Abstract
Accumulating evidence indicates that the synaptic activation of N-methyl-d-aspartate receptors (NMDARs) has a neuroprotective effect on neurons. Our previous study demonstrated that APPL1 (adaptor protein containing pleckstrin homology domain, phosphotyrosine-binding domain, and leucine zipper motif) mediates the synaptic activity-dependent activation of PI3K-Akt signaling via coupling this pathway with NMDAR-PSD95 (postsynaptic density protein 95) complexes. However, the molecular mechanism underlying this process is still unknown. In the present study, we investigated the interaction of APPL1 with PSD95 using co-immunocytochemical staining and western blotting. We found that the PDZ2 domain of PSD95 is a binding partner of APPL1. Furthermore, we identified serine 707 of APPL1, a predicted phosphorylation site within the PDZ-binding motif at the C-terminus, as critical for the binding of APPL1 to PSD95, as well as for activation of the Akt signaling pathway during synaptic activity. This suggests that serine 707 of APPL1 is a potential phosphorylation site and may be involved in regulating the neuroprotective Akt signaling pathway that depends on synaptic NMDAR activity.
Keywords: APPL1, PSD95, Akt, NMDA receptors, Neuroprotection
Introduction
APPL1 (adaptor protein containing pleckstrin homology domain, phosphotyrosine-binding domain, and leucine zipper motif) is a 709-amino-acid membrane-associated protein that is widely and highly expressed in the heart, ovary, pancreas, skeletal muscle, and brain [1].
APPL1 was initially identified as an Akt2-binding protein in a yeast-two hybrid screen [2], and now it is emerging as a critical regulator of various cellular processes in non-neuronal cells. APPL1 has been reported to bind to several membrane receptors and signaling molecules, including the epidermal growth factor receptor [3], the neurotrophin receptor TrkA [4], tumor suppressor gene DCC (deleted in colorectal cancer), serine/threonine kinase Akt [2], and the adiponectin receptors AdipoR1 and AdipoR2 [5]. Through its multiple interactions, APPL1 plays a key role in the regulation of apoptosis [6], cell proliferation and survival [7], vesicular trafficking [8], and chromatin remodeling [3]. In addition, it is indispensable for the signaling and insulin-sensitizing effects of adiponectin [1, 5, 9].
APPL1 is also highly expressed in the central nervous system (CNS) [2], where it is localized on the dendritic spines and synapses of hippocampal neurons [10]. Although APPL1 function in non-neuronal cells has been intensively investigated, so far, there are few reports on its function in the CNS. It has been shown to regulate spine and synapse development in hippocampal neurons. Overexpression of exogenous APPL1 increases the spine and synaptic density and the amount of surface GluA1-containing α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs), while knockdown of endogenous APPL1 leads to a significant decrease in the number of spines and synapses. Akt is critical for these processes, since a decrease in endogenous Akt or expression of dominant-negative Akt significantly impairs spine and synapse formation. APPL1 promotes spine formation and synaptic activity by increasing the activity of Akt and the surface level of AMPARs [10].
In a previous study, we also reported a key role of APPL1 in the CNS [11]. We found that APPL1 is partially co-localized with PSD95 and N-methyl-d-aspartate receptors (NMDARs) in cultured neurons, and couples synaptic NMDARs with the PI3K/Akt pathway through its direct interaction with PSD95. In this process, a key step is the recruitment of APPL1 to the NMDAR-PSD95 complex in a synaptic NMDAR activation-dependent manner. However, the molecular mechanism underlying this recruitment is still unknown.
In this study, we mapped the PDZ2 domain of PSD95 as a binding domain with APPL1, and identified APPL1 serine 707, a putative phosphorylation site at its PDZ-binding motif, as a critical site for the synaptic NMDAR-mediated Akt neuroprotective signaling pathway.
Materials and Methods
Animals
All animal experiments were approved by Zhejiang University School of Medicine Ethics Committee and performed in accordance with the guidelines of the Zhejiang University Animal Experimentation Committee and were in complete compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. C57BL/6 mice and Sprague-Dawley rats were used throughout the study. Every effort was made to minimize the number and suffering of the animals.
Plasmids, Drugs, and Antibodies
Cyan fluorescent protein (CFP)-tagged APPL1 (CFP-APPL1), C-terminal 4-amino-acid-truncated APPL1 (CFP-APPL1Δ4), and mCherry-tagged PSD95 (mCherry-PSD95) have been described in a previous study [11]. CFP- or green fluorescent protein (GFP)-tagged APPL1 with serine 707 replaced by alanine (CFP-APPL1S707A and GFP-APPL1S707A), mCherry-tagged PDZ1- or both PDZ1- and 2-deleted PSD95 (mCherry-PSD95ΔPDZ1 or mCherry-PSD95ΔPDZ1/2) were constructed by conventional molecular cloning. All constructs were verified by DNA sequencing.
Bicuculline and 4-aminopyridine (4-AP) were from Sigma-Aldrich (St Louis, MO).
The polyclonal antibody against APPL1 (H-96, 1:1000) was from Santa Cruz (San Jose, CA), that against mCherry (5993-100, 1:1000) was from Bio Vision (San Jose, CA), that against PSD95 (ab18258, 1:1000) was from Abcam (Cambridge, UK), and that against phospho-Akt (9271, 1:100) was from Cell Signaling Technology (Boston, MA). The anti-GFP rabbit antibody was home-made.
Primary Neuron Culture
The primary neuron culture method was as described previously [11–14]. Briefly, hippocampal and cortical tissues were removed from rat pups on embryonic day 17 under a dissecting microscope in ice-cold Hanks’ balanced salt solution (Gibco, Grand Island, NY). The tissue was digested in 0.25% trypsin (Invitrogen, Carlsbad, CA) at 37°C in an incubator for ~16 min. Digestion was stopped by adding Neurobasal medium containing 5% fetal bovine serum (Gibco), and the tissue was dissected by further gentle aspiration through a pipette. After centrifugation for 3 min at 1,100 rpm, cells were re-suspended and plated onto poly-D-lysine-coated dishes and coverslips at 6–10 × 104 cells/cm2 in Neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen), penicillin/streptomycin (Invitrogen), and glutamine (Gibco) and kept at 37°C in a 5% CO2 humidified incubator. Half of the culture medium was replaced with fresh medium at 4 days in vitro (DIV4) and then at 3-day intervals. At DIV5, cytosine arabinofuranoside (Invitrogen) was added to a final concentration of 2.5 μmol/L to stop glial growth.
Co-immunoprecipitation (Co-IP)
The protocol for co-IP was as previously described [11, 15]. The cortex was dissociated from adult C57BL/6 mice, homogenized quickly on ice, and lysed in buffer containing 50 mmol/L Tris (pH 7.4), 150 mmol/L NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mmol/L PMSF, and 1 μg/mL aprotinin. After centrifugation at 37,000 g at 4°C for 15 min, IP antibody was added to the supernatant and incubated overnight at 4°C. Protein A-Sepharose (GE Healthcare, Piscataway, NJ) was added on the following day. After incubation for 1 h at 4°C, the mixtures were washed four times with lysis buffer, and the immunoprecipitates were eluted with 1.5 × SDS-PAGE loading buffer by boiling at 100°C for 5 min.
The co-IP protocol for transfected HEK293T cells was similar to that for cortex. After transfection with the indicated plasmids for 24 h, HEK293T cells were lysed in buffer containing 50 mmol/L Tris (pH 7.4), 150 mmol/L NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mmol/L phenylmethylsulfonyl fluoride, and 1 μg/mL aprotinin. After centrifugation at 14,000 g at 4°C for 15 min, antibodies were added to the supernatant and incubated overnight at 4°C.
Cell Transfection
At DIV 6, cultured neurons were transfected with the indicated plasmids using calcium phosphate transfection (Clontech, Mountain View, CA). A precipitate containing calcium phosphate and plasmid DNAs was formed by slowly mixing HEPES-buffered saline with a solution containing calcium chloride and plasmid. This mixture was then added to the dishes, incubated for 1–3 h, and the remaining precipitate was digested in 10% CO2 saturated Neurobasal medium for 7 min. Finally, the neurons were cultured for another 3 days in the original culture medium.
For HEK293T cell transfection, we followed the LipofectamineTM 2000 manual (Invitrogen). Briefly, the indicated plasmids and liposomes were mixed with OPTI-MEM medium (Gibco), added to the dishes, and incubated for 3 h. The transfection medium was replaced with Dulbecco’s modified Eagle’s medium and the cells were incubated at 37°C for another 24 h.
Image Acquisition by Structural Illumination Microscopy (SIM) and Analysis
To analyze APPL1 and PSD95 co-localization in cultured neurons, three-dimensional (3D)-SIM images of immunostained neurons were acquired on the DeltaVision OMXV3 imaging system (Applied Precision, Issaquaah, WA) with a 100× 1.4 oil objective (Nikon, Tokyo, Japan), solid-state multimode lasers (488 and 593 nm) and electron-multiplying CCD (charge-coupled device) cameras (Evolve 512_512, Photometrics, Tucson, AZ). Serial Z-stack sectioning was done at 200-nm intervals. The microscope was routinely calibrated with 100-nm fluorescent spheres to calculate both the lateral and axial limits of image resolution. SIM image stacks were reconstructed using softWoRx 5.0 (Applied Precision) with the following settings: pixel size 39.5 nm, channel-specific optical transfer functions, Wiener filter 0.001000, discard negative intensities background, drift correction with respect to first angle, and custom K0 guess angles for camera positions. Pixel registration was corrected to be <1 pixel for all channels using 100-nm Tetraspeck beads (Thermo Fisher, Waltham, MA). Reconstructed images were rendered in 3D using Imaris version 7.7.2 (Bitplane, Zurich, Switzerland). For clarity of display, small linear changes to brightness and contrast were performed on 3D reconstructions throughout the entire image. NIS and Imaris were used to analyze the cluster density and co-localization of APPL1 and PSD95.
Immunocytochemistry
The protocol for neuron staining was as described previously [11, 12]. Briefly, cultured neurons were first fixed in 4% paraformaldehyde for 10 min, and then incubated with the indicated primary antibodies for 1 h at room temperature. After washing 3 times with PBS, the neurons were incubated with secondary antibodies for another 1 h. After washing another 3 times with PBS, the stained neurons were mounted. Images were acquired with a confocal microscope (Fluoview FV1000; Olympus, Japan). MetaMorph 7.5 software (Universal Imaging, NY, NY) was used to analyze the co-localization of clusters and phosphorylated Akt intensity.
Statistics
Data are presented as mean ± SEM. Statistical significance was determined using Student’s unpaired t-test when comparing 2 groups and one-way ANOVA when comparing ≥3 groups. P ≤0.05 was considered to indicate a significant difference.
Results and Discussion
APPL1 is Associated with PSD95 in the Brain and Cultured Hippocampal Neurons
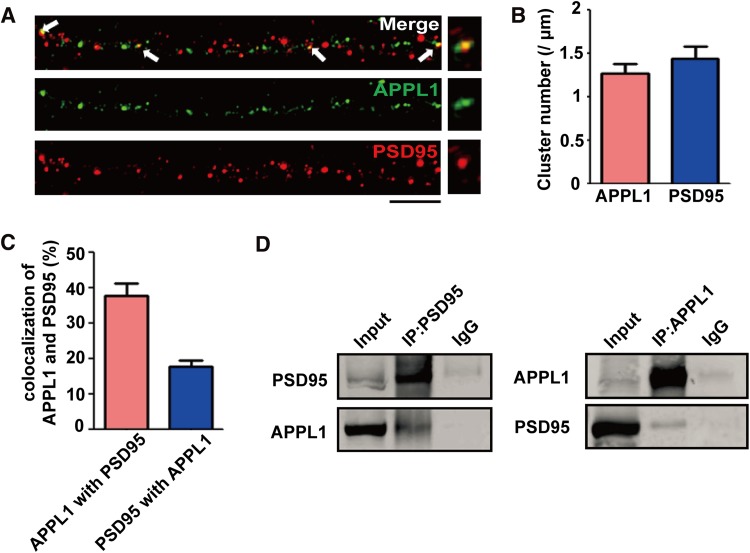
In a previous study, we found that APPL1 couples the PI3K/Akt neuroprotective signaling pathway with synaptic NMDARs, and PSD95 serves as a bridge between APPL1 and synaptic NMDARs [11]. Here, we further examined the co-localization between APPL1 and PSD95 using 3D-SIM in cultured hippocampal neurons (Fig. 1A). Our results showed that the mean number of APPL1 clusters was ~1.3 per μm dendrite, and that of PSD95 was ~1.4 per μm (Fig. 1B). About 38% of APPL1 was co-localized with PSD95, and ~22% of PSD95 was co-localized with APPL1 in cultured hippocampal neurons. Next, we detected the interaction between APPL1 and PSD95 in mouse brain tissue (Fig. 1D), and found that the antibody against APPL1 immunoprecipitated endogenous APPL1 as well as PSD95, indicating that APPL1 and PSD95 are associated in the complex.
Fig. 1.
APPL1 is physically associated with PSD95 in the brain and cultured neurons. (A) Representative 3D-SIM images showing co-localization of APPL1 with PSD95 in cultured hippocampal neurons. Scale bar, 5 μm. (B) Statistics for APPL1 and PSD95 cluster density in dendrites. The mean density of APPL1 clusters was 1.3 ± 0.12 per μm (total 38 neurons, n = 3) and of PSD95 clusters was 1.4 ± 0.14 per μm (total 45 neurons, n = 3). (C) Statistics for APPL1 and PSD95 co-localization. The percentage of APPL1 co-localized with PSD95 was 38 ± 3.61% (total 40 neurons, n = 3), and the that of PSD95 co-localized with APPL1 was 18 ± 1.8% (total 40 neurons, n = 3). (D) APPL1 and PSD95 coexist in the same protein complex in brain tissue. Preparations from adult mouse hippocampus were immunoprecipitated (IP) and blotted with antibodies against PSD95 or APPL1. Three independent co-IP tests were performed for each group.
APPL1 Binds with the PDZ2 Domain of PSD95
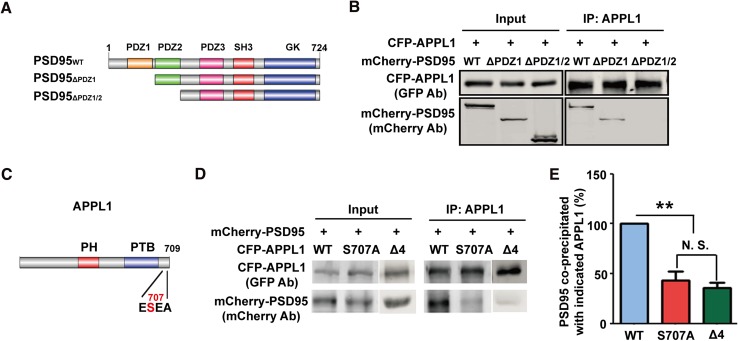
We previously reported that APPL1 interacts with PSD95 through its C-terminal PDZ-binding motif ESEA using fluorescence resonance energy transfer and co-immunoprecipitation [11]. Since there are three PDZ domains in PSD95, in order to further map the binding domain, we constructed two plasmids, mCherry-PSD95△PDZ1 and mCherry-PSD95△PDZ1/2, representing mCherry-tagged PDZ1-deleted or both PDZ1 and PDZ2-deleted PSD95 (Fig. 2A). Following co-transfection of CFP-APPL1 with mCherry-PSD95 or its mutants into HEK293T cells, the co-IP results showed that CFP-APPL1 associated with mCherry-PSD95 and mCherry-PSD95△PDZ1, but not mCherry-PSD95△PDZ1/2 (Fig. 2B), indicating that APPL1 interacts with the PDZ2 domain of PSD95.
Fig. 2.
Mapping of interaction between APPL1 and PSD95. (A) Schematic of the PSD95 construct and its mutants. (B) APPL1 interacts with the PDZ2 domain of PSD95 in HEK293T cells co-transfected with GFP-APPL1 and mCherry-PSD95, mCherry-PSD95△PDZ1, or mCherry-PSD95△PDZ1/2. Three independent co-IP tests were performed. (C) Schematic of the APPL1 construct with serine 707 replaced by alanine. PH, pleckstrin homology domain; PTB, phosphotyrosine-binding domain. (D) APPL1S707A had a significantly decreased interaction with PSD95 in HEK293T cells co-transfected with mCherry-PSD95 and CFP-APPL1 or CFP-APPL1S707A or CFP-APPL1△4. (E) Statistics for (D), **P <0.01, N.S.: no significant difference, n = 4, one-way ANOVA.
We also constructed the plasmid CFP-APPL1S707A with the serine 707 replaced by alanine (Fig. 2C). Co-IP results showed that the interaction of PSD95 and CFP-APPL1S707A was significantly lower than with CFP-APPL1 (Fig. 2D, E), suggesting that serine 707 of APPL1 is critical for its interaction with PSD95. In addition, we repeated the co-IP and showed that there was no interaction between APPL1△4 and PSD95 (Fig. 2D, E), consistent with our previous results [11].
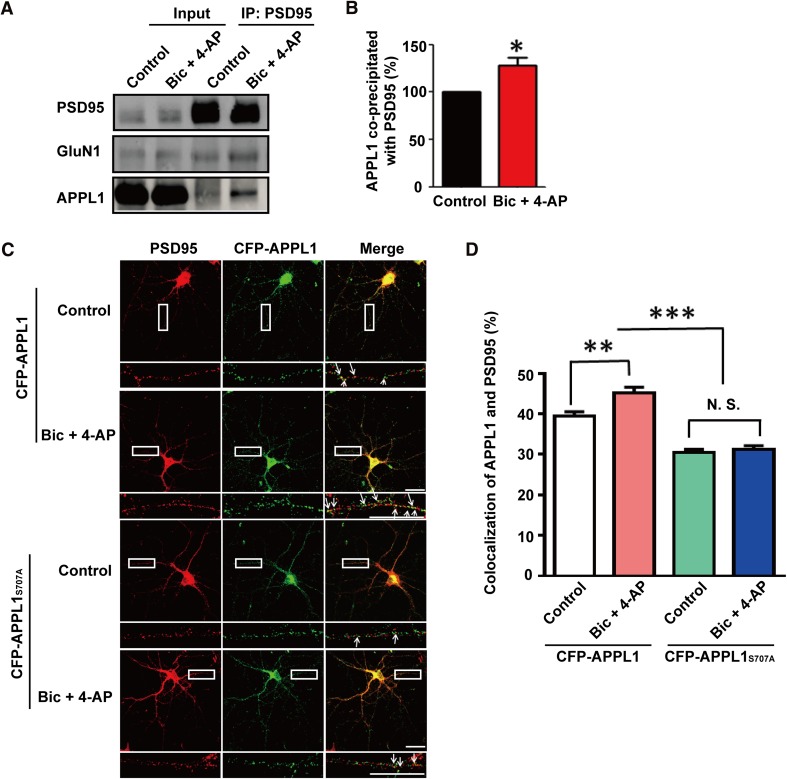
APPL1S707A Interrupts the Increased Association of APPL1 with PSD95 Induced by Synaptic Activity
In a previous study, we revealed that synaptic NMDAR activation enhances the association of APPL1 with GluN1 [11]. However, the change of interaction between APPL1 and PSD95 remained to be determined. We treated cultured neurons with bicuculline (50 μmol/L) and 4-AP (250 μmol/L) for 30 min to activate synaptic NMDARs. Co-IP results showed that synaptic activity significantly increased the association of APPL1 with PSD95 (Fig. 3A, B), further confirming that PSD95 is an important mediator of the recruitment of APPL1 to NMDAR complexes. We further investigated whether synaptic activity increases the APPL1 and PSD95 interaction using immunostaining in hippocampal neurons transfected with CFP-APPL1 or CFP-APPL1S707A. The results showed that synaptic activity significantly increased the co-localization of PSD95 and APPL1 in the neurons transfected with CFP-APPL1 (control: 39.40 ± 1.13%, Bic + 4-AP: 45.11% ± 1.49, P <0.01, 60 neurons each group, 3 independent cultures; Fig. 3D), while co-localization of PSD95 and CFP-APPL1S707A decreased under basal conditions and during synaptic activity (control: CFP-APPL1: 39.40 ± 1.13%, CFP-APPL1S707A: 30.50 ± 0.76%, P <0.0001, 60 neurons each group, 3 independent cultures; Bic + 4-AP: CFP-APPL1: 45.11 ± 1.49%, CFP-APPL1S707A: 31.28 ± 0.89%, P <0.0001, 60 neurons each group, 3 independent cultures; Fig. 3D). However, no significant difference was detected in the co-localization of APPL1 and PSD95 in neurons transfected with CFP-APPL1S707A between the untreated control and synaptic activity groups (control: 30.50 ± 0.76, Bic + 4-AP: 31.28 ± 0.89%, P >0.05, 60 neurons each group, 3 independent cultures; Fig. 3D). Taken together, the APPL1S707A mutant lost its ability for increased association with PSD95 in response to synaptic activity, suggesting that the APPL1 S707 site plays a crucial role in this process.
Fig. 3.
APPL1 serine 707 is critical for synaptic activity-mediated recruitment of APPL1 to PSD95 in cultured neurons. (A) Representative western blot showing that synaptic NMDAR activation significantly increases APPL1 interaction with PSD95 in cultured neurons. (B) Statistics for (A), *P <0.05, n = 3 independent blots, t-test. (C) CFP-APPL1, but not CFP-APPL1S707A, had significantly increased co-localization with PSD95 in response to synaptic activity in cultured neurons. Scale bars, 20 μm. (D) Statistics for co-localizaiton of APPL1 with PSD95. **P <0.01, ***P <0.001, N.S.: no significant difference, 60 neurons each group, 3 independent cultures, one-way ANOVA.
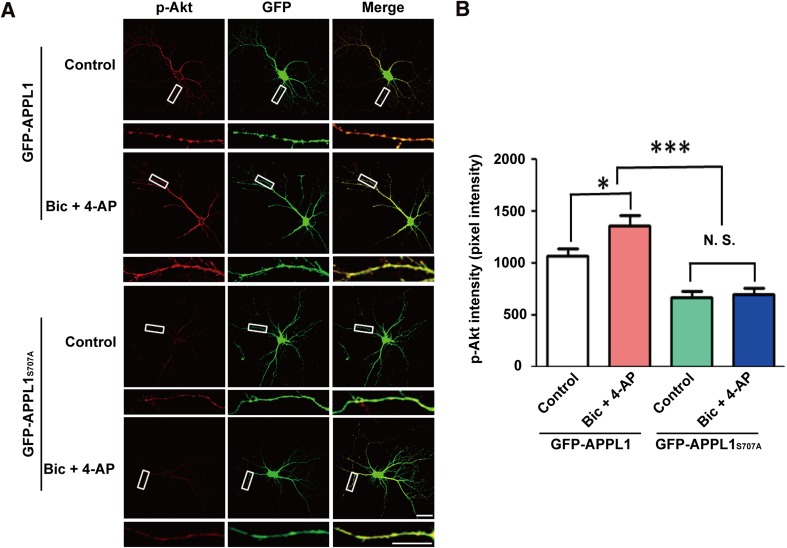
APPL1S707A Disrupts Synaptic Activity-Mediated Activation of Akt
We have reported that APPL1 couples synaptic NMDARs with the PI3K/Akt neuroprotective pathway [11]. Here, we further determined whether the APPL1S707A mutation affected this process. The results showed that Akt phosphorylated at S473 was dramatically increased in cultured hippocampal neurons transfected with GFP-APPL1 when incubated with bicuculline and 4-AP ((in pixel intensity units) control: 1069 ± 67.52, n = 45 neurons; Bic + 4-AP: 1360 ± 99.07, n = 46 neurons, P <0.05; Fig. 4); while phosphorylated Akt in the neurons transfected with GFP-APPL1S707A was significantly decreased under basal conditions and during synaptic activity (control: GFP-APPL1S707A: 670.3 ± 55.11, n = 50 neurons; Bic + 4-AP: GFP-APPL1S707A: 699.2 ± 56.26, n = 46 neurons, P <0.0001; Fig. 4). Notably, synaptic activity failed to increase the phosphorylation level of Akt at S473 in neurons transfected with GFP-APPL1S707A (Fig. 4). These results showed that serine 707 in APPL1 is not only critical for its binding to PSD95 but also for synaptic activity-mediated Akt activation.
Fig. 4.
APPL1S707A mutant disrupts synaptic activity-mediated increase in Akt activity. (A) GFP-APPL1S707A transfection decreased the level of Akt phosphorylation at S473 in response to synaptic activity in cultured neurons. Scale bars, 20 μm. (B) Statistics for intensity of phospho-Akt. *P <0.05, N.S.: no significant difference, 45~50 neurons each group, 3 independent cultures, one-way ANOVA.
Taken together, we have identified PDZ2 as the PSD95 domain to which APPL1 binds and APPL1 serine 707, a putative phosphorylation site at its PDZ binding motif, as a critical site for the synaptic NMDAR-mediated Akt neuroprotective signaling pathway.
In this study, we have revealed that the APPL1S707A mutation disrupts synaptic NMDAR activity-mediated Akt phosphorylation in cultured neurons. Phosphorylation changes the structural conformation of a protein, modifying its function in many ways, including protein-protein interaction and catalytic activity. It has been suggested that APPL1S707 is a potential regulatory site via phosphorylation. However, since so far there is no data showing that APPL1S707 is phosphorylated in vivo, the possibility cannot be excluded that the S707A mutation itself may change the C-terminal structure of APPL1 and affect the interaction between APPL1 and PSD95. Therefore, it will be of interest to investigate whether APPL1S707 is phosphorylated and regulated during neuronal activity in vivo.
NMDARs regulate neuronal development, survival, and physiology; however, aberrant NMDAR activity contributes to neuronal dysfunction and disease, from neurodegenerative to psychiatric disorders [16, 17]. We previously reported that APPL1 couples synaptic NMDAR activity with the Akt neuroprotective pathway through its recruitment to PSD95. Here we have further revealed that APPL1 binds to the PDZ2 domain of PDD95. Interestingly, the PDZ2 domain of PSD95 interacts with a large number of proteins, including NMDARs, Sema 4f, APC, and nNOS [18]. Accumulating evidence has demonstrated that recruitment of the Ca2+-dependent enzyme nNOS via PSD95 is a key contributor to neuronal dysfunction [19–24]. Our result showed that interaction of APPL1 with PSD95 through the PDZ2 domain results in activation of the neuroprotective Akt pathway. Therefore, it is possible that these two proteins, nNOS and APPL1, compete for association with PSD95 via the PDZ2 domain, and this may provide a mechanism for switching between the opposing signaling pathways activated by NMDARs under different conditions. Considering the pro-death effect on neurons mediated by nNOS binding to PSD95, enhancement of APPL1 binding to PSD95 as a competitor for the nNOS interaction with PSD95 with a pro-survival effect may provide a potential means of interfering with neurodegenerative or ischemic brain diseases.
Clearly, it is important to determine whether or how APPL1 recruitment to PSD95 is regulated during neuronal activity. Several phosphorylation sites in APPL1 have been identified, including threonine 399 and serine 401, 459, 691, 693, and 696 by mass spectrometric analysis [25] in non-neural tissues, but there are no reports on the regulation of APPL1 phosphorylation in the CNS. We predicted APPL1 serine 707 to be a putative phosphorylation site by using the online bioinformatics tools GPS 2.0 and DISPHOS. Our results reveal that the APPL1 serine 707 site is critical for its binding to PSD95 and activation of Akt, suggesting it is a potential phosphorylation site in vivo. It will be intriguing to further explore whether APPL1 is phosphorylated in vivo and its regulation during synaptic activity.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (91232303, 81221003, and 81561168).
References
- 1.Deepa SS, Dong LQ. APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab. 2009;296:E22–36. doi: 10.1152/ajpendo.90731.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitsuuchi Y, Johnson SW, Sonoda G, Tanno S, Golemis EA, Testa JR. Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene. 1999;18:4891–4898. doi: 10.1038/sj.onc.1203080. [DOI] [PubMed] [Google Scholar]
- 3.Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/S0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 4.Lin DC, Quevedo C, Brewer NE, Bell A, Testa JR, Grimes ML, et al. APPL1 associates with TrkA and GIPC1 and is required for nerve growth factor-mediated signal transduction. Mol Cell Biol. 2006;26:8928–8941. doi: 10.1128/MCB.00228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, Ramos FJ, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Yao F, Wu R, Morgan M, Thorburn A, Finley RL, Jr, et al. Mediation of the DCC apoptotic signal by DIP13 alpha. J Biol Chem. 2002;277:26281–26285. doi: 10.1074/jbc.M204679200. [DOI] [PubMed] [Google Scholar]
- 7.Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, et al. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 8.Varsano T, Dong MQ, Niesman I, Gacula H, Lou X, Ma T, et al. GIPC is recruited by APPL to peripheral TrkA endosomes and regulates TrkA trafficking and signaling. Mol Cell Biol. 2006;26:8942–8952. doi: 10.1128/MCB.00305-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng KK, Lam KS, Wang Y, Huang Y, Carling D, Wu D, et al. Adiponectin-induced endothelial nitric oxide synthase activation and nitric oxide production are mediated by APPL1 in endothelial cells. Diabetes. 2007;56:1387–1394. doi: 10.2337/db06-1580. [DOI] [PubMed] [Google Scholar]
- 10.Majumdar D, Nebhan CA, Hu L, Anderson B, Webb DJ. An APPL1/Akt signaling complex regulates dendritic spine and synapse formation in hippocampal neurons. Mol Cell Neurosci. 2011;46:633–644. doi: 10.1016/j.mcn.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang YB, Wang JJ, Wang SH, Liu SS, Cao JY, Li XM, et al. Adaptor protein APPL1 couples synaptic NMDA receptor with neuronal prosurvival phosphatidylinositol 3-kinase/Akt pathway. J Neurosci. 2012;32:11919–11929. doi: 10.1523/JNEUROSCI.3852-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XM, Lv XY, Tang Y, Zhu LJ, Luo JH. Cysteine residues 87 and 320 in the amino terminal domain of NMDA receptor GluN2A govern its homodimerization but do not influence GluN2A/GluN1 heteromeric assembly. Neurosci Bull. 2013;29:671–684. doi: 10.1007/s12264-013-1335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu W, Ai H, Peng L, Wang JJ, Zhang B, Liu X, et al. A novel phosphorylation site of N-methyl-d-aspartate receptor GluN2B at S1284 is regulated by Cdk5 in neuronal ischemia. Exp Neurol. 2015;271:251–258. doi: 10.1016/j.expneurol.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Lu W, Fang W, Li J, Zhang B, Yang Q, Yan X, et al. Phosphorylation of Tyrosine 1070 at the GluN2B Subunit Is Regulated by Synaptic Activity and Critical for Surface Expression of N-methyl-d-aspartate (NMDA) Receptors. J Biol Chem. 2015;290:22945–22954. doi: 10.1074/jbc.M115.663450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB. The majority of N-methyl-d-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B) Mol Pharmacol. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- 16.Kemp JA, McKernan RM. NMDA receptor pathways as drug targets. Nat Neurosci. 2002;5(Suppl):1039–1042. doi: 10.1038/nn936. [DOI] [PubMed] [Google Scholar]
- 17.Salter MW, Pitcher GM. Dysregulated Src upregulation of NMDA receptor activity: a common link in chronic pain and schizophrenia. FEBS J. 2012;279:2–11. doi: 10.1111/j.1742-4658.2011.08390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doucet MV, Harkin A, Dev KK. The PSD-95/nNOS complex: new drugs for depression? Pharmacol Ther. 2012;133:218–229. doi: 10.1016/j.pharmthera.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Cao J, Viholainen JI, Dart C, Warwick HK, Leyland ML, Courtney MJ. The PSD95-nNOS interface: a target for inhibition of excitotoxic p38 stress-activated protein kinase activation and cell death. J Cell Biol. 2005;168:117–126. doi: 10.1083/jcb.200407024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toro C, Deakin JF. NMDA receptor subunit NRI and postsynaptic protein PSD-95 in hippocampus and orbitofrontal cortex in schizophrenia and mood disorder. Schizophr Res. 2005;80:323–330. doi: 10.1016/j.schres.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Li F, Xu HB, Luo CX, Wu HY, Zhu MM, et al. Treatment of cerebral ischemia by disrupting ischemia-induced interaction of nNOS with PSD-95. Nat Med. 2010;16:1439–1443. doi: 10.1038/nm.2245. [DOI] [PubMed] [Google Scholar]
- 22.Li LL, Ginet V, Liu X, Vergun O, Tuittila M, Mathieu M, et al. The nNOS-p38MAPK pathway is mediated by NOS1AP during neuronal death. J Neurosci. 2013;33:8185–8201. doi: 10.1523/JNEUROSCI.4578-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Courtney MJ, Li LL, Lai YY. Mechanisms of NOS1AP action on NMDA receptor-nNOS signaling. Front Cell Neurosci. 2014;8:252. doi: 10.3389/fncel.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maher A, El-Sayed NS, Breitinger HG, Gad MZ. Overexpression of NMDAR2B in an inflammatory model of Alzheimer’s disease: modulation by NOS inhibitors. Brain Res Bull. 2014;109:109–116. doi: 10.1016/j.brainresbull.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Gant-Branum RL, Broussard JA, Mahsut A, Webb DJ, McLean JA. Identification of phosphorylation sites within the signaling adaptor APPL1 by mass spectrometry. J Proteome Res. 2010;9:1541–1548. doi: 10.1021/pr901043e. [DOI] [PMC free article] [PubMed] [Google Scholar]