Abstract
Self-replicating gene drives that can spread deleterious alleles through animal populations have been promoted as a much needed but controversial ‘silver bullet’ for controlling invasive alien species. Homing-based drives comprise an endonuclease and a guide RNA (gRNA) that are replicated during meiosis via homologous recombination. However, their efficacy for controlling wild populations is threatened by inherent polymorphic resistance and the creation of resistance alleles via non-homologous end-joining (NHEJ)-mediated DNA repair. We used stochastic individual-based models to identify realistic gene-drive strategies capable of eradicating vertebrate pest populations (mice, rats and rabbits) on islands. One popular strategy, a sex-reversing drive that converts heterozygous females into sterile males, failed to spread and required the ongoing deployment of gene-drive carriers to achieve eradication. Under alternative strategies, multiplexed gRNAs could overcome inherent polymorphic resistance and were required for eradication success even when the probability of NHEJ was low. Strategies causing homozygotic embryonic non-viability or homozygotic female sterility produced high probabilities of eradication and were robust to NHEJ-mediated deletion of the DNA sequence between multiplexed endonuclease recognition sites. The latter two strategies also purged the gene drive when eradication failed, therefore posing lower long-term risk should animals escape beyond target islands. Multiplexing gRNAs will be necessary if this technology is to be useful for insular extirpation attempts; however, precise knowledge of homing rates will be required to design low-risk gene drives with high probabilities of eradication success.
Keywords: gene drive, homing, non-homologous end joining, resistance allele, population eradication, island conservation
1. Introduction
The concept of using ‘selfish’ genetic elements, with biased inheritance, to spread phenotypic traits through wild populations has been discussed for over 50 years [1]. Inspired by the self-replication mechanism of naturally occurring homing endonuclease genes, Burt [2] was the first to propose that synthetic gene drives could be engineered to target essential host genes for the purpose of population control. Self-replication (or ‘homing’) of gene drives is achieved by expression of a site-specific endonuclease (from the gene-drive transgene) that cleaves the partner chromosome at the same genomic site in which the gene drive is integrated. Repair of the double-stranded break via homologous recombination (HR) generates a second copy of the gene drive, resulting in homozygosity. Gene drives are predicted to spread rapidly through a given population because greater than 50% of progeny will inherit the genetic element. Importantly, by restricting the homing event to the germline, it should be possible to propagate a recessive mutation that causes infertility or lethality, which would eventually lead to population decline [3]. Alternatively, population suppression could be achieved by adding genetic ‘cargo’ that causes all offspring that inherit the gene drive to develop as a single sex.
While proof-of-concept gene drives, using naturally occurring and modular endonucleases, have been developed in insects [4,5], these platforms lack the requisite flexibility and stability for field applications. By contrast, recent research demonstrates that the CRISPR/Cas9 genome editing system could provide a mechanism for effective population control by gene drives [6]. These advances have generated tremendous excitement among the agricultural, conservation and health science communities because they offer potential solutions to costly, long-standing problems such as the control or eradication of invasive species [7,8] and the suppression of animal vectors of human disease [9]. Some now view gene drives as a ‘silver bullet’ for conservation science and question not whether this technology is viable but whether it should be used [10], citing the risks associated with the dispersal or human-mediated transport of gene-drive carriers beyond the laboratory or the population targeted for management [11–13]. Here, we critically examine the feasibility of suppressing or eradicating large populations of pest vertebrates using CRISPR/Cas9 technology.
The viability of gene drives for population control depends, in part, on the technical efficacy of the CRISPR/Cas9 system, which comprises two components: Cas9, which functions as a ‘programmable’ endonuclease, and a guide RNA (gRNA) containing a 20 bp sequence that is complementary to the target site within the genome. Recently, germline-homing CRISPR/Cas9 gene drives have been published for the fruit fly Drosophila melanogaster [14], the mosquitoes Anopheles stephensi [15] and Anopheles gambiae [16], and the yeast Saccharomyces cerevisiae [12]. Remarkably, homing rates in these studies are extremely high (greater than 95%) in most cases and maintained over several generations, indicating that CRISPR/Cas9 gene drives are stable. Although HR repair predominates, a competing repair pathway termed non-homologous end-joining (NHEJ) can also repair gene-drive-mediated DNA cleavage. Importantly, NHEJ repair can generate small insertions or deletions (indels) at the gRNA binding site and/or associated PAM (protospacer adjacent motif) sequence that will be resistant to subsequent cleavage by the encoded endonuclease. Even though the probability of NHEJ appears to be low [12,13,15,16], the creation of resistance alleles through NHEJ, and the subsequent selection favouring their spread, provides one mechanism whereby a targeted population might rebound following an initial decline in abundance [15,17]. To achieve eradication success, it is clear that the target population must be extirpated before the proportion of resistance alleles becomes sufficient to produce a positive population growth rate once more. To increase the probability of successful homing and reduce the rate of resistance-allele creation, gene drives could be designed with multiplexed gRNAs that target adjacent DNA recognition sites for cleavage [6,18–20].
As the CRISPR/Cas9 system is an efficient genome editing tool in a wide variety of species, including mice [21,22], it seems likely that gene drives will soon be developed in mammals. Furthermore, deployments on islands are most likely in the first instance to minimize the risks associated with the unplanned dispersal or transport of gene-drive carriers [23]. Invasive alien vertebrates are one of the major threats to island biodiversity [7,23,24] and exotic rodents are likely responsible for the greatest number of extinctions and ecosystem changes on islands [25,26]. Exotic vertebrates have also been devastating for human agriculture [27], costing individual economies millions of dollars in lost production. In Australia, for example, annual agricultural losses due to the house mouse (Mus musculus) and European rabbit (Oryctolagus cuniculus) are estimated at AU$148 million and these pests also have severe environmental and social impacts [28]. Although the success of vertebrate eradication attempts has increased in recent decades, particularly on islands [29], these attempts are typically extremely costly. In addition, current control methods (e.g. the broad-scale delivery of toxic baits used to kill invasive rodents) need to address many social, political and ethical considerations [30]. The eradication of alien rodents on islands therefore provides the perfect model system for testing the efficacy of introducing a novel technology into the toolbox of vertebrate eradication science.
Here, we use in silico experiments to test the efficacy of different gene-drive strategies for eradicating exotic vertebrates from islands. Initially, we focus on mice, which we believe will be among the first vertebrate species targeted for gene-drive development because: (i) mice are widely used as an experimental model organism; (ii) the molecular genetic control of sex determination and fertility in mice is well understood and can therefore be exploited for gene-drive design strategies; and (iii) mice have frequently been transported beyond their native ranges and have severe environmental and economic consequences in their exotic ranges. Specifically, we test four realistic CRISPR/Cas9 gene-drive strategies that could be readily developed based on existing literature (figure 1). To replicate the trajectory of island populations inoculated with gene-drive carriers as realistically as possible, we develop an individual-based model (IBM) that simulates genetic inheritance while incorporating the impacts of demographic stochasticity on the outcome of the gene-drive inoculation. Using this system, we explicitly consider the potential for evolution to fight back through the formation and spread of resistance alleles, and explore the conditions under which this mechanism could allow populations of mice to ‘escape’ extirpation. Having identified gene-drive strategies that could feasibly achieve the eradication of mice on islands, we compare the efficacy of these designs for two other vertebrate pests, black rats (Rattus rattus) and European rabbits.
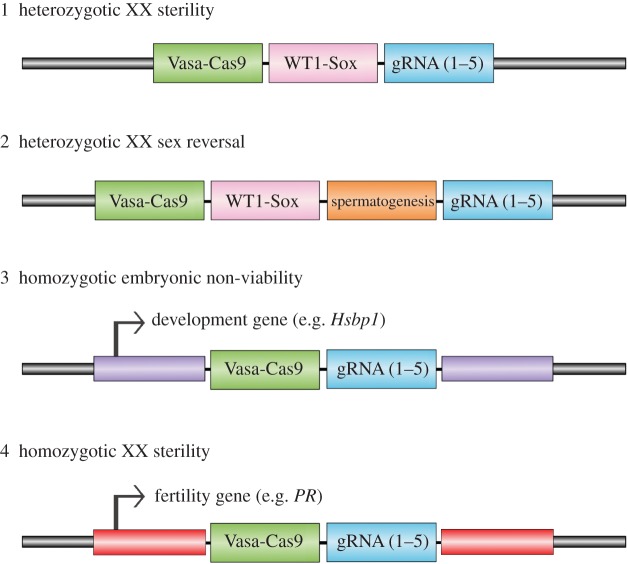
Figure 1.
Schematic representation of potential gene-drive constructs for each of the four strategies modelled in this study. Expression cassettes for each of the gene-drive components are indicated by coloured boxes. The purple (construct 3) and red (construct 4) indicate endogenous genes that are essential for embryonic development and fertility, respectively. Arrows indicate translational start sites. Not drawn to scale.
2. Material and methods
(a). Model overview
We developed a bespoke IBM to explore the probability of achieving the eradication of vertebrate pest species on islands using gene-drive technology (for details of the model structure, see electronic supplementary material, figure S1). The model assumed a discrete-time, pre-breeding census design [31] for populations of different sizes that were inoculated by a number of ‘introduced’ gene-drive carriers. Simulated pest populations were represented as a collection of individuals characterized by the following state variables: age, maternal and paternal allosomes (X/Y), the status of maternal and paternal alleles targeted by the gene drive (gene-drive-positive, or the number of susceptible gRNA recognition sites remaining for gene-drive-negative alleles), genotypic sex (male/female), phenotypic sex (male/female) and fertility status (sterile/fertile). We tracked individuals as they transitioned through the following stages of each breeding cycle: mate allocation, reproduction with autosome and allosome inheritance, gene-drive homing and resistance allele formation in the germlines of heterozygous individuals, phenotypic sex allocation, density-dependent mortality and ageing. Annual supplementation with additional gene-drive carriers was considered, as was the possibility that some individuals of a target population might possess some innate resistance to the gene-drive homing mechanism due to nucleotide polymorphisms that alter the gene-drive PAM or gRNA target sequence [32,33]. Our IBM explicity accounted for random demographic and genetic factors, allowing for realistic simulations of the fate of populations for which gene-drive eradication is attempted. We coded the model using the R computing environment [34].
(b). Gene-drive strategies
We use the term gene-drive strategy to refer to the demographic impact on individuals that are homozygous or heterozygous for the gene drive. We tested the following gene-drive strategies that could potentially be developed based on existing literature:
Strategy 1—Heterozygotic XX sterility. The aim of this ‘daughterless’ strategy is to drive an XX male sex-reversing transgene through the population to generate a deficiency of female breeding stock. Several members of the Sox gene family have been demonstrated to induce male development in XX mice, including Sry (the Y-linked mammalian testis determining factor; [35]) and Sox9 (an Sry target gene; [36]). The latter option is attractive because Sox9 has an evolutionarily conserved role in vertebrate sex determination and has been validated using the relatively compact (WT-1) promoter. Importantly, XX male carriers of the gene drive would be sterile, in part because they lack Y-linked spermatogenesis genes. The gene drive would not be expected to affect the reproductive fitness of XY males [37], thereby enabling gene-drive transmission. This gene-drive construct could be positioned autosomally in an intergenic region or at a known ‘safe harbour’ locus such as Rosa26 [38].
Strategy 2—Heterozygotic XX sex reversal. This strategy is similar to (1) but contains additional cargo that enables XX males to transmit the gene drive. The approach is based on recent transgenic studies, indicating that expression of Sry and the Y-linked spermatogenesis gene Eif2s3y (or Sox9 and its X-linked homologue Eif2s3x) in XO mice is sufficient to generate reproductively competent males [39,40]. Therefore, through incorporation of multiple Y-linked spermatogenesis genes (or their X chromosome homologues) as gene-drive cargo, it may be possible to restore normal fertility to XX sex-reversed gene-drive carrier mice. However, we stress that spermatogenesis in the male XO transgenic mice is not normal and that they require assisted reproduction to generate offspring. Furthermore, meiotic block of XX germ cells in the testis would need to be circumvented [41]. Therefore, from a practical standpoint, this strategy would require considerable work-up before deployment.
Strategy 3—Homozygotic embryonic non-viability. For this strategy, the gene-drive cassette does not carry cargo but instead generates a loss-of-function mutation in a gene that causes recessive embryonic lethality. Provided that the mutation is haplo-sufficient and the gene drive is propagated through germline homing (see below), this approach should initially allow rapid spread of the gene drive and an accumulation of (somatic) heterozygous individuals. However, as mating between heterozygous carriers becomes increasingly likely, the population should decline due to embryonic lethality of their progeny arising from gene-drive homozygosity. Positioning the gene-drive element in an exon would confer loss-of-function for the gene-drive allele via premature termination of the open reading frame. Deletion-containing alleles created by NHEJ would also result in loss-of-function if a frameshift mutation were generated [3,17]. Given that recessive embryonic or perinatal lethality occurs in about one-third of KO mice [42], there is a plethora of target loci that could be considered for this approach, provided development of null oocytes or spermatogonia is not compromised by germline homing. To limit the fitness cost of carrying moribund embryos in pregnant females, it would be prudent to select a gene-drive target gene that causes embryonic lethality prior to implantation (e.g. Hsbp1; [43]).
Strategy 4—Homozygotic XX sterility. This strategy again assumes exonic placement and is similar to (3) except that population suppression is achieved through infertility of homozygous females instead of recessive embryonic lethality. While mutations in several genes are known to cause recessive infertility in female mice, a significant caveat here is that homozygosity in the germline (induced by germline homing) must not compromise oogenesis. One attractive candidate locus for gene-drive localization is the progesterone receptor (PR; [44]), mutation of which has been shown to cause recessive female infertility. However, before embarking on this strategy, confirmation that germline null females are fertile is required (using Vasa-Cre and a PR floxed allele; [45]).
(c). Multiplexed guide RNA expression
All of the CRIPSR/Cas9 gene drives published to date have used a single gRNA to direct site-specific homing [12,14–16]. A disadvantage of this strategy is that indels generated by NHEJ-mediated DNA repair will very likely become resistant to homing due to modification of the gRNA binding sequence and/or the associated PAM. A potential solution to this issue is to employ multiple gRNAs that cleave at several closely spaced sites across the target region [6,18]. From a practical standpoint, this could be achieved using U6 promoters from different species (to avoid recombination-mediated instability) [46] or possibly via expression of a polycistronic transcript containing multiple gRNAs using an RNA polymerase II promoter [47,48]. Instability caused by conserved gRNA sequences could potentially be avoided using diverse gRNA scaffolds [49]. Methodology for sequential gRNA expression in the germline requires development.
(d). Genetics
We assumed Mendelian inheritance of autosomes and allosomes and modelled the stochastic transfer of chromosomes from parents to offspring with Bernoulli distributions. We assumed germline-specific homing for all gene-drive strategies tested such that homing occurred prior to meiosis in the germlines of males and females that are heterozygous for the gene drive (i.e. dual-germline homing) [17]. From a practical perspective, germline-specific homing could potentially be achieved by driving Cas9 expression with the previously validated Vasa promoter, although modification of regulatory sequences may be required to avoid Cas9 activity in oocytes that lack the transgene [45]. As ubiquitous robust Cas9 expression does not cause any overt phenotypic impact [50], we assumed there is no fitness cost for gene-drive carriers.
Within each gene-drive strategy simulated, we tested different numbers (1–5) of gRNAs that were assumed to target unique recognition (cutting) sequences, thereby influencing the probabilities of successful homing and resistance-allele creation. The outcome of a homing event at each individual cutting site is governed by the probability of cutting occurring (PC) and the probability of NHEJ conditional on cutting having occurred (PN). Assuming S gRNAs targeting S unique cutting sites, the state into which a wild-type allele can move during homing is conditional on the number of susceptible sites it currently possesses. If we assume that Cas9-mediated cutting occurs sequentially at each recognition site, then each cutting event is independent and the probability of a wild-type allele moving from s to j susceptible sites (Psj) is:
where the allele is left unchanged when j = s because cutting has failed at all s susceptible sites, while a resistance allele is created when j = 0 because cutting and NHEJ has occurred at all s sites. The probability of successful homing is simply 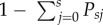 . Based on data for non-vertebrate species, we assumed a cutting probability of 95% (i.e. PC = 0.95) for each gRNA simulated, and a realistic estimate of the probability of NHEJ conditional on cutting having occurred equal to 2% (i.e. PN = 0.02) [16]. Using this parametrization, we tested gene-drive strategies against a reference scenario with a single gRNA and no NHEJ (PN = 0). For the two strategies that used exonic placement of gene drives, we assumed that each indel created carried a two-thirds probability of disrupting the target gene [20].
. Based on data for non-vertebrate species, we assumed a cutting probability of 95% (i.e. PC = 0.95) for each gRNA simulated, and a realistic estimate of the probability of NHEJ conditional on cutting having occurred equal to 2% (i.e. PN = 0.02) [16]. Using this parametrization, we tested gene-drive strategies against a reference scenario with a single gRNA and no NHEJ (PN = 0). For the two strategies that used exonic placement of gene drives, we assumed that each indel created carried a two-thirds probability of disrupting the target gene [20].
By contrast, if multiplexed gRNAs are expressed simultaneously, then deletions of intervening sequences might also occur due to NHEJ. Assuming that the probability of NHEJ-mediated DNA repair is independent of the distance between target sites, the probability of successful homing is:
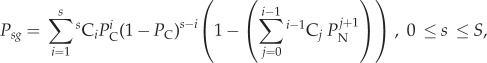 |
while calculations for all Psj are more complex (see the electronic supplementary material, appendix S1). We compared simulation results for multiplexed gRNAs under the assumption of sequential or simultaneous cutting. For the latter, we assumed that any deletion of intervening sequence between two recognition sites resulted in disruption (i.e. loss of function) of the target gene, and that at least one functional allele was required for embryonic viability (Strategy 3) or XX fertility (Strategy 4).
(e). Demography
For simplicity, we assumed constant survival and fertility rates across all age classes, an equal sex ratio at birth and a fixed generation time. We also assumed a polygynous breeding system such that males could mate with multiple females while fertile females could only mate with a single male per breeding cycle. To ensure stable pest populations when at carrying capacity (K), we set fertility rates based on empirical litter-size data per breeding cycle for wild populations of each species, and then calculated the survival rates required to produce a population growth rate (r) of zero. To allow for improvements in species vital rates at low densities, we assumed θ-logistic population growth mediated through variation in the survival rate p such that:
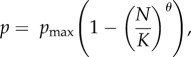 |
where N is the current population size, and pmax is the maximum survival rate at low population density which we set to produce the maximum population growth rate (rmax) estimated for the species. We assumed a shape parameter θ equal to 2 to ensure that the population growth rate was only reduced substantially as the population size approached K. To incorporate the effects of demographic stochasticity (i.e. variation in the population growth rate that occurs even if the mean demographic rates remain constant), we modelled the outcome of all survival probabilities with Bernoulli distributions and sampled the number of offspring produced by each reproducing female from Poisson distributions. Full details of the demographic parameters used are found in electronic supplementary material, table S1.
(f). Initiation and model output
We initiated each simulated population at K, assuming an equal number of mature males and females. We then simulated the eradication attempt as the addition of 100 males that were somatic heterozygous for the gene-drive cassette. To account for demographic and genetic stochasticity, we performed 1000 replicate simulations of the model for each parametrization tested. Using these replicates, we compared gene-drive strategies by calculating the probability of population eradication and the mean time to eradication. We defined successful eradication as the reduction in the target population size to zero or the complete loss of one sex.
(g). Sensitivity analyses
Although PC for CRISPR/Cas9 gene drives has been shown to be very high for different taxa, two parameters are likely critical determinants of the viability of gene drives for pest eradication: (i) the probability of NHEJ (PN); and (ii) the probability of inherent gene-drive resistance (PR) due to nucleotide polymorphism at sequences targeted by the gRNAs. Assuming a simulated pest mouse population of 50 000 individuals, we conducted a sensitivity analysis on PN, PR and the number of multiplexed gRNAs, generating 100 000 unique combinations of these three parameters using latin hypercube sampling [51]. We then ran a single simulation per parameter set [52] and examined the influence of these parameters on the probability of successful eradication. We assumed that PR at all cutting sites was equal and independent, and simulated initial resistance by sampling the number of susceptible sites (s) on each autosome in the population from binomial distributions with probability parameter equal to (1−PR) and size parameter equal to the number of gRNAs.
For the homozygotic embryonic non-viability strategy, we also tested the possible influence of reproductive compensation following the mating of two gene-drive carriers, whereby embryonic survival post-implantation was increased when the number of implants was low following the death of homozygous embryos. To achieve this, we modified the reproduction component of the original model and assumed instead that each mating resulted in 10 embryos [53]. After accounting for homozygotic non-viability prior to implantation, the embryonic survival rate was modified as a function of the number of implants, to compare the output of models that included: (i) no compensation (i.e. a constant embryonic survival rate of 0.6, resulting in six offspring per female mouse on average); (ii) linear compensation; or θ-logistic compensation (see electronic supplementary material, figure S2 for details).
3. Results and discussion
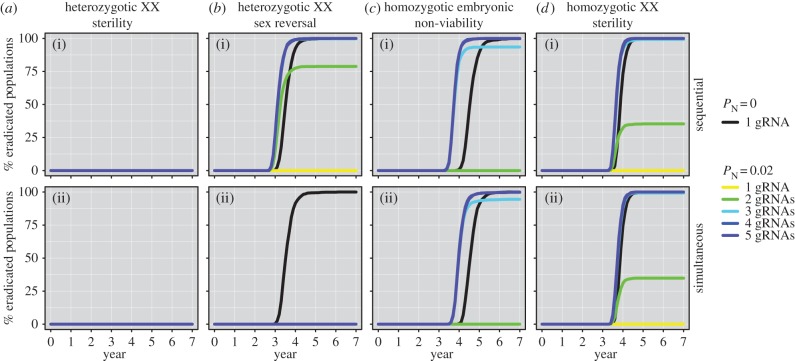
Our results illustrate clear differences in efficacy between different gene-drive eradication strategies, and highlight that selection favouring the spread of resistance alleles could allow populations to escape gene-drive suppression. Assuming an island carrying capacity of K = 50 000 mice, the heterozygotic XX sterility (‘daughterless’) strategy failed to cause a single eradication (figure 2a), nor indeed successfully suppress the population size. By contrast, the remaining three strategies are capable of causing rapid population decline to eradication, despite only 100 gene-drive carriers being introduced initially (figure 2b–d). However, in all cases, multiplexed gRNAs were required for eradication even when the probability of NHEJ was assumed to be low (PN = 0.02).
Figure 2.
The percentage of eradicated mouse populations over time for different gene-drive strategies, assuming a starting population size of 50 000 individuals and either sequential (independent) or simultaneous gRNA activity on multiple DNA recognition sites. The gene-drive strategies are: (a) heterozygotic XX sterility; (b) heterozygotic XX sex reversal; (c) homozygotic embryonic non-viability; and (d) homozygotic XX sterility. Within each strategy, different numbers of multiplexed mRNA guides were tested. Results are shown for simulations that assume the probability of NHEJ occurring following cutting is 0 or 2% (i.e. PN = 0 or 0.02). Note that no simulated eradications occurred for the first gene-drive strategy, so all lines overlap in a.
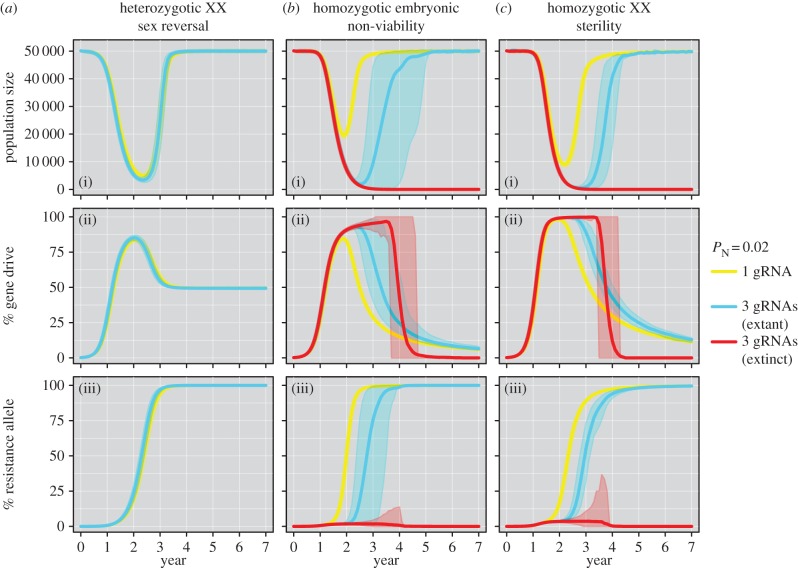
For the three scenarios capable of spreading the gene drive (heterozygotic sex reversal, homozygotic embryonic non-viability and homozygotic XX sterility), initial population suppression can be followed by recovery if resistance alleles develop and spread throughout the population (figure 3). With PN = 0.02 [16] and a single gRNA, population recovery was guaranteed (i.e. occurred in 100% of iterations) for all simulated mouse populations (figures 2 and 3). This result is consistent with that of Eckhoff et al. [17], who recently modelled gene-drive spread in mosquitoes and found that if resistance alleles formed by NHEJ have wild-type fecundity then it is unlikely that gene-drive suppression will succeed. They reasoned (although they did not test) that multiple DNA cleavage targets could solve this problem. Our results agree with this hypothesis, showing that as the number of multiplexed (and independently expressed) gRNAs was increased, the rate of resistance-allele creation was slowed and eradication became more likely (figure 2). However, the efficacy of payload gene drives positioned within introns (e.g. Strategies 1 and 2) will be severely compromised if multiplexed gRNA expression is simultaneous such that the NHEJ-mediated deletion of sequence spanning recognition sites is possible (figure 2b). Under this assumption, the probability of successful homing can decrease as additional gRNAs are incorporated and thereby promote the creation of resistance alleles that are favoured by selection (figure 3a; electronic supplementary material, S3).
Figure 3.
Mean mouse population trajectories for gene-drive strategies and two example designs employing one or three mRNA guides, assuming simultaneous gRNA activity on multiple DNA recognition sites. The gene-drive strategies are: (a) heterozygotic XX sex reversal; (b) homozygotic embryonic non-viability; and (c) homozygotic XX sterility. These trajectories assume an island carrying capacity (and starting population size) of 50 000 individuals and illustrate means derived from stochastic simulations for: (i) the population size; (ii) the percentage of the population with at least one gene-drive allele; and (iii) the percentage of the population with at least one resistance allele (i.e. at least one allele with no susceptible gRNA recognition sites remaining). Ribbons represent 95% CIs.
By contrast, strategies involving the exonic placement of disruptive gene drives are robust to inter-site deletions because this produces non-functional alleles (i.e. functionally equivalent to a successful homing event) that will be eliminated by selection (figure 2c,d). For the strategy of homozygotic XX sterility under simultaneous gRNA expression, for example, the percentage of eradicated populations increased from 35% with two gRNAs, to 99% with three gRNAs, and then to 100% with four and five gRNAs (figure 2d). For the homozygotic embryonic non-viability strategy, simulating reproductive compensation post-implantation did not impact model outputs substantially (electronic supplementary material, figure S2) because the majority of embryos produced by crossing heterozygotic gene-drive carriers are non-viable. It is noteworthy that eradication success was not guaranteed for many scenarios, which partly reflects the importance of demographic and genetic stochasticity once populations have been suppressed by the gene drive. Once a small population size is achieved, random increases in the frequency of resistance alleles can allow a population to ‘escape’ eradication and grow once more (figure 3). This effect could potentially be mitigated by targeting coding sequences that are essential for protein function such that in-frame deletions also confer loss-of-function.
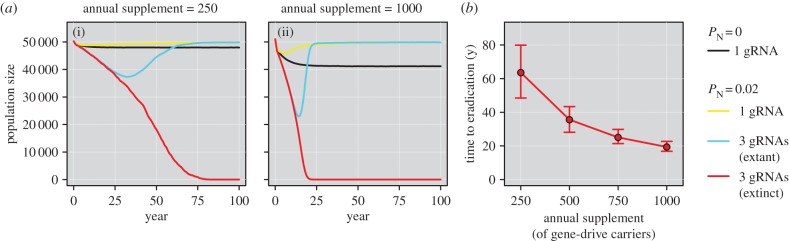
The poor performance of a gene-drive strategy that causes heterozygotic XX sterility (figure 2a) is explained by the failure of the gene drive to spread because heterozygous females cannot pass on the construct. Consequently, the gene drive can also be lost from the population due to demographic and genetic stochasticity. Although previous modelling suggests this approach (employing a naturally occurring gene drive) could work for vertebrates, it relies on the regular continued release of gene-drive carriers into the pest population [54]. Our simulations confirm this result and demonstrate that large numbers of gene-drive carriers are required to achieve eradication with this strategy, even under the optimistic assumption of independent gRNA expression (figure 4a). For example, assuming a starting mouse population size of 50 000, annual supplementation of 250 gene-drive carriers and three gRNAs for the gene drive, our results indicate only a 2% probability of eradication. The mean time to extirpation for these same populations was also very long (64 years; figure 4b) and corresponds to a total introduction effort of 16 000 individuals. We conclude that, with the exception of extremely small island populations, this gene-drive strategy will not be feasible due to the prohibitively large laboratory costs required for success and the long expected times to eradication. A further problem is that the regular introduction of large numbers of gene-drive carriers would render the evaluation of eradication success very difficult. However, this strategy might be suitable for a low-risk, proof-of-concept field trial on an island with a very small pest vertebrate population.
Figure 4.
The influence of annual supplementation of gene-drive carriers on simulated mouse populations assuming the ‘daughterless’ strategy causing heterozygotic XX sterility, and a starting population size of 50 000 individuals. (a) Mean population trajectories for annual supplements of (i) 250 individuals and (ii) 1000 individuals. Confidence intervals are omitted to improve clarity. (b) Mean time to eradication (±95% CIs) as a function of annual supplement size assuming an NHEJ probability of 0.02 and three gRNAs.
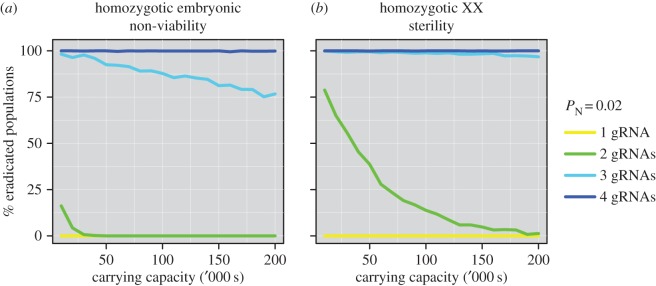
For the exonic, disruptive drives capable of eradicating a population assuming simultaneous gRNA expression (Strategies 3 and 4), there is an interaction between the probability of eradication success and the size of the target pest population (figure 5). The larger the population, the more generations that are required to achieve eradication and therefore the more opportunity for resistance alleles to be created and subsequently spread. However, our results suggest that using four or more gRNAs could produce high probabilities of eradication up to the maximum population size tested of 200 000 individuals (figure 5). This result held across all three species tested (mice, rats and rabbits) and, although mean times to eradication were inversely correlated with the pace of life history, again there was little to distinguish these two gene-drive strategies (electronic supplementary material, figure S4). Similarly, as more gRNAs were multiplexed, the mean time to eradication success changed little (electronic supplementary material, figure S4).
Figure 5.
The interaction between gene-drive design and the carrying capacity of an island for mice. The percentage of eradicated simulated mouse populations assuming simultaneous gRNA expression and different carrying capacities is shown for: (a) homozygotic embryonic non-viability; and (b) homozygotic XX sterility.
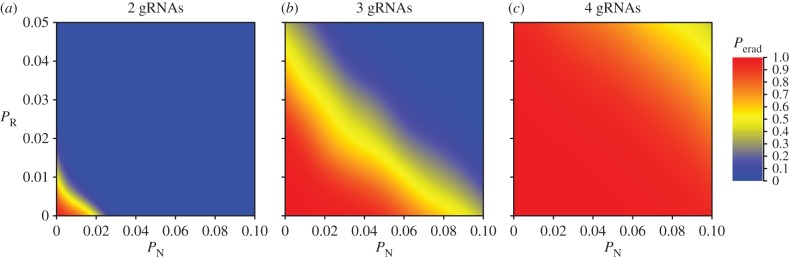
Increasing PN above 0.02 drastically reduced the probability of eradication (figure 6). For simulated mouse populations and a gene drive causing homozygotic XX sterility, for example, even three gRNAs were insufficient to guarantee success with PN as high as 0.1 (figure 6c). Clearly, empirical research is required to derive accurate estimates of this critical parameter in target species and to inform future modelling efforts. While it is also possible that an eradication attempt might fail due to naturally occurring resistance alleles that are carried by a small proportion of individuals [32,33], our results suggest that multiplexing gRNAs could overcome this problem. Assuming PR at each site is independent, multiplexing gRNAs could guard against polymorphic resistance by reducing the effective number of resistance alleles (i.e. alleles with no susceptible sites) that exist in the target population.
Figure 6.
The impact of the probability of NHEJ (PN) and existing polymorphic resistance (PR) on the probability of successful mouse eradication (Perad) under the homozygotic XX sterility gene-drive strategy. The results shown assume an island carrying capacity of 50 000 mice, 100 gene-drive carriers used for inoculation, simultaneous gRNA expression and (a–c) two, three or four gRNAs. The plotted probabilities are derived from a binomial spatial spline fitted to the sensitivity-analysis output separately for each panel.
In practice, the design of gene drives for pest control is likely to trade off eradication efficacy against the genetic consequences for the target population should eradication fail. If this occurs, an efficient purging mechanism will be beneficial to reduce the risk of dispersal or transport of gene-drive positive animals to other regions [6]. If eradication were attempted with the heterozgotic XX sex reversal strategy but failed subsequently, the gene-drive construct would persist in males because there is no negative effect of the drive and hence no effective purging mechanism (figure 3a(ii)). By contrast, following eradication failure, the gene drive is slowly purged from the population under both the homozygotic embryonic non-viability and XX sterility scenarios. A trade-off exists that must be considered when choosing a gene-drive strategy—gene-drive spread is best achieved by strategies that have little effect on, or potentially increase [54], individual fitness whereas gene-drive purging is promoted if the fitness of gene-drive carriers is reduced. However, even a successful gene-drive eradication attempt would likely require gene-drive-positive animals to be present on islands for many years (electronic supplementary material, figure S4). Therefore, the risk of unintended or deliberate transport of the gene drive beyond target islands is likely to be substantial. Although not modelled here, ‘daisy drive’ systems with limited self-replication potential may prove useful in this respect [49].
A requirement of the homozygotic embryonic non-viability and XX sterility strategies is that homing must occur in the germline. While this has not been demonstrated in mammals, efficient germline homing has been demonstrated in insects using the Vasa promoter. A mammalian version of this promoter has previously been demonstrated to drive CRE-recombinase expression in mice efficiently, suggesting that a Vasa-Cas9 transgene could be efficacious for germline homing. As indicated in Material and methods, a number of candidate loci for each of these strategies could be readily identified, which should also facilitate rapid validation.
To date, many of the published gene-drive modelling approaches have been deterministic ([2,33,55,56], but see [15]). By contrast, the importance of stochastic models of extinction risk has long been recognized in the conservation literature [57]. Our individual-based modelling approach accounted for the effects of random demographic and genetic events, which are particularly important as populations get small, and therefore provides more realistic estimates of eradication probabilities for pest vertebrate populations. However, we parametrized models for different mammal species based on typical values reported in the literature and assumed year-round breeding. We stress that to use such an in silico approach to estimate times to eradication in the field would require accurate knowledge of the rate of homing and NHEJ-mediated repair, as well as detailed empirical data on the demography, mating strategy and dispersal characteristics of the target pest population on any given island. Further, our results might not hold when the assumption of a single panmictic population is violated; e.g. the spread of gene drives could be inhibited in metapopulations consisting of multiple subpopulations linked by limited dispersal. However, the spatial dynamics of pest populations will likely be less important for eradications attempted on islands than continental land masses.
In summary, until now, there has been no conclusive demonstration that gene-drive technology presents a viable solution to controlling existing populations of invasive vertebrates. Our results suggest that multiplexing gRNAs within gene-drive constructs could mitigate the existence or production of resistance alleles for exonically placed disruptive gene drives, and achieve the eradication of realistic pest population sizes. For example, we simulated eradication success for populations of up to 200 000 individuals which represents the estimated mouse population on the main island (approx. 2000 ha) of the Antipodes Islands group [58]. Although such multiplexing is theoretically possible, this strategy has not yet been used in the context of gene drives. Further empirical research is therefore critical to develop an understanding of how the probability of successful homing is affected by the number, expression and target sequence of multiple guides.
Supplementary Material
Data accessibility
R code is available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.t78gv [59].
Authors' contributions
All authors designed the research. T.A.A.P. developed the computer code and produced the results. T.A.A.P. and P.T. drafted the manuscript. All authors provided comments and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
We thank The University of Adelaide's Environment Institute for funding this work.
References
- 1.Hamilton WD. 1967. Extraordinary sex ratios. Science 156, 477–488. ( 10.1126/science.156.3774.477) [DOI] [PubMed] [Google Scholar]
- 2.Burt A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R. Soc. Lond. B 270, 921–928. ( 10.1098/rspb.2002.2319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deredec A, Burt A, Godfray HCJ. 2008. The population genetics of using homing endonuclease genes in vector and pest management. Genetics 179, 2013–2026. ( 10.1534/genetics.108.089037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simoni A, Siniscalchi C, Chan Y-S, Huen DS, Russell S, Windbichler N, Crisanti A. 2014. Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster. Nucleic. Acids. Res. 42, 7461–7472. ( 10.1093/nar/gku387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan Y-S, Huen DS, Glauert R, Whiteway E, Russell S. 2013. Optimising homing endonuclease gene drive performance in a semi-refractory species: the Drosophila melanogaster experience. PLoS ONE 8, e54130 ( 10.1371/journal.pone.0054130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 ( 10.7554/eLife.03401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JA, Altwegg R, Evans DM, Ewen JG, Gordon IJ, Pettorelli N, Young JK. 2016. Is there a future for genome-editing technologies in conservation? Anim. Conserv. 19, 97–101. ( 10.1111/acv.12273) [DOI] [Google Scholar]
- 8.Lindholm AK. et al 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326. ( 10.1016/j.tree.2016.02.001) [DOI] [PubMed] [Google Scholar]
- 9.Sinkins SP, Gould F. 2006. Gene drive systems for insect disease vectors. Nat. Rev. Genet. 7, 427–435. ( 10.1038/nrg1870) [DOI] [PubMed] [Google Scholar]
- 10.Webber BL, Raghu S, Edwards OR. 2015. Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc. Natl Acad. Sci. USA 112, 10 565–10 567. ( 10.1073/pnas.1514258112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esvelt K. 2016. Gene editing can drive science to openness. Nature 534, 153 ( 10.1038/534153a) [DOI] [PubMed] [Google Scholar]
- 12.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. 2015. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255. ( 10.1038/nbt.3412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbari OS. et al 2015. Safeguarding gene drive experiments in the laboratory. Science 349, 927–929. ( 10.1126/science.aac7932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gantz VM, Bier E. 2015. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444. ( 10.1126/science.aaa5945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743. ( 10.1073/pnas.1521077112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond A. et al 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechol. 34, 78–83. ( 10.1038/nbt.3439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckhoff PA, Wenger EA, Godfray HCJ, Burt A. 2016. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl Acad. Sci. USA 114, E255–E264. ( 10.1073/pnas.1611064114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Champer J, Buchman A, Akbari OS. 2016. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146–159. ( 10.1038/nrg.2015.34) [DOI] [PubMed] [Google Scholar]
- 19.Xie K, Minkenberg B, Yang Y. 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl Acad. Sci. USA 112, 3570–3575. ( 10.1073/pnas.1420294112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noble C, Olejarz J, Esvelt KM, Church GM, Nowak MA. 2017. Evolutionary dynamics of CRISPR gene drives. Sci. Adv. 3, e1601964 ( 10.1126/sciadv.1601964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P, Schimenti JC, Bolcun-Filas E. 2015. A mouse geneticist's practical guide to CRISPR applications. Genetics 199, 1–15. ( 10.1534/genetics.114.169771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Wang H, Shivalila Chikdu S, Cheng Albert W, Shi L, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. ( 10.1016/j.cell.2013.08.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell KJ, et al. 2015. The next generation of rodent eradications: innovative technologies and tools to improve species specificity and increase their feasibility on islands. Biol. Conserv. 185, 47–58. ( 10.1016/j.biocon.2014.10.016) [DOI] [Google Scholar]
- 24.Blackburn TM, Cassey P, Duncan RP, Evans KL, Gaston KJ. 2004. Avian extinction and mammalian introductions on oceanic islands. Science 305, 1955–1958. ( 10.1126/science.1101617) [DOI] [PubMed] [Google Scholar]
- 25.Harris DB. 2009. Review of negative effects of introduced rodents on small mammals on islands. Biol. Invasions 11, 1611–1630. ( 10.1007/s10530-008-9393-0) [DOI] [Google Scholar]
- 26.Doherty TS, Glen AS, Nimmo DG, Ritchie EG, Dickman CR. 2016. Invasive predators and global biodiversity loss. Proc. Natl Acad. Sci. USA 113, 11 261–11 265. ( 10.1073/pnas.1602480113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenseth NC. et al 2003. Mice, rats, and people: the bio-economics of agricultural rodent pests. Front. Ecol. Environ. 1, 367–375. ( 10.1890/1540-9295(2003)001%5B0367:MRAPTB%5D2.0.CO;2) [DOI] [Google Scholar]
- 28.McLeod R, Norris A. 2004. Counting the cost: impact of invasive animals in Australia. Canberra, Australia: Cooperative Research Centre for Pest Animal Control. [Google Scholar]
- 29.Gregory S, Henderson W, Smee E, Cassey P.. 2014. Eradications of vertebrate pests in Australia. Canberra, Australia: Invasive Animals Cooperative Research Centre. [Google Scholar]
- 30.Russell JC. et al 2016. Importance of lethal control of invasive predators for island conservation. Conserv. Biol. 30, 670–672. ( 10.1111/cobi.12666) [DOI] [PubMed] [Google Scholar]
- 31.Caswell H. 2001. Matrix population models: construction, analysis, and interpretation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 32.Estoup A, Ravigné V, Hufbauer R, Vitalis R, Gautier M, Facon B. 2016. Is there a genetic paradox of biological invasion? Annu. Rev. Ecol. Syst. 47, 51–72. ( 10.1146/annurev-ecolsys-121415-032116) [DOI] [Google Scholar]
- 33.Unckless RL, Messer PW, Connallon T, Clark AG. 2015. Modeling the manipulation of natural populations by the mutagenic chain reaction. Genetics 201, 425–431. ( 10.1534/genetics.115.177592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org/. [Google Scholar]
- 35.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121. ( 10.1038/351117a0) [DOI] [PubMed] [Google Scholar]
- 36.Vidal VPI, Chaboissier M-C, de Rooij DG, Schedl A. 2001. Sox9 induces testis development in XX transgenic mice. Nat. Genet. 28, 216–217. ( 10.1038/90046) [DOI] [PubMed] [Google Scholar]
- 37.Sutton E. et al 2011. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J. Clin. Invest. 121, 328–341. ( 10.1172/JCI42580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu VT, Weber T, Graf R, Sommermann T, Petsch K, Sack U, Volchkov P, Rajewsky K, Kühn R. 2016. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16, 4 ( 10.1186/s12896-016-0234-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. 2014. Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science 343, 69–72. ( 10.1126/science.1242544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamauchi Y, Riel JM, Ruthig VA, Ortega EA, Mitchell MJ, Ward MA. 2016. Two genes substitute for the mouse Y chromosome for spermatogenesis and reproduction. Science 351, 514–516. ( 10.1126/science.aad1795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgoyne PS. 1987. The role of the mammalian Y chromosome in spermatogenesis. Development 101(Suppl.), 133–141. [DOI] [PubMed] [Google Scholar]
- 42.Mohun T. et al 2013. Deciphering the mechanisms of developmental disorders (DMDD): a new programme for phenotyping embryonic lethal mice. Dis. Model. Mech. 6, 562–566. ( 10.1242/dmm.011957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eroglu B, Min J-N, Zhang Y, Szurek E, Moskophidis D, Eroglu A, Mivechi NF. 2014. An essential role for heat shock transcription factor binding protein 1 (HSBP1) during early embryonic development. Dev. Biol. 386, 448–460. ( 10.1016/j.ydbio.2013.12.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Shyamala G, Conneely OM, O'Malley BW. 1995. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Gene Dev. 9, 2266–2278. ( 10.1101/gad.9.18.2266) [DOI] [PubMed] [Google Scholar]
- 45.Gallardo T, Shirley L, John GB, Castrillon DH. 2007. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417. ( 10.1002/dvg.20310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roelz R, Pilz IH, Mutschler M, Pahl HL. 2010. Of mice and men: human RNA polymerase III promoter U6 is more efficient than its murine homologue for shRNA expression from a lentiviral vector in both human and murine progenitor cells. Exp. Hematol. 38, 792–797. ( 10.1016/j.exphem.2010.05.005) [DOI] [PubMed] [Google Scholar]
- 47.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. 2014. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54, 698–710. ( 10.1016/j.molcel.2014.04.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Port F, Bullock SL. 2016. Augmenting CRISPR applications in Drosophila with tRNA-flanked Cas9 and Cpf1 sgRNAs. Nat. Methods 13, 852–854. ( 10.1038/nmeth.3972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noble C, et al. 2016. Daisy-chain gene drives for the alteration of local populations. bioRxiv. ( 10.1101/057307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Platt RJ. et al 2014. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. ( 10.1016/j.cell.2014.09.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang K-T, Li R, Sudjianto A. 2006. Design and modeling for computer experiments. Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- 52.Prowse TAA, Bradshaw CJA, Delean S, Cassey P, Lacy RC, Wells K, Aiello-Lammens ME, Akcakaya HR, Brook BW. 2016. An efficient protocol for the global sensitivity analysis of stochastic ecological models. Ecosphere 7, e01238 ( 10.1002/ecs2.1238) [DOI] [Google Scholar]
- 53.Nagy A, Gertsenstein M, Vintersten K, Behringer R. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd edn Cold Spring Harbour, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 54.Backus GA, Gross K. 2016. Genetic engineering to eradicate invasive mice on islands: modeling the efficiency and ecological impacts. Ecosphere 7, e01589 ( 10.1002/ecs2.1589) [DOI] [Google Scholar]
- 55.Huang YX, Lloyd AL, Legros M, Gould F. 2009. Gene-drive in age-structured insect populations. Evol. App. 2, 143–159. ( 10.1111/j.1752-4571.2008.00049.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teem JL, Gutierrez JB. 2014. Combining the Trojan Y chromosome and daughterless carp eradication strategies. Biol. Invasions 16, 1231–1240. ( 10.1007/s10530-013-0476-1) [DOI] [Google Scholar]
- 57.Gilpin ME, Soulé ME. 1986. Minimum viable populations: processes of species extinction. In Conservation biology: the science of scarcity and diversity (ed. Soule ME.), pp. 19–34. Sunderland, MA: Sinauer. [Google Scholar]
- 58.Government of New Zealand. 2016. Million dollar mouse: eradicating mice from antipodes. Wellington, NZ: Department of Conservation. [Google Scholar]
- 59.Prowse TAA, Cassey P, Ross JV, Pfitzner C, Wittmann TA, Thomas P. 2017. Data from: Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Dryad Digital Repository. ( 10.5061/dryad.t78gv) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Prowse TAA, Cassey P, Ross JV, Pfitzner C, Wittmann TA, Thomas P. 2017. Data from: Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Dryad Digital Repository. ( 10.5061/dryad.t78gv) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
R code is available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.t78gv [59].