Our recent estimation of the divergence time and isolation of Death Valley pupfishes, including the iconic Devil's Hole pupfish (DHP), rewrote widespread assumptions about this group. These species were previously assumed to be relic populations isolated over millions of years; our genomic analyses indicated recent colonization of Devil's Hole within the past 105–830 years and frequent gene flow among Death Valley populations [1]. These results understandably attracted substantial attention given the iconic battle for conservation and intense management of DHP [2]; nonetheless, a young age for this species should not diminish its conservation value. Indeed, we argue that the unique natural history of this species makes it a prime candidate for exhibiting one of the fastest mutation rates observed in any vertebrate [3].
Sağlam et al. [4] argue that we overestimated the substitution rate, the rate at which mutations occur and fix between lineages over time, in pupfishes due to our analysis of a concatenated dataset of RADseq loci and therefore underestimated the age of DHP. Specifically, Sağlam et al. argue that a multi-species coalescent analysis would remove our bias and provide strikingly different results. Here, we test this assumption by reanalysing our original RADseq dataset under a multi-species coalescent model and comparing the estimated substitution rates to an analysis of concatenated RADseq loci. It is well known that divergence times estimated from concatenated, multi-gene analyses can overestimate species divergence times due to older gene-tree divergence compared with the true species-tree divergence time, a fact that we both cited [5] and demonstrated in our study: our concatenated gene analysis estimated DHP divergence at 2500–6500 years (fig. 2 of [1]) versus our coalescent analysis using dadi [6] at 105–830 years (fig. 3 and table S2 of [1]). However, the effect of concatenation on substitution rate estimation, as opposed to divergence time, is a new area of inquiry (contra Sağlam et al.'s claims). This is supported by the fact that two recent papers introducing this topic and coining a new term for a similar phenomenon (SPILS: substitutions produced by incomplete lineage sorting) [7,8] were published after our study was published in 2016. To the best of our knowledge, no study has yet estimated the bias due to concatenation of loci into a supermatrix on the estimation of substitution rates.
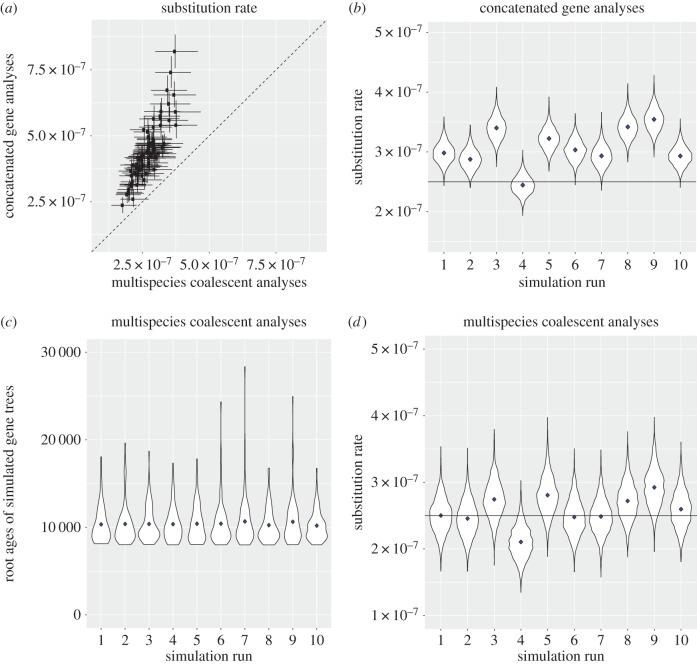
Here, we estimate that gene concatenation results in no more than twofold overestimation of substitution rates in our dataset using 80 subsets of 100 loci each from our original dataset (figure 1a; see data and scripts: doi:10.5061/dryad.9727v [9]). For each dataset, we compared substitution rates under concatenation and a multi-species coalescent approach using the flexible RevBayes software [10].
Figure 1.
(a) Empirical clock rate estimates compared between concatenated supermatrix and multi-species coalescent approaches from 80 subsets of 100 ddRADseq loci each from [1], pruned to the Laguna Chichancanab species and outgroup coastal C. artifrons. (b,d) Substitution rate simulations compared between concatenated supermatrix and multi-species coalescent approaches in 10 datasets simulated on the fixed species tree with Ne = 1000, estimated from the empirical dataset, and a Jukes–Cantor model of nucleotide substitution. (c) Distribution of root ages in the simulated gene trees (n = 1000) across 10 independent runs.
We also estimated substitution rates in 10 simulated datasets using either concatenation or a multi-species coalescent approach (figure 1b,d; see the electronic supplementary material). Each simulated dataset contained 100 loci of 100 bp each simulated under a multi-species coalescent model with an effective population size (Ne) of 1000 constant across all branches of the tree and a substitution rate of 2.5 × 10−7 (i.e. close to our estimate for the Laguna Chichancanab species). We observed the same minimal overestimation of substitution rate that never exceeded twofold in each simulated dataset and the expected increase in substitution rate variance under the multi-species coalescent (figure 1b,d). Most of the gene trees in our simulated analysis diverged at similar times relative to the true species tree age (8000 years) while very few exhibited substantial bias (figure 1c). This is expected given the approximately exponential prior on the distribution of coalescent events (with rate = 1/2Ne) and small Ne. Furthermore, our original concatenated analysis included 16 567 loci (table S2 of [1]) and multi-species coalescent analysis remains impractical for more than 100 loci [10,11]. We also performed simulations assuming an effective population size of 2000 and 5000 and found that twofold overestimation due to concatenation was robust to a realistic range of variation in this parameter (electronic supplementary material, figure S1).
We conclude that our original estimates of DHP divergence time may be revised to no more than 210–1660 years (95% confidence interval across a range of models). These results still rewrite the history of this species as extremely recent and do not challenge any of the conclusions in our original study. Nonetheless, we caution that these inferences are based on a biased reduced-representation sample of the Cyprinodon genome [12] and limited population sampling. For example, excluding loci with missing data disproportionately removes loci with higher mutation rates, resulting in a bias towards underestimation of substitution rate [13].
In contrast to the simplified framing of Sağlam et al. [4], the effect of concatenation on substitution rates is complex and depends on the demographic history of the group under consideration. Both concatenated gene and multi-species coalescent analyses assume a strict bifurcating model of population divergence with no gene flow [5,10,11]. By contrast, we found evidence for substantial secondary gene flow among species in Death Valley, including DHP [1], and have found similar patterns in Caribbean pupfishes [14] and other fish groups [15]. Colonization of Laguna Chichancanab after its initial formation or secondary gene flow from coastal populations would result in underestimation, not overestimation, of substitution rates in concatenated gene and multi-species coalescent phylogenetic analyses [16].
Sağlam et al. [4] use a simplified point calculation to quantify the expected discrepancy in divergence times between concatenated gene and multi-species coalescent analyses. However, their point calculation ignores the enormous amount of variance present in this estimate which has subsequently been addressed by 17 years of investigation in this field [5,6,10,11]. Although we do not advocate for this approach, phylogenetic analyses of concatenated supermatrices are still a common method for estimation of substitution rates in new taxa [7,8].
Furthermore, Sağlam et al. [4] assume an unrealistic Ne of 50 000 in C. artifrons based on a single study of microsatellite variation within a much more widely distributed species; reported Ne ranged from 3500 to 30 000 based on an assumed microsatellite mutation rate which is unknown for this species [17]. Using our multi-species coalescent analysis, we estimated ancestral Ne in the coastal C. artifrons population to be 2000 individuals, similar to estimates for other Caribbean pupfishes which underwent a recent population bottleneck [18,19]. If we use Sağlam et al.'s point estimate, this would result in a calibration of 12 000 years and consequently a 0.5 slower substitution rate and median divergence time of 383 years for DHP. This result does not challenge the major conclusions in our original study.
Finally, Sağlam et al. [4] argue that ‘the typical vertebrate mutation rate’ of 1 × 10−8 mutations/site/generation is appropriate for DHP. This is the rate for humans [20,21], not teleosts [22], and ignores the negative scaling of mutation rate with Ne over more than an order of magnitude demonstrated in the reference Sağlam et al. cite for their claim [20]. Population genetic principles predict that smaller populations are expected to exhibit higher mutation rates due to the weaker strength of purifying selection relative to genetic drift in removing alleles that increase the mutation rate [20]. Indeed, DHP inhabits the smallest species range known, with the smallest Ne ever observed in a natural population [1], and may reasonably exhibit one of the fastest mutation rates observed in a vertebrate. Moreover, the unique life-history characteristics of the DHP—small size, short generation time, high metabolism, high temperatures and severe environmental stressors, such as starvation—are each associated with increased mutation rates [23–25]. These characteristics, which are often shared across Cyprinodon species, suggest that we should expect higher mutation rates in pupfishes, and DHP in particular, than the typical teleost [3].
In conclusion, we have demonstrated that our original concatenated gene analysis may have overestimated substitution rate, and subsequently underestimated the age of DHP, by a maximum of no more than twofold. Substitution rate may also have been underestimated due to widespread gene flow and stringent filtering. These results do not in any way challenge our original major conclusions of a rapid time scale for speciation, genetic assimilation and the evolution of intrinsic reproductive incompatibilities in pupfishes [1]. Furthermore, nearly all unique life-history characteristics of DHP suggest that its mutation rate may indeed be unprecedented among vertebrates. We consider this line of inquiry to deserve greater attention, rather than dismissal.
Supplementary Material
Footnotes
The accompanying comment can be viewed at http://dx.doi.org/10.1098/rspb.2017.0021.
Data accessibility
All data and scripts are available from Dryad Digital Repository (doi:10.5061/dryad.9727v).
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Martin CH, Crawford JE, Turner BJ, Simons LH. 2016. Diabolical survival in Death Valley: recent pupfish colonization, gene flow, and genetic assimilation in the smallest species range on earth. Proc. R. Soc. B 283, 20152334 ( 10.1098/rspb.2015.2334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoike SL, Pister EP. 2010. Threatened fishes of the world: Cyprinodon diabolis (Wales, 1930). Environ. Biol. Fishes 88, 399–400. ( 10.1007/s10641-010-9651-8) [DOI] [Google Scholar]
- 3.Martin C, Höhna S. In revision New evidence for the recent divergence of Devil's Hole pupfish and the plausibility of elevated mutation rates in endangered taxa. Mol. Ecol. [DOI] [PubMed] [Google Scholar]
- 4.Sağlam İK, Baumsteiger J, Miller MR. 2017. Failure to differentiate between divergence of species and their genes can result in over-estimation of mutation rates in recently diverged species. Proc. R. Soc. B 284, 20170021 ( 10.1098/rspb.2017.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794. ( 10.1016/j.cell.2014.03.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 ( 10.1371/journal.pgen.1000695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendes FK, Hahn MW. 2016. Gene tree discordance causes apparent substitution rate variation. Syst. Biol. 65, 711–721. ( 10.1093/sysbio/syw018) [DOI] [PubMed] [Google Scholar]
- 8.Ogilvie HA, Drummond AJ. 2017. StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. msx126 ( 10.1093/molbev/msx126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin CH, Höhna S, Crawford JE, Turner BJ, Richards EJ, Simons LH. 2017. Data from: The complex effects of demographic history on the estimation of substitution rate: concatenated gene analysis results in no more than twofold overestimation. Dryad Digital Repository. ( 10.5061/dryad.9727v) [DOI] [PMC free article] [PubMed]
- 10.Höhna S, Landis M, Heath T, Boussau B, Lartillot N, Moore B, Huelsenbeck J, Ronquist F. 2016. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 65, 726–736. ( 10.1093/sysbio/syw021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heled J, Drummond AJ. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580. ( 10.1093/molbev/msp274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold B, Corbett-Detig RB, Hartl D, Bomblies K. 2013. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol. Ecol. 22, 3179–3190. ( 10.1111/mec.12276) [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Knowles LL. 2016. Unforeseen consequences of excluding missing data from next-generation sequences: simulation study of RAD sequences. Syst. Biol. 65, 357–365. ( 10.1093/sysbio/syu046) [DOI] [PubMed] [Google Scholar]
- 14.Martin CH. 2016. The cryptic origins of evolutionary novelty: 1000-fold faster trophic diversification rates without increased ecological opportunity or hybrid swarm. Evolution 70, 2504–2519. ( 10.1111/evo.13046) [DOI] [PubMed] [Google Scholar]
- 15.Martin CH, Cutler JS, Friel JP, Dening Touokong C, Coop G, Wainwright PC. 2015. Complex histories of repeated gene flow in Cameroon crater lake cichlids cast doubt on one of the clearest examples of sympatric speciation. Evolution 69, 1406–1422. ( 10.1111/evo.12674) [DOI] [PubMed] [Google Scholar]
- 16.Ho SYW, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087–3101. ( 10.1111/j.1365-294X.2011.05178.x) [DOI] [PubMed] [Google Scholar]
- 17.Duvernell DD, Lindmeier JB, Faust KE, Whitehead A. 2008. Relative influences of historical and contemporary forces shaping the distribution of genetic variation in the Atlantic killifish, Fundulus heteroclitus. Mol. Ecol. 17, 1344–1360. ( 10.1111/j.1365-294X.2007.03648.x) [DOI] [PubMed] [Google Scholar]
- 18.McGirr JA, Martin CH. 2017. Novel candidate genes underlying extreme trophic specialization in Caribbean pupfishes. Mol. Biol. Evol. 34, 873–888. ( 10.1093/molbev/msw286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards E, Martin C. In press. Adaptive introgression from distant Caribbean islands contributed to the diversification of a microendemic radiation of trophic specialist pupfishes. bioRxiv. ( 10.1101/115055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M. 2010. Evolution of the mutation rate. Trends Genet. 26, 345–352. ( 10.1016/j.tig.2010.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smeds L, Qvarnstrom A, Ellegren H. 2016. Direct estimate of the rate of germline mutation in a bird. Genome Res. 26, 1211–1218. ( 10.1101/gr.204669.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaillon O, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957. ( 10.1038/nature03025) [DOI] [PubMed] [Google Scholar]
- 23.Baer CF, Miyamoto MM, Denver DR. 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8, 619–631. ( 10.1038/nrg2158) [DOI] [PubMed] [Google Scholar]
- 24.Martin AP, Palumbi SR. 1993. Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA 90, 4087–4091. ( 10.1073/pnas.90.9.4087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji J, et al. 2012. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells 30, 435–440. ( 10.1002/stem.1011) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Martin CH, Höhna S, Crawford JE, Turner BJ, Richards EJ, Simons LH. 2017. Data from: The complex effects of demographic history on the estimation of substitution rate: concatenated gene analysis results in no more than twofold overestimation. Dryad Digital Repository. ( 10.5061/dryad.9727v) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data and scripts are available from Dryad Digital Repository (doi:10.5061/dryad.9727v).