech R&D Program (863 Program) of China (SQ2015AA0200064); the Doctoral Fund of the Ministry of Education of China (20120092120068); and the Graduate Candidate Research Innovation Program of Jiangsu Province, China (KYLX15_0188).
References
- 1.Golde TE, Petrucelli L, Lewis J. Targeting Abeta and tau in Alzheimer’s disease, an early interim report. Exp Neurol. 2010;223:252–266. doi: 10.1016/j.expneurol.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonda DJ, Wang X, Lee HG, Smith MA, Perry G, Zhu X. Neuronal failure in Alzheimer’s disease: a view through the oxidative stress looking-glass. Neurosci Bull. 2014;30:243–252. doi: 10.1007/s12264-013-1424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorey E, Chang N, Liu QY, Yang Z, Zhang W. Apolipoprotein E, amyloid-beta, and neuroinflammation in Alzheimer’s disease. Neurosci Bull. 2014;30:317–330. doi: 10.1007/s12264-013-1422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cagnin A, Brooks DJ, Kennedy AM, Gunn RN, Myers R, Turkheimer FE, et al. In-vivo measurement of activated microglia in dementia. Lancet. 2001;358:461–467. doi: 10.1016/S0140-6736(01)05625-2. [DOI] [PubMed] [Google Scholar]
- 5.Yasojima K, Schwab C, McGeer EG, McGeer PL. Up-regulated production and activation of the complement system in Alzheimer’s disease brain. Am J Pathol. 1999;154:927–936. doi: 10.1016/S0002-9440(10)65340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swardfager W, Lanctot K, Rothenburg L, Wong A, Cappell J, Herrmann N. A meta-analysis of cytokines in Alzheimer’s disease. Biol Psychiatry. 2010;68:930–941. doi: 10.1016/j.biopsych.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Sui X, Liu J, Yang X. Cerebrospinal fluid biomarkers of Alzheimer’s disease. Neurosci Bull. 2014;30:233–242. doi: 10.1007/s12264-013-1412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlad SC, Miller DR, Kowall NW, Felson DT. Protective effects of NSAIDs on the development of Alzheimer disease. Neurology. 2008;70:1672–1677. doi: 10.1212/01.wnl.0000311269.57716.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang DF, Li J, Wu H, Cui Y, Bi R, Zhou HJ, et al. CFH variants affect structural and functional brain changes and genetic risk of Alzheimer’s disease. Neuropsychopharmacology. 2016;41:1034–1045. doi: 10.1038/npp.2015.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HZ, Bi R, Hu QX, Xiang Q, Zhang C, Zhang DF, et al. Validating GWAS-identified risk Loci for Alzheimer’s disease in Han Chinese populations. Mol Neurobiol. 2016;53:379–390. doi: 10.1007/s12035-014-9015-z. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Song D, Jiang Y, Wang J, Feng R, Zhang L, et al. CR1 rs3818361 polymorphism contributes to Alzheimer’s disease susceptibility in Chinese population. Mol Neurobiol 2016, 53: 4054–4059. [DOI] [PubMed]
- 13.Li X, Shen N, Zhang S, Liu J, Jiang Q, Liao M, et al. CD33 rs3865444 polymorphism contributes to Alzheimer’s disease susceptibility in Chinese, European, and North American populations. Mol Neurobiol. 2015;52:414–421. doi: 10.1007/s12035-014-8880-9. [DOI] [PubMed] [Google Scholar]
- 14.Qin X, Peng Q, Zeng Z, Chen Z, Lin L, Deng Y, et al. Interleukin-1A -889C/T polymorphism and risk of Alzheimer’s disease: a meta-analysis based on 32 case-control studies. J Neurol. 2012;259:1519–1529. doi: 10.1007/s00415-011-6381-6. [DOI] [PubMed] [Google Scholar]
- 15.Hua Y, Zhao H, Kong Y, Lu X. Meta-analysis of the association between the interleukin-1A -889C/T polymorphism and Alzheimer’s disease. J Neurosci Res. 2012;90:1681–1692. doi: 10.1002/jnr.23062. [DOI] [PubMed] [Google Scholar]
- 16.Hayes A, Green EK, Pritchard A, Harris JM, Zhang Y, Lambert JC, et al. A polymorphic variation in the interleukin 1A gene increases brain microglial cell activity in Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2004;75:1475–1477. doi: 10.1136/jnnp.2003.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan H, Xia Q, Ge P, Wu S. Genetic polymorphism of interleukin 1beta -511C/T and susceptibility to sporadic Alzheimer’s disease: a meta-analysis. Mol Biol Rep. 2013;40:1827–1834. doi: 10.1007/s11033-012-2237-0. [DOI] [PubMed] [Google Scholar]
- 18.Di Bona D, Plaia A, Vasto S, Cavallone L, Lescai F, Franceschi C, et al. Association between the interleukin-1beta polymorphisms and Alzheimer’s disease: a systematic review and meta-analysis. Brain Res Rev. 2008;59:155–163. doi: 10.1016/j.brainresrev.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Ehl C, Kolsch H, Ptok U, Jessen F, Schmitz S, Frahnert C, et al. Association of an interleukin-1beta gene polymorphism at position -511 with Alzheimer’s disease. Int J Mol Med. 2003;11:235–238. [PubMed] [Google Scholar]
- 20.Ravaglia G, Paola F, Maioli F, Martelli M, Montesi F, Bastagli L, et al. Interleukin-1beta and interleukin-6 gene polymorphisms as risk factors for AD: a prospective study. Exp Gerontol. 2006;41:85–92. doi: 10.1016/j.exger.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Sciacca FL, Ferri C, Licastro F, Veglia F, Biunno I, Gavazzi A, et al. Interleukin-1B polymorphism is associated with age at onset of Alzheimer’s disease. Neurobiol Aging. 2003;24:927–931. doi: 10.1016/S0197-4580(03)00011-3. [DOI] [PubMed] [Google Scholar]
- 22.Licastro F, Veglia F, Chiappelli M, Grimaldi LM, Masliah E. A polymorphism of the interleukin-1 beta gene at position +3953 influences progression and neuro-pathological hallmarks of Alzheimer’s disease. Neurobiol Aging. 2004;25:1017–1022. doi: 10.1016/j.neurobiolaging.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Flex A, Giovannini S, Biscetti F, Liperoti R, Spalletta G, Straface G, et al. Effect of proinflammatory gene polymorphisms on the risk of Alzheimer’s disease. Neurodegener Dis. 2014;13:230–236. doi: 10.1159/000353395. [DOI] [PubMed] [Google Scholar]
- 24.Qi HP, Qu ZY, Duan SR, Wei SQ, Wen SR, Bi S. IL-6-174 G/C and -572 C/G polymorphisms and risk of Alzheimer’s disease. PLoS One. 2012;7:e37858. doi: 10.1371/journal.pone.0037858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua Y, Guo X, Huang Q, Kong Y, Lu X. Association between interleukin-6 -174G/C polymorphism and the risk of Alzheimer’s disease: a meta-analysis. Int J Neurosci. 2013;123:626–635. doi: 10.3109/00207454.2013.784286. [DOI] [PubMed] [Google Scholar]
- 26.Licastro F, Grimaldi LM, Bonafe M, Martina C, Olivieri F, Cavallone L, et al. Interleukin-6 gene alleles affect the risk of Alzheimer’s disease and levels of the cytokine in blood and brain. Neurobiol Aging. 2003;24:921–926. doi: 10.1016/S0197-4580(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Jia J. The interleukin-6 gene -572C/G promoter polymorphism modifies Alzheimer’s risk in APOE epsilon 4 carriers. Neurosci Lett. 2010;482:260–263. doi: 10.1016/j.neulet.2010.07.051. [DOI] [PubMed] [Google Scholar]
- 28.He MX, Yang WL, Zhang MM, Lian YJ, Hua HY, Zeng JS, et al. Association between interleukin-6 gene promoter -572C/G polymorphism and the risk of sporadic Alzheimer’s disease. Neurol Sci. 2010;31:165–168. doi: 10.1007/s10072-009-0199-3. [DOI] [PubMed] [Google Scholar]
- 29.Di Bona D, Candore G, Franceschi C, Licastro F, Colonna-Romano G, Camma C, et al. Systematic review by meta-analyses on the possible role of TNF-alpha polymorphisms in association with Alzheimer’s disease. Brain Res Rev. 2009;61:60–68. doi: 10.1016/j.brainresrev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Lee YH, Choi SJ, Ji JD, Song GG. Association between TNF-alpha promoter -308 A/G polymorphism and Alzheimer’s disease: a meta-analysis. Neurol Sci. 2015;36:825–832. doi: 10.1007/s10072-015-2102-8. [DOI] [PubMed] [Google Scholar]
- 31.Lio D, Annoni G, Licastro F, Crivello A, Forte GI, Scola L, et al. Tumor necrosis factor-alpha -308A/G polymorphism is associated with age at onset of Alzheimer’s disease. Mech Ageing Dev. 2006;127:567–571. doi: 10.1016/j.mad.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 32.Liu SY, Zeng FF, Chen ZW, Wang CY, Zhao B, Li KS. Vascular endothelial growth factor gene promoter polymorphisms and Alzheimer’s disease risk: a meta-analysis. CNS Neurosci Ther. 2013;19:469–476. doi: 10.1111/cns.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bossu P, Ciaramella A, Moro ML, Bellincampi L, Bernardini S, Federici G, et al. Interleukin 18 gene polymorphisms predict risk and outcome of Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2007;78:807–811. doi: 10.1136/jnnp.2006.103242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu JT, Tan L, Song JH, Sun YP, Chen W, Miao D, et al. Interleukin-18 promoter polymorphisms and risk of late onset Alzheimer’s disease. Brain Res. 2009;1253:169–175. doi: 10.1016/j.brainres.2008.11.083. [DOI] [PubMed] [Google Scholar]
- 35.Bossu P, Ciaramella A, Salani F, Bizzoni F, Varsi E, Di Iulio F, et al. Interleukin-18 produced by peripheral blood cells is increased in Alzheimer’s disease and correlates with cognitive impairment. Brain Behav Immun. 2008;22:487–492. doi: 10.1016/j.bbi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Chapuis J, Hot D, Hansmannel F, Kerdraon O, Ferreira S, Hubans C, et al. Transcriptomic and genetic studies identify IL-33 as a candidate gene for Alzheimer’s disease. Mol Psychiatry. 2009;14:1004–1016. doi: 10.1038/mp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu XC, Tan L, Jiang T, Tan MS, Zhang W, Yu JT. Association of IL-12A and IL-12B polymorphisms with Alzheimer’s disease susceptibility in a Han Chinese population. J Neuroimmunol. 2014;274:180–184. doi: 10.1016/j.jneuroim.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Ribizzi G, Fiordoro S, Barocci S, Ferrari E, Megna M. Cytokine polymorphisms and Alzheimer disease: possible associations. Neurol Sci. 2010;31:321–325. doi: 10.1007/s10072-010-0221-9. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Qian X, Teng H, Ding Y, Zhang L. Association of interleukin-4 genetic polymorphisms with sporadic Alzheimer’s disease in Chinese Han population. Neurosci Lett. 2014;563:17–21. doi: 10.1016/j.neulet.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Zhang J, Tian C, Xiao Y, Li X, He C, et al. The -1082G/A polymorphism in IL-10 gene is associated with risk of Alzheimer’s disease: a meta-analysis. J Neurol Sci. 2011;303:133–138. doi: 10.1016/j.jns.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Di Bona D, Rizzo C, Bonaventura G, Candore G, Caruso C. Association between interleukin-10 polymorphisms and Alzheimer’s disease: a systematic review and meta-analysis. J Alzheimers Dis. 2012;29:751–759. doi: 10.3233/JAD-2012-111838. [DOI] [PubMed] [Google Scholar]
- 42.Moraes CF, Benedet AL, Souza VC, Lins TC, Camargos EF, Naves JO, et al. Cytokine gene polymorphisms and Alzheimer’s disease in Brazil. Neuroimmunomodulation. 2013;20:239–246. doi: 10.1159/000350368. [DOI] [PubMed] [Google Scholar]
- 43.Arosio B, Bergamaschini L, Galimberti L, La Porta C, Zanetti M, Calabresi C, et al. +10 T/C polymorphisms in the gene of transforming growth factor-beta1 are associated with neurodegeneration and its clinical evolution. Mech Ageing Dev. 2007;128:553–557. doi: 10.1016/j.mad.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Caraci F, Bosco P, Signorelli M, Spada RS, Cosentino FI, Toscano G, et al. The CC genotype of transforming growth factor-beta1 increases the risk of late-onset Alzheimer’s disease and is associated with AD-related depression. Eur Neuropsychopharmacol. 2012;22:281–289. doi: 10.1016/j.euroneuro.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Hamaguchi T, Okino S, Sodeyama N, Itoh Y, Takahashi A, Otomo E, et al. Association of a polymorphism of the transforming growth factor-beta1 gene with cerebral amyloid angiopathy. J Neurol Neurosurg Psychiatry. 2005;76:696–699. doi: 10.1136/jnnp.2003.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peila R, Yucesoy B, White LR, Johnson V, Kashon ML, Wu K, et al. A TGF-beta1 polymorphism association with dementia and neuropathologies: the HAAS. Neurobiol Aging. 2007;28:1367–1373. doi: 10.1016/j.neurobiolaging.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Luedecking EK, DeKosky ST, Mehdi H, Ganguli M, Kamboh MI. Analysis of genetic polymorphisms in the transforming growth factor-beta1 gene and the risk of Alzheimer’s disease. Hum Genet. 2000;106:565–569. doi: 10.1007/s004390000313. [DOI] [PubMed] [Google Scholar]
- 48.Griffin WS, Stanley LC, Ling C, White L, MacLeod V, Perrot LJ, et al. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci USA. 1989;86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Liu L, Kang J, Sheng JG, Barger SW, Mrak RE, et al. Neuronal-glial interactions mediated by interleukin-1 enhance neuronal acetylcholinesterase activity and mRNA expression. J Neurosci. 2000;20:149–155. doi: 10.1523/JNEUROSCI.20-01-00149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Griffin WS, Mrak RE. Interleukin-1 in the genesis and progression of and risk for development of neuronal degeneration in Alzheimer’s disease. J Leukoc Biol. 2002;72:233–238. [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng JG, Jones RA, Zhou XQ, McGinness JM, Van Eldik LJ, Mrak RE, et al. Interleukin-1 promotion of MAPK-p38 overexpression in experimental animals and in Alzheimer’s disease: potential significance for tau protein phosphorylation. Neurochem Int. 2001;39:341–348. doi: 10.1016/S0197-0186(01)00041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Combarros O, Sanchez-Guerra M, Infante J, Llorca J, Berciano J. Gene dose-dependent association of interleukin-1A [-889] allele 2 polymorphism with Alzheimer’s disease. J Neurol. 2002;249:1242–1245. doi: 10.1007/s00415-002-0819-9. [DOI] [PubMed] [Google Scholar]
- 53.Combarros O, Llorca J, Sanchez-Guerra M, Infante J, Berciano J. Age-dependent association between interleukin-1A (-889) genetic polymorphism and sporadic Alzheimer’s disease. A meta-analysis. J Neurol. 2003;250:987–989. doi: 10.1007/s00415-003-1136-7. [DOI] [PubMed] [Google Scholar]
- 54.Yildiz SH, Erdogan MO, Artan S, Solak M, Yaman M, Ozbabalik BD, et al. Association of Alzheimer’s disease with APOE and IL-1alpha gene polymorphisms. Am J Alzheimers Dis Other Demen. 2015;30:756–761. doi: 10.1177/1533317512461557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serretti A, Olgiati P, Politis A, Malitas P, Albani D, Dusi S, et al. Lack of association between interleukin-1 alpha rs1800587 polymorphism and Alzheimer’s disease in two Independent European samples. J Alzheimers Dis. 2009;16:181–187. doi: 10.3233/JAD-2009-0946. [DOI] [PubMed] [Google Scholar]
- 56.Wei X, Chen X, Fontanilla C, Zhao L, Liang Z, Dodel R, et al. C/T conversion alters interleukin-1A promoter function in a human astrocyte cell line. Life Sci. 2007;80:1152–1156. doi: 10.1016/j.lfs.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dominici R, Cattaneo M, Malferrari G, Archi D, Mariani C, Grimaldi LM, et al. Cloning and functional analysis of the allelic polymorphism in the transcription regulatory region of interleukin-1 alpha. Immunogenetics. 2002;54:82–86. doi: 10.1007/s00251-002-0445-9. [DOI] [PubMed] [Google Scholar]
- 58.Murphy GM, Jr, Claassen JD, DeVoss JJ, Pascoe N, Taylor J, Tinklenberg JR, et al. Rate of cognitive decline in AD is accelerated by the interleukin-1 alpha -889 *1 allele. Neurology. 2001;56:1595–1597. doi: 10.1212/WNL.56.11.1595. [DOI] [PubMed] [Google Scholar]
- 59.Yucesoy B, Peila R, White LR, Wu KM, Johnson VJ, Kashon ML, et al. Association of interleukin-1 gene polymorphisms with dementia in a community-based sample: the Honolulu-Asia Aging Study. Neurobiol Aging. 2006;27:211–217. doi: 10.1016/j.neurobiolaging.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Hedley R, Hallmayer J, Groth DM, Brooks WS, Gandy SE, Martins RN. Association of interleukin-1 polymorphisms with Alzheimer’s disease in Australia. Ann Neurol. 2002;51:795–797. doi: 10.1002/ana.10196. [DOI] [PubMed] [Google Scholar]
- 61.Dundar NO, Aktekin B, Ekinci NC, Sahinturk D, Yavuzer U, Yegin O, et al. Interleukin-1beta secretion in hippocampal sclerosis patients with mesial temporal lobe epilepsy. Neurol Int. 2013;5:e17. doi: 10.4081/ni.2013.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santtila S, Savinainen K, Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 1998;47:195–198. doi: 10.1046/j.1365-3083.1998.00300.x. [DOI] [PubMed] [Google Scholar]
- 63.Payao SL, Goncalves GM, de Labio RW, Horiguchi L, Mizumoto I, Rasmussen LT, et al. Association of interleukin 1beta polymorphisms and haplotypes with Alzheimer’s disease. J Neuroimmunol. 2012;247:59–62. doi: 10.1016/j.jneuroim.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 64.Ma SL, Tang NL, Lam LC, Chiu HF. Lack of association of the interleukin-1beta gene polymorphism with Alzheimer’s disease in a Chinese population. Dement Geriatr Cogn Disord. 2003;16:265–268. doi: 10.1159/000072811. [DOI] [PubMed] [Google Scholar]
- 65.Eriksson UK, Pedersen NL, Reynolds CA, Hong MG, Prince JA, Gatz M, et al. Associations of gene sequence variation and serum levels of C-reactive protein and interleukin-6 with Alzheimer’s disease and dementia. J Alzheimers Dis. 2011;23:361–369. doi: 10.3233/JAD-2010-101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strauss S, Bauer J, Ganter U, Jonas U, Berger M, Volk B. Detection of interleukin-6 and alpha 2-macroglobulin immunoreactivity in cortex and hippocampus of Alzheimer’s disease patients. Lab Invest. 1992;66:223–230. [PubMed] [Google Scholar]
- 67.Quintanilla RA, Orellana DI, Gonzalez-Billault C, Maccioni RB. Interleukin-6 induces Alzheimer-type phosphorylation of tau protein by deregulating the cdk5/p35 pathway. Exp Cell Res. 2004;295:245–257. doi: 10.1016/j.yexcr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 68.Pola R, Flex A, Gaetani E, Lago AD, Gerardino L, Pola P, et al. The -174 G/C polymorphism of the interleukin-6 gene promoter is associated with Alzheimer’s disease in an Italian population [corrected] Neuroreport. 2002;13:1645–1647. doi: 10.1097/00001756-200209160-00015. [DOI] [PubMed] [Google Scholar]
- 69.Fontalba A, Gutierrez O, Llorca J, Mateo I, Vazquez-Higuera JL, Berciano J, et al. Gene-gene interaction between CARD8 and interleukin-6 reduces Alzheimer’s disease risk. J Neurol. 2009;256:1184–1186. doi: 10.1007/s00415-009-5080-z. [DOI] [PubMed] [Google Scholar]
- 70.Capurso C, Solfrizzi V, Colacicco AM, D’Introno A, Frisardi V, Imbimbo BP, et al. Interleukin 6-174 G/C promoter and variable number of tandem repeats (VNTR) gene polymorphisms in sporadic Alzheimer’s disease. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:177–182. doi: 10.1016/j.pnpbp.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 71.Han XM, Wang CH, Sima X, Liu SY. Interleukin-6 -174G/C polymorphism and the risk of Alzheimer’s disease in Caucasians: a meta-analysis. Neurosci Lett. 2011;504:4–8. doi: 10.1016/j.neulet.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 72.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Oijen M, Arp PP, de Jong FJ, Hofman A, Koudstaal PJ, Uitterlinden AG, et al. Polymorphisms in the interleukin 6 and transforming growth factor beta1 gene and risk of dementia. The Rotterdam Study. Neurosci Lett. 2006;402:113–117. doi: 10.1016/j.neulet.2006.03.050. [DOI] [PubMed] [Google Scholar]
- 74.Deming Y, Xia J, Cai Y, Lord J, Holmans P, Bertelsen S, et al. A potential endophenotype for Alzheimer’s disease: cerebrospinal fluid clusterin. Neurobiol Aging. 2016;37(208):e201–e209. doi: 10.1016/j.neurobiolaging.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishimura M, Sakamoto T, Kaji R, Kawakami H. Influence of polymorphisms in the genes for cytokines and glutathione S-transferase omega on sporadic Alzheimer’s disease. Neurosci Lett. 2004;368:140–143. doi: 10.1016/j.neulet.2004.06.076. [DOI] [PubMed] [Google Scholar]
- 76.Rasmussen L, Delabio R, Horiguchi L, Mizumoto I, Terazaki CR, Mazzotti D, et al. Association between interleukin 6 gene haplotype and Alzheimer’s disease: a Brazilian case-control study. J Alzheimers Dis. 2013;36:733–738. doi: 10.3233/JAD-122407. [DOI] [PubMed] [Google Scholar]
- 77.Janelsins MC, Mastrangelo MA, Park KM, Sudol KL, Narrow WC, Oddo S, et al. Chronic neuron-specific tumor necrosis factor-alpha expression enhances the local inflammatory environment ultimately leading to neuronal death in 3xTg-AD mice. Am J Pathol. 2008;173:1768–1782. doi: 10.2353/ajpath.2008.080528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tobinick E, Gross H, Weinberger A, Cohen H. TNF-alpha modulation for treatment of Alzheimer’s disease: a 6-month pilot study. MedGenMed. 2006;8:25. [PMC free article] [PubMed] [Google Scholar]
- 79.McAlpine FE, Lee JK, Harms AS, Ruhn KA, Blurton-Jones M, Hong J, et al. Inhibition of soluble TNF signaling in a mouse model of Alzheimer’s disease prevents pre-plaque amyloid-associated neuropathology. Neurobiol Dis. 2009;34:163–177. doi: 10.1016/j.nbd.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montgomery SL, Mastrangelo MA, Habib D, Narrow WC, Knowlden SA, Wright TW, et al. Ablation of TNF-RI/RII expression in Alzheimer’s disease mice leads to an unexpected enhancement of pathology: implications for chronic pan-TNF-alpha suppressive therapeutic strategies in the brain. Am J Pathol. 2011;179:2053–2070. doi: 10.1016/j.ajpath.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng X, Yang L, He P, Li R, Shen Y. Differential activation of tumor necrosis factor receptors distinguishes between brains from Alzheimer’s disease and non-demented patients. J Alzheimers Dis. 2010;19:621–630. doi: 10.3233/JAD-2010-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He P, Zhong Z, Lindholm K, Berning L, Lee W, Lemere C, et al. Deletion of tumor necrosis factor death receptor inhibits amyloid beta generation and prevents learning and memory deficits in Alzheimer’s mice. J Cell Biol. 2007;178:829–841. doi: 10.1083/jcb.200705042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li R, Yang L, Lindholm K, Konishi Y, Yue X, Hampel H, et al. Tumor necrosis factor death receptor signaling cascade is required for amyloid-beta protein-induced neuron death. J Neurosci. 2004;24:1760–1771. doi: 10.1523/JNEUROSCI.4580-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Montgomery SL, Narrow WC, Mastrangelo MA, Olschowka JA, O’Banion MK, Bowers WJ. Chronic neuron- and age-selective down-regulation of TNF receptor expression in triple-transgenic Alzheimer disease mice leads to significant modulation of amyloid- and Tau-related pathologies. Am J Pathol. 2013;182:2285–2297. doi: 10.1016/j.ajpath.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Manoochehri M, Kamali K, Rahgozar M, Ohadi M, Farrokhi H, Khorshid HR. Lack of association between tumor necrosis factor-alpha -308 G/A polymorphism and risk of developing late-onset Alzheimer’s disease in an Iranian population. Avicenna J Med Biotechnol. 2009;1:193–197. [PMC free article] [PubMed] [Google Scholar]
- 86.Vural P, Degirmencioglu S, Parildar-Karpuzoglu H, Dogru-Abbasoglu S, Hanagasi HA, Karadag B, et al. The combinations of TNFalpha-308 and IL-6 -174 or IL-10 -1082 genes polymorphisms suggest an association with susceptibility to sporadic late-onset Alzheimer’s disease. Acta Neurol Scand. 2009;120:396–401. doi: 10.1111/j.1600-0404.2009.01230.x. [DOI] [PubMed] [Google Scholar]
- 87.Maggioli E, Boiocchi C, Zorzetto M, Sinforiani E, Cereda C, Ricevuti G, et al. The human leukocyte antigen class III haplotype approach: new insight in Alzheimer’s disease inflammation hypothesis. Curr Alzheimer Res. 2013;10:1047–1056. doi: 10.2174/15672050113106660169. [DOI] [PubMed] [Google Scholar]
- 88.Ardebili SM, Yeghaneh T, Gharesouran J, Rezazadeh M, Farhoudi M, Ayromlou H, et al. Genetic association of TNF-alpha-308 G/A and -863 C/A polymorphisms with late onset Alzheimer’s disease in Azeri Turk population of Iran. J Res Med Sci. 2011;16:1006–1013. [PMC free article] [PubMed] [Google Scholar]
- 89.Sarajarvi T, Helisalmi S, Antikainen L, Makinen P, Koivisto AM, Herukka SK, et al. An association study of 21 potential Alzheimer’s disease risk genes in a Finnish population. J Alzheimers Dis. 2010;21:763–767. doi: 10.3233/JAD-2010-100597. [DOI] [PubMed] [Google Scholar]
- 90.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lau S, Bates KA, Sohrabi HR, Rodrigues M, Martins G, Dhaliwal SS, et al. Functional effects of genetic polymorphism in inflammatory genes in subjective memory complainers. Neurobiol Aging. 2012;33:1054–1056. doi: 10.1016/j.neurobiolaging.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 92.Tang H, Mao X, Xie L, Greenberg DA, Jin K. Expression level of vascular endothelial growth factor in hippocampus is associated with cognitive impairment in patients with Alzheimer’s disease. Neurobiol Aging. 2013;34:1412–1415. doi: 10.1016/j.neurobiolaging.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kalaria RN, Cohen DL, Premkumar DR, Nag S, LaManna JC, Lust WD. Vascular endothelial growth factor in Alzheimer’s disease and experimental cerebral ischemia. Brain Res Mol Brain Res. 1998;62:101–105. doi: 10.1016/S0169-328X(98)00190-9. [DOI] [PubMed] [Google Scholar]
- 94.Sanchez A, Tripathy D, Luo J, Yin X, Martinez J, Grammas P. Neurovascular unit and the effects of dosage in VEGF toxicity: role for oxidative stress and thrombin. J Alzheimers Dis. 2013;34:281–291. doi: 10.3233/JAD-121636. [DOI] [PubMed] [Google Scholar]
- 95.Del Bo R, Ghezzi S, Scarpini E, Bresolin N, Comi GP. VEGF genetic variability is associated with increased risk of developing Alzheimer’s disease. J Neurol Sci. 2009;283:66–68. doi: 10.1016/j.jns.2009.02.318. [DOI] [PubMed] [Google Scholar]
- 96.He D, Lu W, Chang K, Liu Y, Zhang J, Zeng Z. Vascular endothelial growth factor polymorphisms and risk of Alzheimer’s disease: a meta-analysis. Gene. 2013;518:296–302. doi: 10.1016/j.gene.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 97.Shahbazi M, Fryer AA, Pravica V, Brogan IJ, Ramsay HM, Hutchinson IV, et al. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J Am Soc Nephrol. 2002;13:260–264. doi: 10.1681/ASN.V131260. [DOI] [PubMed] [Google Scholar]
- 98.Cao L, Jiao X, Zuzga DS, Liu Y, Fong DM, Young D, et al. VEGF links hippocampal activity with neurogenesis, learning and memory. Nat Genet. 2004;36:827–835. doi: 10.1038/ng1395. [DOI] [PubMed] [Google Scholar]
- 99.Felderhoff-Mueser U, Schmidt OI, Oberholzer A, Buhrer C, Stahel PF. IL-18: a key player in neuroinflammation and neurodegeneration? Trends Neurosci. 2005;28:487–493. doi: 10.1016/j.tins.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 100.Ojala J, Alafuzoff I, Herukka SK, van Groen T, Tanila H, Pirttila T. Expression of interleukin-18 is increased in the brains of Alzheimer’s disease patients. Neurobiol Aging. 2009;30:198–209. doi: 10.1016/j.neurobiolaging.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 101.Segat L, Milanese M, Arosio B, Vergani C, Crovella S. Lack of association between Interleukin-18 gene promoter polymorphisms and onset of Alzheimer’s disease. Neurobiol Aging. 2010;31:162–164. doi: 10.1016/j.neurobiolaging.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 102.Yu JT, Song JH, Wang ND, Wu ZC, Zhang Q, Zhang N, et al. Implication of IL-33 gene polymorphism in Chinese patients with Alzheimer’s disease. Neurobiol Aging. 1014;2012(33):e1011–e1014. doi: 10.1016/j.neurobiolaging.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 103.Vom Berg J, Prokop S, Miller KR, Obst J, Kalin RE, Lopategui-Cabezas I, et al. Inhibition of IL-12/IL-23 signaling reduces Alzheimer’s disease-like pathology and cognitive decline. Nat Med 2012, 18: 1812–1819. [DOI] [PubMed]
- 104.Tan MS, Yu JT, Jiang T, Zhu XC, Guan HS, Tan L. IL12/23 p40 inhibition ameliorates Alzheimer’s disease-associated neuropathology and spatial memory in SAMP8 mice. J Alzheimers Dis. 2014;38:633–646. doi: 10.3233/JAD-131148. [DOI] [PubMed] [Google Scholar]
- 105.Liu L, Xu Y, Liu Z, Chen J, Zhang Y, Zhu J, et al. IL12 polymorphisms, HBV infection and risk of hepatocellular carcinoma in a high-risk Chinese population. Int J Cancer. 2011;128:1692–1696. doi: 10.1002/ijc.25488. [DOI] [PubMed] [Google Scholar]
- 106.Stanilova S, Miteva L. Taq-I polymorphism in 3’UTR of the IL-12B and association with IL-12p40 production from human PBMC. Genes Immun. 2005;6:364–366. doi: 10.1038/sj.gene.6364213. [DOI] [PubMed] [Google Scholar]
- 107.Reale M, Iarlori C, Feliciani C, Gambi D. Peripheral chemokine receptors, their ligands, cytokines and Alzheimer’s disease. J Alzheimers Dis. 2008;14:147–159. doi: 10.3233/jad-2008-14203. [DOI] [PubMed] [Google Scholar]
- 108.Shimizu E, Kawahara K, Kajizono M, Sawada M, Nakayama H. IL-4-induced selective clearance of oligomeric beta-amyloid peptide(1-42) by rat primary type 2 microglia. J Immunol. 2008;181:6503–6513. doi: 10.4049/jimmunol.181.9.6503. [DOI] [PubMed] [Google Scholar]
- 109.Kawahara K, Suenobu M, Yoshida A, Koga K, Hyodo A, Ohtsuka H, et al. Intracerebral microinjection of interleukin-4/interleukin-13 reduces beta-amyloid accumulation in the ipsilateral side and improves cognitive deficits in young amyloid precursor protein 23 mice. Neuroscience. 2012;207:243–260. doi: 10.1016/j.neuroscience.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 110.Arai N, Nomura D, Villaret D, DeWaal Malefijt R, Seiki M, Yoshida M, et al. Complete nucleotide sequence of the chromosomal gene for human IL-4 and its expression. J Immunol. 1989;142:274–282. [PubMed] [Google Scholar]
- 111.Marsh DG, Neely JD, Breazeale DR, Ghosh B, Friedhoff LR, Schou C, et al. Total serum IgE levels and chromosome 5q. Clin Exp Allergy. 1995;25(Suppl 2):79–83. doi: 10.1111/j.1365-2222.1995.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 112.Kang HJ, Kim JM, Kim SW, Shin IS, Park SW, Kim YH, et al. Associations of cytokine genes with Alzheimer’s disease and depression in an elderly Korean population. J Neurol Neurosurg Psychiatry. 2015;86:1002–1007. doi: 10.1136/jnnp-2014-308469. [DOI] [PubMed] [Google Scholar]
- 113.Shibata N, Ohnuma T, Takahashi T, Baba H, Ishizuka T, Ohtsuka M, et al. The effect of IL4 +33C/T polymorphism on risk of Japanese sporadic Alzheimer’s disease. Neurosci Lett. 2002;323:161–163. doi: 10.1016/S0304-3940(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 114.Strle K, Zhou JH, Shen WH, Broussard SR, Johnson RW, Freund GG, et al. Interleukin-10 in the brain. Crit Rev Immunol. 2001;21:427–449. doi: 10.1615/CritRevImmunol.v21.i5.20. [DOI] [PubMed] [Google Scholar]
- 115.Kiyota T, Ingraham KL, Swan RJ, Jacobsen MT, Andrews SJ, Ikezu T. AAV serotype 2/1-mediated gene delivery of anti-inflammatory interleukin-10 enhances neurogenesis and cognitive function in APP + PS1 mice. Gene Ther. 2012;19:724–733. doi: 10.1038/gt.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chakrabarty P, Li A, Ceballos-Diaz C, Eddy JA, Funk CC, Moore B, et al. IL-10 alters immunoproteostasis in APP mice, increasing plaque burden and worsening cognitive behavior. Neuron. 2015;85:519–533. doi: 10.1016/j.neuron.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lio D, Licastro F, Scola L, Chiappelli M, Grimaldi LM, Crivello A, et al. Interleukin-10 promoter polymorphism in sporadic Alzheimer’s disease. Genes Immun. 2003;4:234–238. doi: 10.1038/sj.gene.6363964. [DOI] [PubMed] [Google Scholar]
- 118.Bagnoli S, Cellini E, Tedde A, Nacmias B, Piacentini S, Bessi V, et al. Association of IL10 promoter polymorphism in Italian Alzheimer’s disease. Neurosci Lett. 2007;418:262–265. doi: 10.1016/j.neulet.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 119.Ma SL, Tang NL, Lam LC, Chiu HF. The association between promoter polymorphism of the interleukin-10 gene and Alzheimer’s disease. Neurobiol Aging. 2005;26:1005–1010. doi: 10.1016/j.neurobiolaging.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 120.Medway C, Combarros O, Cortina-Borja M, Butler HT, Ibrahim-Verbaas CA, de Bruijn RF, et al. The sex-specific associations of the aromatase gene with Alzheimer’s disease and its interaction with IL10 in the Epistasis Project. Eur J Hum Genet. 2014;22:216–220. doi: 10.1038/ejhg.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rees LE, Wood NA, Gillespie KM, Lai KN, Gaston K, Mathieson PW. The interleukin-10-1082 G/A polymorphism: allele frequency in different populations and functional significance. Cell Mol Life Sci. 2002;59:560–569. doi: 10.1007/s00018-002-8448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Culpan D, Prince JA, Matthews S, Palmer L, Hughes A, Love S, et al. Neither sequence variation in the IL-10 gene promoter nor presence of IL-10 protein in the cerebral cortex is associated with Alzheimer’s disease. Neurosci Lett. 2006;408:141–145. doi: 10.1016/j.neulet.2006.08.068. [DOI] [PubMed] [Google Scholar]
- 123.Tesseur I, Zou K, Esposito L, Bard F, Berber E, Can JV, et al. Deficiency in neuronal TGF-beta signaling promotes neurodegeneration and Alzheimer’s pathology. J Clin Invest. 2006;116:3060–3069. doi: 10.1172/JCI27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Caraci F, Battaglia G, Busceti C, Biagioni F, Mastroiacovo F, Bosco P, et al. TGF-beta 1 protects against Abeta-neurotoxicity via the phosphatidylinositol-3-kinase pathway. Neurobiol Dis. 2008;30:234–242. doi: 10.1016/j.nbd.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 125.Yokota M, Ichihara S, Lin TL, Nakashima N, Yamada Y. Association of a T29– > C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation. 2000;101:2783–2787. doi: 10.1161/01.CIR.101.24.2783. [DOI] [PubMed] [Google Scholar]
- 126.Chang WW, Zhang L, Jin YL, Yao YS. Meta-analysis of the transforming growth factor-beta1 polymorphisms and susceptibility to Alzheimer’s disease. J Neural Transm (Vienna) 2013;120:353–360. doi: 10.1007/s00702-012-0850-7. [DOI] [PubMed] [Google Scholar]
- 127.Shawkatova I, Javor J, Parnicka Z, Vrazda L, Novak M, Buc M. No association between cytokine gene polymorphism and risk of Alzheimer’s disease in Slovaks. Acta Neurobiol Exp (Wars) 2010;70:303–307. doi: 10.55782/ane-2010-1802. [DOI] [PubMed] [Google Scholar]
- 128.Araria-Goumidi L, Lambert JC, Mann DM, Lendon C, Frigard B, Iwatsubo T, et al. Association study of three polymorphisms of TGF-beta1 gene with Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2002;73:62–64. doi: 10.1136/jnnp.73.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rodriguez-Rodriguez E, Sanchez-Juan P, Mateo I, Llorca J, Infante J, Garcia-Gorostiaga I, et al. Serum levels and genetic variation of TGF-beta1 are not associated with Alzheimer’s disease. Acta Neurol Scand. 2007;116:409–412. doi: 10.1111/j.1600-0404.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 130.Singhal G, Jaehne EJ, Corrigan F, Toben C, Baune BT. Inflammasomes in neuroinflammation and changes in brain function: a focused review. Front Neurosci. 2014;8:315. doi: 10.3389/fnins.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mrak RE, Griffin WS. Glia and their cytokines in progression of neurodegeneration. Neurobiol Aging. 2005;26:349–354. doi: 10.1016/j.neurobiolaging.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 132.Su F, Bai F, Zhou H, Zhang Z. Microglial toll-like receptors and Alzheimer’s disease. Brain Behav Immun. 2016;52:187–198. doi: 10.1016/j.bbi.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 133.Colton C, Wilcock DM. Assessing activation states in microglia. CNS Neurol Disord Drug Targets. 2010;9:174–191. doi: 10.2174/187152710791012053. [DOI] [PubMed] [Google Scholar]
- 134.Grammas P, Ovase R. Inflammatory factors are elevated in brain microvessels in Alzheimer’s disease. Neurobiol Aging. 2001;22:837–842. doi: 10.1016/S0197-4580(01)00276-7. [DOI] [PubMed] [Google Scholar]
- 135.Cacabelos R, Barquero M, Garcia P, Alvarez XA. Varela de Seijas E. Cerebrospinal fluid interleukin-1 beta (IL-1 beta) in Alzheimer’s disease and neurological disorders. Methods Find Exp Clin Pharmacol. 1991;13:455–458. [PubMed] [Google Scholar]
- 136.Li Y, Liu L, Barger SW, Griffin WS. Interleukin-1 mediates pathological effects of microglia on tau phosphorylation and on synaptophysin synthesis in cortical neurons through a p38-MAPK pathway. J Neurosci. 2003;23:1605–1611. doi: 10.1523/JNEUROSCI.23-05-01605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sheng JG, Zhu SG, Jones RA, Griffin WS, Mrak RE. Interleukin-1 promotes expression and phosphorylation of neurofilament and tau proteins in vivo. Exp Neurol. 2000;163:388–391. doi: 10.1006/exnr.2000.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nemeth H, Toldi J, Vecsei L. Role of kynurenines in the central and peripheral nervous systems. Curr Neurovasc Res. 2005;2:249–260. doi: 10.2174/1567202054368326. [DOI] [PubMed] [Google Scholar]
- 139.Rahman A, Ting K, Cullen KM, Braidy N, Brew BJ, Guillemin GJ. The excitotoxin quinolinic acid induces tau phosphorylation in human neurons. PLoS One. 2009;4:e6344. doi: 10.1371/journal.pone.0006344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 141.Caraci F, Battaglia G, Bruno V, Bosco P, Carbonaro V, Giuffrida ML, et al. TGF-beta1 pathway as a new target for neuroprotection in Alzheimer’s disease. CNS Neurosci Ther. 2011;17:237–249. doi: 10.1111/j.1755-5949.2009.00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen JM, Jiang GX, Li QW, Zhou ZM, Cheng Q. Increased serum levels of interleukin-18, -23 and -17 in Chinese patients with Alzheimer’s disease. Dement Geriatr Cogn Disord. 2014;38:321–329. doi: 10.1159/000360606. [DOI] [PubMed] [Google Scholar]