Abstract
As the control center of organisms, the brain remains little understood due to its complexity. Taking advantage of imaging methods, scientists have found an accessible approach to unraveling the mystery of neuroscience. Among these methods, optical imaging techniques are widely used due to their high molecular specificity and single-molecule sensitivity. Here, we overview several optical imaging techniques in neuroscience of recent years, including brain clearing, the micro-optical sectioning tomography system, and deep tissue imaging.
Keywords: Brain imaging, Tissue clearing, MOST, Optical microscopy, Deep tissue imaging
Introduction
The brain is a complex organ composed of neurons, glia, microglia, and vascular tissues. By exploring brain structure and function, scientists can not only understand the mechanisms of emotion, sleep, and cognition, but also bring relief to patients who suffer from nervous system diseases, such as depression, autism, Alzheimer’s disease, and Parkinson’s disease. Observation is the most straightforward way to understanding. Thus, brain imaging has turned out to be an effective technique to carve a path for neuroscience. Compared with general methods such as the molecular biological techniques, brain imaging has the advantage that it presents the real physical features and functions (in vivo imaging) of neurons.
Brain imaging has developed over centuries. At the turn of the last century, Camillo Golgi and Santiago Ramón y Cajal did the pioneering work of neuronal tracing using light microscopy [1, 2]. Subsequently, scientists opened the gate of this newborn field [3]. In the last decades, many techniques have been developed, including patch-clamp recording [4], electroencephalography [5], magnetic resonance imaging [6], positron emission tomography [7], and optical imaging, the latter being the main concern of this review.
Optical methods such as confocal [8] and two-photon microscopy [9, 10] have been widely used in brain imaging for a long time. Svoboda et al. [11] achieved in vivo brain imaging with two-photon excitation microscopy in the open-skull mouse [12–17]. However, the surgery probably induced inflammatory reactions [16, 18], resulting in impaired image quality. What is more, the axons and dendrites of neurons extend in many directions, some interneurons crossing several millimeters through a large volume of brain tissue. However, brain tissue is a strong scattering medium, which makes it difficult to focus the excitation light on a small target point and detect the emitted signal. Therefore, to reconstruct the three-dimensional (3D) morphology of neurons, deep tissue imaging techniques are needed.
Here, we review the state-of-the-art optical techniques used to achieve deep brain imaging. First, brain-clearing treatments such as CLARITY [19] and CUBIC are presented [20]. Then, we introduce micro-optical sectioning tomography (MOST) [21], which can achieve whole-brain imaging at submicron resolution. Finally, we discuss the in vivo deep tissue optical imaging methods developed recently.
Brain Clearing
Brain-clearing techniques make the whole brain transparent by dramatically reducing light scattering and improving imaging depth.
From the perspective of optics, the brain remains opaque because of the mismatching of the refractive index. The lipids widely distributed throughout the brain cause the inhomogeneity of scattering. Therefore, removing the lipids, or replacing them with other molecules can effectively rematch the refractive index. Based on this, there are several promising methods, including Scale [22], BABB [23], SeeDB [24], Clear (T) or Clear (T2) [25], and 3DISCO [26–28], which are compared in Table 1.
Table 1.
Pros and cons of major brain clearing methods.
| Techniques | Brief | Clearing Time | PROS | CONS | References |
|---|---|---|---|---|---|
| BABB | Clearing reagent containing several organic solvents | Days | First tissue clearing method | Severe fluorescence quenching; tissue shrinkage; toxicity |
Becker et al. [23] |
| 3DISCO | BABB-based reagent improvement | Days | Strong clearing capability | Fluorescence quenching; tissue shrinkage | Erturk et al. [26–28] |
| SeeDB | Simple immersion method to rematch RI with fructose | Days | Fluorescence and neuron morphology preserved | Incomplete clearing; no immunostaining | Ke et al. [24, 29] |
| FRUIT | SeeDB method improved by adding urea | Days | Better clearing than SeeDB | Minimal tissue expansion | Hou et al. [30] |
| Clear (T)/Clear (T2) | SeeDB method improved by replacing fructose with formamide | Hours- days | Less time-consuming than SeeDB | Incomplete clearing; no immunostaining | Kuwajima et al. [25] |
| Scale | Hyperhydration method by denaturing protein with urea | Weeks | Strong clearing; fluorescence preserved | Severe tissue swelling; no immunostaining | Hama et al. [22] |
| CUBIC | Scale method improved with additional RI rematch | Weeks | Less expansion than Scale; immunostaining available | Protein loss during clearing | Susaki et al. [20], Tainaka et al. [31] |
| CLARITY [19, 32–35] | Hydrogel embedding method with SDS clearing | One week | Strong clearing; neuron morphology preserved | Electrophoresis equipment required | Chung et al. [19], Lee et al. [32], Poguzhelskaya et al. [33], Pointer et al. [34], Tomer et al. [35] |
| PACT, RIMS, and PARS | Improved CLARITY methods | Days | Less time-consuming than CLARITY | Tissue structure damage; partial fluorescence quenching | Yang et al. [36] |
BABB, SeeDB, Clear (T), Clear (T2), Scale, 3DISCO, and CLARITY, names of the methods.
CUBIC clear, unobstructed brain imaging cocktails and computational analysis, PACT passive clarity technique, PARS perfusion-assisted agent release in situ, RIMS refractive index matching solution.
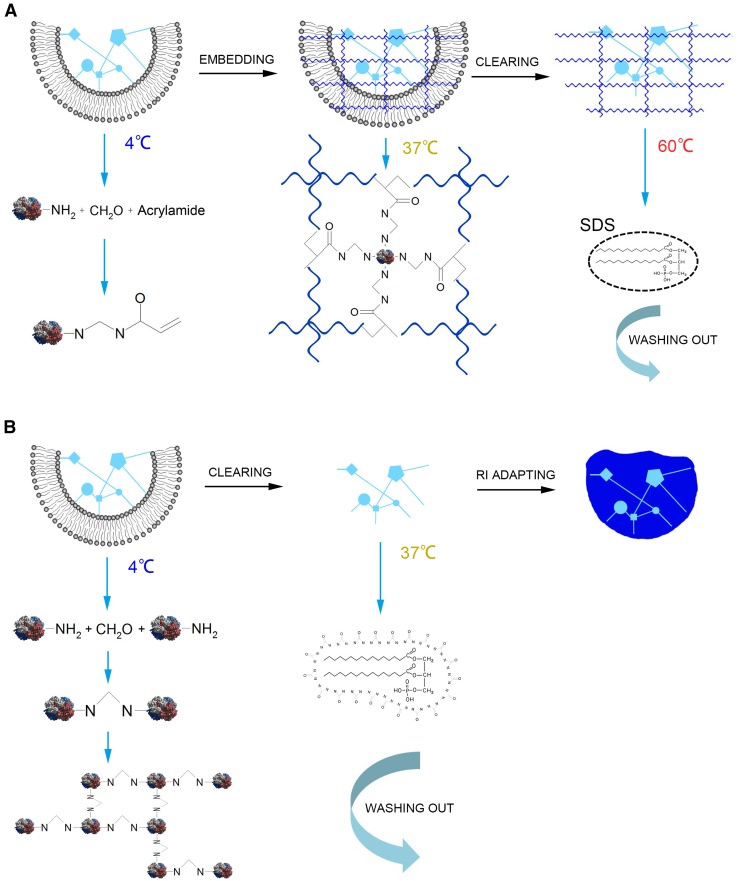
CLARITY, first reported in 2013 by Chung et al. [19], was a revolutionary method to transform the brain into a transparent tissue. It removes lipids by electrophoresis, and is suitable for light-sheet fluorescence microscopy [19, 20] to achieve whole-brain imaging [34]. CLARITY first uses a hydrogel as a protein preserver by infusing the brain with acrylamide and bisacrylamide together with formaldehyde and thermally triggers initiators at 4 °C [19, 37]. The formaldehyde in the mixed reagent helps to crosslink the brain constituents as well as establishing covalent bonds between the hydrogel monomers and biomolecules [19]. After infusion, the treated brain is incubated at 37 °C for 3 h to make a hybrid construct of gel and brain [19]. Then, aiming at efficient clearing, Chung and colleagues chose an ionic extraction technique (electrophoretic tissue clearing) instead of hydrophobic organic solubilization [19]. However, this improvement resulted in other problems, such as the difficulty of selecting the optimal parameters for different samples [20], and the electrophoretic tissue clearing procedure may induce tissue degradation when heated [36]. The principle of CLARITY is illustrated in Fig. 1A. Using CLARITY, the imaging depth with traditional optical microscopes reaches the micron level [19]. Nonetheless, there is no doubt that CLARITY challenges common views. Using CLARITY, labeling specific cells in the brain reversibly is never a problem [37], and researchers can view to depths that are only limited by the working distance of the microscope objective [34]. What is more, there is less loss of proteins, fluorescence signal, and structure of axons and dendrites when using CLARITY compared with conventional methods [19, 34]. Besides, Chung et al., using postmortem human brains as samples, demonstrated that CLARITY is also suitable for the human brain [19].
Fig. 1.
Principles of CLARITY and CUBIC. A Principle of CLARITY. The amino groups from brain tissue protein covalently combine with formaldehyde and acrylamides at 4 °C. Then, the whole brain tissue is embedded in acrylamide gel so the residual acrylamide-connected proteins bind tightly to the framework of the gel at 37 °C. Finally, lipids are completely washed out with sodium dodecyl sulfate (SDS) at 60 °C. B Principle of CUBIC. The amino groups in brain tissue protein covalently combine with each other via PFA. Then the lipids are completely washed out with CUBIC Reagent 1.
Since CLARITY was published, a growing number of researchers have devoted themselves to improving this method [32, 33, 35]. In April 2014, Tomer et al. described an advanced protocol based on CLARITY to achieve simpler lipid removal [35]. The revised protocol allows deep imaging of clarified brains, taking advantage of advanced confocal microscopes and light-sheet microscopes (CLARITY optimized light-sheet microscopy) [35]. Besides, the time required is reduced and the imaging depth is extended to millimeters (a maximum of 5.78 mm) [35]. Poguzhelskaya et al. have also slightly amended the original CLARITY protocol, naming it CLARITY2 [33]. They inserted a cutting step after hydrogel fixation to improve the clearing efficiency [33]. However, the depth of imaging was not increased. What is worse, the cutting procedure may damage samples and thereby influence the 3D reconstruction. At the end of 2014, Lee et al. revised this protocol, making it suitable for many organs [32]. Although CLARITY has been improved, many deficiencies still need to be addressed.
Apart from CLARITY, Susaki et al. invented a protocol of whole-brain imaging using chemical cocktails named CUBIC (clear, unobstructed brain imaging cocktails and computational analysis) [20] and its principle is shown in Fig. 1B. This protocol can be combined with immunofluorescence, and its scale ranges from the whole brain to subcellular structures. The most groundbreaking feature is that CUBIC also allows profiling of the time-course of expression [20]. Researchers first comprehensively screened the CUBIC reagents, resulting in the optimal reagents and procedure. In this procedure, it was confirmed that CUBIC could be completed within 2 weeks and with the preservation of fluorescence [20]. The depth of the immunostained signal that can be detected is > 750 μm [20]. Later, they extended CUBIC to whole-body imaging in both infant and adult mice. As a result, CUBIC is suitable for whole-organ imaging and the resolution can reach the single-cell level. More interestingly, taking diabetic pancreas samples, they found that CUBIC is able to reconstruct the 3D pathology of islets of Langerhans [31].
Other brain-clearing protocols include the passive clarity technique (PACT), refractive index matching solution (RIMS), and perfusion-assisted agent release in situ (PARS) [36]. The PACT reagents are applicable to transgenic tissue with fluorescent protein, and enhance the signal-to-noise ratio. RIMS is used to match a suitable imaging refractive index, while keeping the cleared tissues for a much longer time. PARS has been applied to whole organs, and the imaging depth can be extended to the millimeter level (maximum depth up to 6 mm in whole-brain imaging). In conclusion, using PACT, RIMS, and PARS, the whole brain can be imaged with single-cell resolution [36].
Brain-clearing techniques have flourished in recent years but still leave much room for improvement, such as the tissue deformation, long specimen treatment times, and the reagent toxicity. But these methods represent a milestone in neuroimaging and provide an effective method for the diagnostic analysis of several certain intractable brain diseases such as the 3D visualization of Aβ plaques and microglia in AD human clinic samples [38].
Micro-Optical Sectioning Tomography
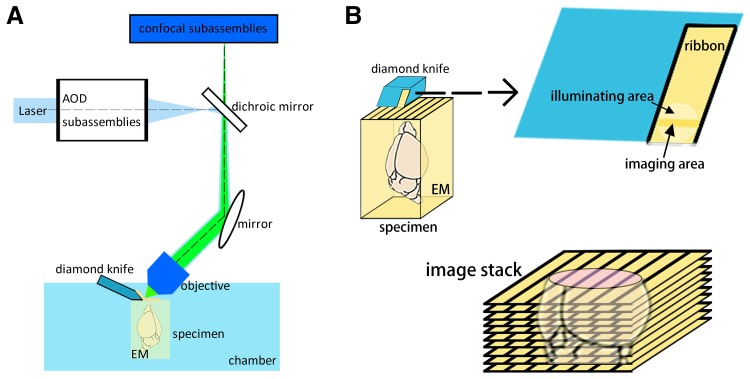
Using conventional optical microscopy to examine a whole brain is challenging. Inspired by brain clearing and tissue sectioning, the micro-optical sectioning tomography (MOST) system was developed to achieve whole mouse-brain imaging at the micron level [21]. With a microtome, optical microscope, and data-processing unit, MOST can simultaneously section and image specimens, collecting ribbons from the microtome, and ultimately reconstructing 3D images of the whole brain. A schematic of this system is shown in Fig. 2 [21, 39, 40]. The resulting images reach mesoscopic resolution, and it is even able to resolve the neurites of neurons [21]. However, the raw images from this system are 8-bit grayscale and need subsequent pseudo-color to help visualization, which limits its visual effectiveness. What is more, it takes a long time (>10 days) to image a centimeter-sized whole mouse brain, collecting more than 15,000 coronal sections to generate a file of 8 terabytes [21]. These workers also indicated that a specific sectioning mode would be helpful for maintaining the accuracy and integrity of brain atlases [41]. Advanced specimen preparation processes, such as fluorescence MOST [21] using transgenic EGFP [42, 43] and EYFP [44, 45] would help in the labeling of specimens. The introduction of confocal techniques and acoustical optical deflectors (Fig. 2) are able to rescue some of the information loss during sectioning and imaging [46], and help to achieve a higher resolution as well as a faster scan imaging [39, 45]. Furthermore, to maintain the fluorescence signals, different embedding approaches during specimen preparation also have diverse effects on both the processes and results of the MOST system; these include glycol methacrylate, Unicryl, and LR White, among which modified glycol methacrylate is relatively more suitable [44]. In addition, the MOST system can provide direct 3D brain reconstruction, showing both individual cells and vessels including capillaries, at a resolution of 1-μm voxels [47–49].
Fig. 2.
Schematic of the fluorescence MOST system [21, 39, 40]. A The system usually consists of a confocal microscope, an acoustical optical deflector scanner, and a microtome with a diamond knife and a moveable chamber. The motion of the chamber matches spatial coordinate axes. The specimen is embedded in embedding materials (EM) and mounted in the chambe. B Using the diamond knife, the system generates a series of coronal sample ribbons, and images each small horizontal stripe simultaneously. Finally, an image stack is collected, and maintains almost all important regions and fine structures of the brain. 3D reconstruction can then be carried out.
In conclusion, the MOST system provides a new path to imaging the whole brain with high resolution and accuracy. However, its long imaging acquisition time, requirement for large storage space, and sectioning methods still leave many challenges.
Deep Tissue Imaging
Imaging deep inside tissue has been a major challenge with the high scattering properties of brain. Both brain clearing and the MOST system aim to bypass the problem of light scattering by using sample treatments of transparent process and muti-sections. So they cannot be applied in vivo and are thus unable to exploit neuronal function in the brain. Furthermore, the sample processing also generates artifacts after slicing or chemical washing. Therefore, scientists have endeavored to develop optical microscopy to break through the depth limit for in vivo imaging [50]. These efforts can be broadly categorized into two groups: the first reduces the light scattering, whereas the second makes use of the scattered light.
Since ultrasonic scattering is 2–3 orders weaker than optical scattering in the brain, ultrasonic imaging can greatly reduce the scattered light and thus provide better penetration depth. Wang et al. developed functional photoacoustic microscopy, which provides multi-wavelength imaging of optical absorption. The imaging depth is >1 mm below the sample surface and the ratio of maximum imaging depth to depth resolution exceeds 100 [51].
Our group reported one-photon optical microscopy for high-resolution molecular imaging in thick biological tissue, called focal modulation microscopy. It uses a focal modulation technique to suppress the out-of-focus fluorescence signal excited by scattered light. We obtained in vivo one-photon fluorescence imaging with diffraction-limited resolution up to 600 μm deep inside highly-scattering media [52, 53]. When this technique is combined with two-photon microscopy, the imaging depth can be further improved [54].
Longer wavelengths undergo less scattering, so they can penetrate deeper inside the brain. By using a fluorescent probe in the 1.3–1.4 μm near-infrared window, Hong et al. reported in vivo imaging to a depth of >2 mm in mouse brain with sub-10-μm resolution [55]. Xu et al. have made an exceptional contribution to multi-photon microscopy. They first improved two-photon microscopy and achieved a 1-mm imaging depth in the adult mouse brain in vivo with an excitation wavelength of 1280 nm [56]. Later, they further extended the excitation wavelength to 1700 nm with three-photon microscopy to image fluorescence-labeled vascular structures and neurons within the hippocampus in vivo [57].
Instead of reducing the light scattering, scientists have also made efforts to use the scattered light. Optical phase conjugation can focus light through millimeter-thick strongly-scattering media by modifying and optimizing the wave-front of the input light field [58]. Our group combined digital optical phase conjugation with ultrasound, and achieved 3D fluorescence imaging with ~38 μm spatial resolution up to 2 mm deep inside brain slices [59]. Later, the resolution was further improved to 10 μm [60].
Another effective method of using scattered light is to measure and correct wave-front aberrations through adaptive optics. Ji et al. proposed a method of adaptive optics based on pupil segmentation and achieved high-resolution two-photon images at 400 μm deep inside mouse brain [61, 62]. Wang et al. demonstrated the adaptive correction of complex optical aberrations at a high numerical aperture. They compensated the rapid spatial aberrations and recovered diffraction-limited images over large volumes up to 240 μm per side [63, 64]. Wang et al. modulated the intensity or phase of light rays with pupil segmentation based on parallel adaptive optics. As a result, they improved the structural and functional imaging of fine neuronal processes over a large imaging volume [65].
There is no doubt that deep tissue imaging is extremely important for brain imaging in vivo. However, the strong scattering properties of the brain pose a major challenge for deep imaging at high resolution. Besides, many deep tissue imaging techniques, such as adaptive optics, need additional processing time for aberration compensation. Therefore, future work may focus on the improvement of imaging depth, spatial resolution, and image acquisition time.
Discussion and Outlook
Optical brain imaging allows an understanding of the structure and function of neurons and this will further help to explain the mechanisms underlying such processes as decision-making, emotion, and memory. Here, we briefly reviewed the current techniques from three points of view: brain clearing, micro-optical sectioning tomography, and deep tissue imaging.
Brain clearing techniques transform the brain from a strong scattering medium to a homogeneous medium. After that, the excitation light can penetrate much deeper so that a larger volume sample can be scanned with a traditional optical microscope. However, brain clearing still faces many challenges. For example, sample deformation is obvious in many tissue-clearing methods. BABB and 3DISCO shrink the brain, while Scale, CUBIC, and CLARITY expand it. Moreover, how the tissue deformation changes the structure of neurons is still unknown. In another aspect, the clearing procedure is inefficient. Most clearing methods are time-consuming. No method can complete clearing of the whole mouse brain within 3 days and most require more than a week. The greatest drawback is that after chemical reagent treatment, imaging information can only be obtained from a dead brain. For in vivo imaging, other means have to be found.
MOST is unique and special in achieving whole-brain imaging. It is widely acceptable for its thorough reconstruction of a whole-brain model. It has become an effective approach to investigating the fundamental structure, spread, and connections of neurons. However, similar to tissue clearing, in vivo signals cannot be obtained and a large data-processing capacity is also required.
We understand that one of the challenges of optical brain imaging is to obtain in vivo signals. The current optical imaging technique for mouse brain requires open-skull or thinned-skull operations [12–18, 66, 67]. The surgery is either severely or mildly traumatic. Light-scattering in brain tissue is the toughest problem. While making use of longer excitation wavelengths can increase imaging depth, special corresponding fluorophores are required. Adaptive optics can significantly increase the imaging depth by compensating wave-front aberration. However, this sacrifices imaging speed.
The development of in vivo brain imaging in the future must be noninvasive, with high resolution, rapid imaging speed, and deep penetration, so it still has a long way to go.
Acknowledgments
This review was supported by the National Basic Research Development Program (973 Program) of China (2015CB352005), the National Natural Science Foundation of China (6142780065, 81527901, and 31571110), the Natural Science Foundation of Zhejiang Province of China (Y16F050002), and Fundamental Research Funds for the Central Universities of China.
Footnotes
Xinpei Zhu and Yanfang Xia have contributed equally to this work.
References
- 1.Golgi C. Sulla struttura della sostanza grigia del cervello. Gazzetta Medica Italiana. Lombardia. 1873;33:244–246. [Google Scholar]
- 2.Ramón S, Cajal S. Textura del Sistema Nervioso del Hombre y de los Vertebrados. Madrid Nicolas Moya, 1904.
- 3.Osten P, Margrie TW. Mapping brain circuitry with a light microscope. Nat Methods. 2013;10:515–523. doi: 10.1038/nmeth.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu W, Sugai T, Yoshimura H, Onoda N. Convergence of olfactory and gustatory connections onto the endopiriform nucleus in the rat. Neuroscience. 2004;126:1033–1041. doi: 10.1016/j.neuroscience.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 5.Sejnowski TJ, Destexhe A. Why do we sleep? Brain Res. 2000;886:208–223. doi: 10.1016/S0006-8993(00)03007-9. [DOI] [PubMed] [Google Scholar]
- 6.Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusnard DA, Raichle ME. Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- 8.Yoder EJ, Kleinfeld D. Cortical imaging through the intact mouse skull using two-photon excitation laser scanning microscopy. Microsc Res Tech. 2002;56:304–305. doi: 10.1002/jemt.10002. [DOI] [PubMed] [Google Scholar]
- 9.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson JM, Dombeck DA, Diaz-Rios M, Harris-Warrick RM, Brownstone RM. Two-photon calcium imaging of network activity in XFP-expressing neurons in the mouse. J Neurophysiol. 2007;97:3118–3125. doi: 10.1152/jn.01207.2006. [DOI] [PubMed] [Google Scholar]
- 11.Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- 12.Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 13.Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, et al. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Holtmaat A, Wilbrecht L, Knott GW, Welker E, Svoboda K. Experience-dependent and cell-type-specific spine growth in the neocortex. Nature. 2006;441:979–983. doi: 10.1038/nature04783. [DOI] [PubMed] [Google Scholar]
- 15.Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hubener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–317. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose CR. Two-photon sodium imaging in dendritic spines. Cold Spring Harb Protoc. 2012;2012:1161–1165. doi: 10.1101/pdb.prot072074. [DOI] [PubMed] [Google Scholar]
- 18.Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–551. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- 19.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 21.Li A, Gong H, Zhang B, Wang Q, Yan C, Wu J, et al. Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science. 2010;330:1404–1408. doi: 10.1126/science.1191776. [DOI] [PubMed] [Google Scholar]
- 22.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 23.Becker K, Jahrling N, Saghafi S, Weiler R, Dodt HU. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One. 2012;7:e33916. doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 25.Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erturk A, Becker K, Jahrling N, Mauch CP, Hojer CD, Egen JG, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 27.Erturk A, Bradke F. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO) Exp Neurol. 2013;242:57–64. doi: 10.1016/j.expneurol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Erturk A, Lafkas D, Chalouni C. Imaging cleared intact biological systems at a cellular level by 3DISCO. J Vis Exp 2014. [DOI] [PMC free article] [PubMed]
- 29.Ke MT, Imai T. Optical clearing of fixed brain samples using SeeDB. Curr Protoc Neurosci 2014, 66: Unit 2 22. [DOI] [PubMed]
- 30.Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Park JH, Seo I, Park SH, Kim S. Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung, and intestine. BMC Dev Biol. 2014;14:48. doi: 10.1186/s12861-014-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poguzhelskaya E, Artamonov D, Bolshakova A, Vlasova O, Bezprozvanny I. Simplified method to perform CLARITY imaging. Mol Neurodegener. 2014;9:19. doi: 10.1186/1750-1326-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pointer K, Kuo JS, Dempsey RJ. CLARITY–a clearer view of the brain. Neurosurgery. 2013;73:N16. doi: 10.1227/01.neu.0000432622.44397.74. [DOI] [PubMed] [Google Scholar]
- 35.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuroscience Underwood E. Tissue imaging method makes everything clear. Science. 2013;340:131–132. doi: 10.1126/science.340.6129.131. [DOI] [PubMed] [Google Scholar]
- 38.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 39.Qi X, Yang T, Li L, Wang J, Zeng S, Lv X. Fluorescence micro-optical sectioning tomography using acousto-optical deflector-based confocal scheme. Neurophotonics. 2015;2:041406. doi: 10.1117/1.NPh.2.4.041406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silvestri L, Bria A, Costantini I, Sacconi L, Peng H, Iannello G, et al. Micron-scale resolution optical tomography of entire mouse brains with confocal light sheet microscopy. J Vis Exp 2013. [DOI] [PMC free article] [PubMed]
- 41.Wang Q, Li A, Gong H, Xu D, Luo Q. Quantitative study on the hygroscopic expansion of spurr resin to obtain a high-resolution atlas of the mouse brain. Exp Biol Med (Maywood) 2012;237:1134–1141. doi: 10.1258/ebm.2012.012142. [DOI] [PubMed] [Google Scholar]
- 42.Zheng T, Yang Z, Li A, Lv X, Zhou Z, Wang X, et al. Visualization of brain circuits using two-photon fluorescence micro-optical sectioning tomography. Opt Express. 2013;21:9839–9850. doi: 10.1364/OE.21.009839. [DOI] [PubMed] [Google Scholar]
- 43.Gong H, Zeng S, Yan C, Lv X, Yang Z, Xu T, et al. Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution. Neuroimage. 2013;74:87–98. doi: 10.1016/j.neuroimage.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Yang Z, Hu B, Zhang Y, Luo Q, Gong H. Development of a plastic embedding method for large-volume and fluorescent-protein-expressing tissues. PLoS One. 2013;8:e60877. doi: 10.1371/journal.pone.0060877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi X, Lv X, Xiong H, Yan C, Chen J, Gong H, et al. Technical considerations on confocal based fluorescence micro-optical sectioning tomography for visualizing brain circuits. Proceedings of SPIE-The International Society for Optical Engineering. 2014;8928:89280T. [Google Scholar]
- 46.Qi X, Xiong H, Lv X, Chen J, Gong H, Luo Q, et al. Improved detectability of neuronal connectivity on mechanical sectioning setup by using confocal detection. J Biomed Opt. 2013;18:50506. doi: 10.1117/1.JBO.18.5.050506. [DOI] [PubMed] [Google Scholar]
- 47.Wu J, He Y, Yang Z, Guo C, Luo Q, Zhou W, et al. 3D BrainCV: simultaneous visualization and analysis of cells and capillaries in a whole mouse brain with one-micron voxel resolution. Neuroimage. 2014;87:199–208. doi: 10.1016/j.neuroimage.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 48.Xue S, Gong H, Jiang T, Luo W, Meng Y, Liu Q, et al. Indian-ink perfusion based method for reconstructing continuous vascular networks in whole mouse brain. PLoS One. 2014;9:e88067. doi: 10.1371/journal.pone.0088067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J, Guo C, Chen S, Jiang T, He Y, Ding W, et al. Direct 3D analyses reveal barrel-specific vascular distribution and cross-barrel branching in the mouse barrel cortex. Cereb Cortex. 2016;26:23–31. doi: 10.1093/cercor/bhu166. [DOI] [PubMed] [Google Scholar]
- 50.Ntziachristos V. Going deeper than microscopy: the optical imaging frontier in biology. Nat Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 51.Zhang HF, Maslov K, Stoica G, Wang LV. Functional photoacoustic microscopy for high-resolution and noninvasive in vivo imaging. Nat Biotechnol. 2006;24:848–851. doi: 10.1038/nbt1220. [DOI] [PubMed] [Google Scholar]
- 52.Chen N, Wong CH, Sheppard CJ. Focal modulation microscopy. Opt Express. 2008;16:18764–18769. doi: 10.1364/OE.16.018764. [DOI] [PubMed] [Google Scholar]
- 53.Gong W, Si K, Chen N, Sheppard CJ. Improved spatial resolution in fluorescence focal modulation microscopy. Opt Lett. 2009;34:3508–3510. doi: 10.1364/OL.34.003508. [DOI] [PubMed] [Google Scholar]
- 54.Si K, Gong W, Chen N, Sheppard CJ. Two-photon focal modulation microscopy in turbid media. Appl Phys Lett. 2011;99:233702. doi: 10.1063/1.3665936. [DOI] [Google Scholar]
- 55.Hong G, Diao S, Chang J, Antaris AL, Chen C, Zhang B, et al. Through-skull fluorescence imaging of the brain in a new near-infrared window. Nat Photonics. 2014;8:723–730. doi: 10.1038/nphoton.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobat D, Durst ME, Nishimura N, Wong AW, Schaffer CB, Xu C. Deep tissue multiphoton microscopy using longer wavelength excitation. Opt Express. 2009;17:13354–13364. doi: 10.1364/OE.17.013354. [DOI] [PubMed] [Google Scholar]
- 57.Horton NG, Wang K, Kobat D, Clark CG, Wise FW, Schaffer CB, et al. three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics 2013, 7. [DOI] [PMC free article] [PubMed]
- 58.Yaqoob Z, Psaltis D, Feld MS, Yang C. Optical phase conjugation for turbidity suppression in biological samples. Nat Photonics. 2008;2:110–115. doi: 10.1038/nphoton.2007.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Si K, Fiolka R, Cui M. Fluorescence imaging beyond the ballistic regime by ultrasound-pulse-guided digital phase conjugation. Nat Photonics. 2012;6:657–661. doi: 10.1038/nphoton.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Si K, Fiolka R, Cui M. Breaking the spatial resolution barrier via iterative sound-light interaction in deep tissue microscopy. Sci Rep 2012, 2. [DOI] [PMC free article] [PubMed]
- 61.Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc Natl Acad Sci U S A. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji N, Milkie DE, Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat Methods. 2010;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 63.Wang K, Milkie DE, Saxena A, Engerer P, Misgeld T, Bronner ME, et al. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat Methods. 2014;11:625–628. doi: 10.1038/nmeth.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K, Sun W, Richie CT, Harvey BK, Betzig E, Ji N. Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat Commun. 2015;6:7276. doi: 10.1038/ncomms8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Liu R, Milkie DE, Sun W, Tan Z, Kerlin A, et al. Multiplexed aberration measurement for deep tissue imaging in vivo. Nat Methods. 2014;11:1037–1040. doi: 10.1038/nmeth.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grutzendler J, Yang G, Pan F, Parkhurst CN, Gan WB. Transcranial two-photon imaging of the living mouse brain. Cold Spring Harb Protoc 2011, 2011. [DOI] [PMC free article] [PubMed]