Abstract
The dopamine transporter (DAT) is involved in the regulation of extracellular dopamine levels. A 40-bp variable-number tandem repeat (VNTR) polymorphism in the 3′-untranslated region (3′UTR) of the DAT has been reported to be associated with various phenotypes that are involved in the aberrant regulation of dopaminergic neurotransmission. In the present study, we found that miR-137 and miR-491 caused a marked reduction of DAT expression, thereby influencing neuronal dopamine transport. Moreover, the regulation of miR-137 and miR-491 on this transport disappeared after the DAT was silenced. The miR-491 seed region that is located on the VNTR sequence in the 3′UTR of the DAT and the regulatory effect of miR-491 on the DAT depended on the VNTR copy-number. These data indicate that miR-137 and miR-491 regulate DAT expression and dopamine transport at the post-transcriptional level, suggesting that microRNA may be targeted for the treatment of diseases associated with DAT dysfunction.
Keywords: Dopamine transporter, Solute carrier family 6 member 3, hsa-miR-137, hsa-miR-491-5p, 3′-Untranslated region, Variable-number tandem repeat, Posttranscriptional regulation
Introduction
Dopamine (DA) is one of the principal neurotransmitters in the central nervous system, participating in motion control, emotion, cognition, drug addiction, neuroendocrine regulation, and other vital activities through the activation of DA receptors [1–7]. Reuptake is a normal mechanism by which neurotransmitters pass through the presynaptic membrane and then are removed from the synaptic cleft [1, 2, 6, 8]. Psychoactive substances block DA reuptake to increase its concentration in the synaptic cleft [2, 8]. With reuptake blocked, the normal effects of neurotransmitters are magnified. Reuptake through the cell membrane that is mediated by the DA transporter (DAT) diminishes DA signaling in a sodium-dependent manner, and this plays a crucial role in maintaining DA homeostasis in neurons [2, 9, 10]. The gene encoding the DAT, solute carrier family 6 member 3 (SLC6A3), belongs to the SLC6 family of transporters [11, 12]. The 3′ untranslated region (UTR) of SLC6A3 mRNA contains a 40-bp tandem repeat, referred to as a variable-number tandem repeat (VNTR) polymorphism (rs28363170) [13], which can be present in 3–11 copies [11, 14]. The 9- and 10-repeat alleles are the most frequently found in the human population [12, 14, 15]. Changes in the number of copies of this region are closely associated with idiopathic epilepsy, attention-deficit/hyperactivity disorder (ADHD), alcohol and cocaine dependence, susceptibility to Parkinson’s disease, and resistance to nicotine dependence [6, 11, 14, 16, 17].
MicroRNAs (miRNAs) are small non-coding RNAs ~21 nt long. They regulate ~30% of the DNA in the genome [18] and can regulate gene expression at many levels, including the transcriptional, post-transcriptional, translational, and epigenetic levels, thus participating in the evolution of a species, embryonic development, metabolism, and the occurrence of disease [18, 19]. The roles of miRNAs in the nervous system have been studied extensively, from physiology to pathology [20, 21]. Most seed regions of miRNAs are located in the 3′UTR of the gene’s mRNA, and miRNAs may regulate the post-transcriptional level of DATs through the 3′UTR of SLC6A3 mRNA. However, still unknown are the miRNAs that target the VNTR seed region of SLC6A3 and the relationship between the VNTR copy-number and this type of regulatory effect on the DAT. The primary objectives of the present study were to investigate the post-transcriptional regulatory effect that underlies the actions of miRNAs on DAT levels and activity.
Using bioinformatics software prediction (TargetScan and miRanda) [22, 23] as well as the analysis of luciferase expression activity, mRNA and protein levels, and activity levels, we found that hsa-miR-137 (miR-137, MIMAT0000429) and hsa-miR-491-5p (miR-491, MIMAT0002807) targeted the SLC6A3 3′UTR (1945 bp, taxonomic ID: 9606, NM_001044:1990..3934) and regulated DAT expression at the post-transcriptional level, and that miR-491 targeted the VNTR seed region of the 3′UTR of SLC6A3. Moreover, both miRNAs were found to regulate DA transport. To our knowledge, this is the first report of the direct regulation of DAT by miR-137 and miR-491.
Materials and Methods
Cell Culture
Human dopaminergic SK-N-SH neuroblastoma cells (ATCC HTB-11) were grown in minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and 0.1 g/L sodium pyruvate at 37 °C under 5% CO2. Human dopaminergic SK-N-BE(2) neuroblastoma cells (ATCC CRL-2271) were grown in MEM:F12 (1:1) supplemented with FBS to a final concentration of 10%, and 0.1 g/L sodium pyruvate at 37 °C under 5% CO2 (all media chemicals were from Invitrogen, Carlsbad, CA). Retinoic acid (10 μmol/L; all-trans-retinoic acid, ATRA; Sigma, St. Louis, MO) was used to induce neural cell differentiation. The expression levels of miR-137 and DAT were significantly increased, while miR-491 was not significantly changed after treatment with retinoic acid. Human hepatoma HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. Cells were grown at 37 °C in a humid atmosphere with 5% CO2. This study was performed using both SK-N-SH cells and SK-N-BE(2) cells. The data for SK-N-BE(2) cells are similar to SK-N-SH cells, which are not shown.
Plasmid Construction and Transfection
We used SK-N-SH cellular genomic DNA as a template and obtained the target gene SLC6A3 3′UTR through PCR amplification. The target gene inserted into the upstream multiple cloning site of a synthetic Renilla luciferase reporter gene (hRluc) in the psiCHECK2 vector (Promega, Madison, WI) at the Xho I and Not I sites, resulted in a psiCHECK2-SLC6A3 3′UTR plasmid. Mutations of the predicted seed regions of miR-137 and miR-212 within the human SLC6A3 3′UTR were generated by overlap extension PCR and then cloned into the psiCHECK2 vector. All of the constructs were confirmed by sequencing (Invitrogen). We used the Lipofectamine 2000 plasmid transfection reagent (Invitrogen) according to the manufacturer’s instructions.
The real-time PCR primers, miRNA seed region mutation primers, and VNTR gene sequences used to construct the plasmids are listed in Table 1.
Table 1.
PCR primers and VNTR sequences (h, human).
| Name | Sequence | Description |
|---|---|---|
| hSLC6A3 forward | 5′-ATGAGTAAGAGCAAATGCTCCG-3′ | Primers for human SLC6A3 mRNA CDS-3′UTR (coding sequence and 3′UTR); template: cDNA synthesis from SK-N-SH cell total RNA; insertion vector of amplification products: pcDNA3.1/NT-GFP-TOPO |
| hSLC6A3 reverse | 5′-GCAGTTTTTCCATTGTGGATGTC-3′ | |
| hSLC6A3 3′UTR forward | 5′-AGAGCGGCCGCGCAGTTTTTCCATTGTGGATGTC-3′ | Primers for human DAT mRNA 3′UTR; restriction sites: Xho I, Not I; insertion vector of amplification products: psiCHECK2 |
| hSLC6A3 3′UTR reverse | 5′-CCGCTCGAGAGGGAGCAGAGACGAAGAC-3′ | |
| hSLC6A3 137MUT forward | 5′-AGTTTTTGTTTACAAGAATAATTACGATATCTGAGTGAAG-3′ | Overlap extension PCR primers for miR-137 seed region mutation |
| hSLC6A3 137MUT reverse | 5′-CTTCACTCAGATATCGTAATTATTCTTGTAAACAAAAACT-3′ | |
| hGAPDH forward | 5′-TGCACCACCAACTGCTTAGC-3′ | Real-time PCR primers for human GAPDH mRNA |
| hGAPDH reverse | 5′-GGCATGGACTGTGGTCATGAG-3′ | |
| hSLC6A3 forward | 5′-TGTGCTGGAAGCTGGTCA-3′ | Real-time PCR primers for human SLC6A3 mRNA |
| hSLC6A3 reverse | 5′-GGGTCTGAAGGTCACAATGC-3′ | |
| Human mutation VNTR | 5′-AGCGTGTACTACCCCAGGACGCATGCAGGGCCGCGAGA-3′ | miR-491 seed region mutation of DAT mRNA VNTR |
| Human wild-type VNTR | 5′-AGCGTGTACTACCCCAGGACGCATGCAGGGCCCCCACA-3′ | Basic unit of DAT mRNA VNTR |
Transfection with Small RNAs
Transfection with miRNA mimics, anti-miRNAs, and small interfering RNAs (siRNAs) was performed using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions. The miRNA mimics (double-stranded RNA oligonucleotides), anti-miRNAs (2′-O-methyl antisense oligonucleotides against the target miRNAs), negative control duplexes, pre-designed siRNAs of the human DAT, and the scrambled negative control siRNA were from Ribobio (Guangzhou, China) and applied at a final concentration of 100 or 150 nmol/L. After incubation of the small RNAs for 24–72 h, the cells were harvested for further analyses [24, 25].
Analysis of Luciferase Reporter Gene Expression
After each purified reporter plasmid (0.2 μg) was transfected into SK-N-SH or SK-N-BE(2) cells using Lipofectamine 2000 for 4 h, miRNA mimics, anti-miRNAs, or negative control duplexes were incubated for another 24–72 h [24, 25].
Luciferase reporter gene expression was analyzed using a Dual-luciferase Reporter Assay System (Promega, E1910) according to the manufacturer’s instructions. Renilla luciferase activity was normalized to the corresponding firefly luciferase activity.
Analysis of Dopamine Transport
Cells were plated in 24-well culture dishes (20,000 cells/well) and transfected with siRNA or miRNA mimics and their control miRNAs for 48 or 72 h. After the cells were incubated with 5 μg/mL DA hydrochloride (Sigma) for 5 h, the cells were washed three times with ice-cold phosphate-buffered saline (PBS) and lysed in RIPA buffer. Finally, intracellular DA concentrations were assayed using a DA ELISA (enzyme-linked immunosorbent assay) kit (Elabscience, Wuhan, China, catalog no. E-EL-0046c; or Rocky Mountain Diagnostics, Colorado Springs, CO, catalog no. BA-E-5300).
Analysis of mRNA Expression Levels
RNA extraction and cDNA synthesis were performed using an SV Total RNA Isolation System (Promega) and a GoScript Reverse Transcription System (Promega) according to the manufacturer’s instructions. After the resultant cDNA was diluted tenfold, real-time quantitative PCR was performed using the CFX96 Real-time System (Bio-Rad, Hercules, CA) and a SYBR Premix Ex Taq Kit (Takara, Dalian, China). GAPDH was used as the normalization control to analyze target gene mRNA expression levels. Relative quantification was performed using the comparative cycle threshold (C t) method after determining the C t values for the reference (GAPDH) and target genes (SLC6A3) in each sample, according to the method [24, 25].
Analysis of Protein Expression Levels
After reacting for 24 h, the cell sample was prepared with RIPA cell lysis buffer (25 mmol/L Tris–HCl, 150 mmol/L NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate, pH 7.6) for 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The cells were then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA) and blocked. The membrane was washed and incubated with primary anti-DAT antibody (1:1000–1:2000, Abcam, Cambridge, UK; catalog nos ab5990a and b111468) and secondary antibodies (1:1000; Cell Signaling Technology, Beverly, MA). Equal parts of luminol and peroxide reagents (Millipore) were mixed for chemiluminescence (Thermo Fisher Scientific). The luminescent liquid was then dropped onto a PVDF membrane and colored using the ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA) for Western blot analysis. The bands underwent grayscale scanning and processing analysis using Image Lab 4.0 imaging software (Bio-Rad) [24, 25].
Fluorescence Microscopy
pcDNA3.1-GFP-DAT-CDS-3′UTR fusion plasmid (0.2 μg) was transfected into HepG2 cells at 70% confluence using Lipofectamine 2000 (Invitrogen) for 4 h before introduction of miRNA mimics, anti-miRNAs, or negative control duplexes for another 72 h [27, 28]. DAT-overexpressing HepG2 cells were next incubated at 37 °C with 10 μg/mL Hoechst 33342 (Sigma) for 15 min, and then visualized and photographed using a fluorescence microscope (Olympus, Tokyo, Japan) with a 20× objective.
Data Analysis
The data are expressed as mean ± SD. Student’s t-test was used for statistical analysis. Values of P < 0.05 were considered statistically significant.
Results
TargetScan [22] (www.targetscan.org) and miRanda (www.microrna.org) [23] were used for miRNA target prediction. Using the bioinformatics prediction, we screened and verified two of the most useful miRNAs: miR-137 and miR-491. Our results confirmed that their effective binding sites were located in the 3′UTR of SLC6A3 mRNA, and the miR-491 seed region was located on the VNTR sequence.
miR-137 Regulates DAT Expression at the Post-transcriptional Level
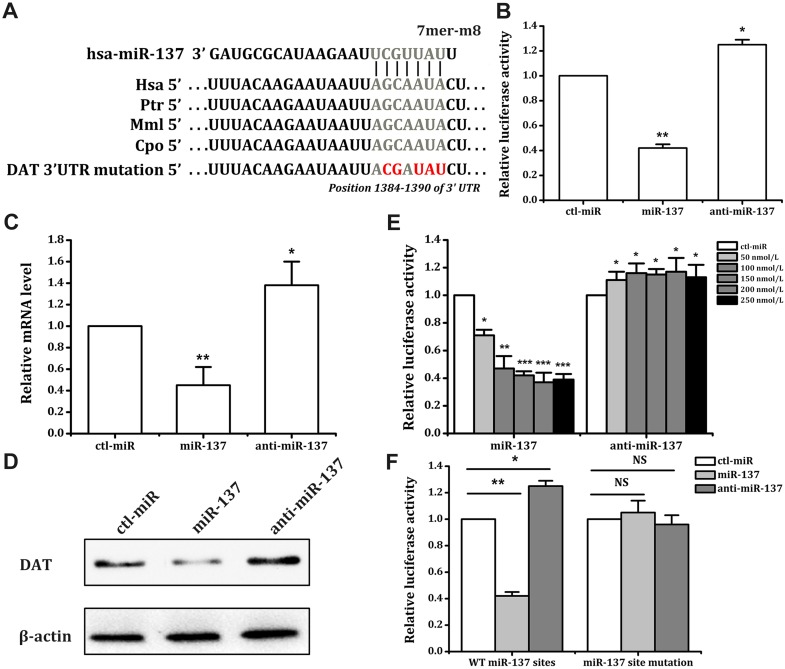
Bioinformatics analysis indicated that the seed region for miR-137 was located in the 3′ UTR of SLC6A3 mRNA. This sequence is highly conserved in primates but not in rodents (Fig. 1A). After constructing the expression vector for luciferase reporter genes, we transferred reporter plasmids and miRNAs into SK-N-SH cells, and tests were performed after 24 h of culture. Our results confirmed that miR-137 reduced the luciferase expression activity of these cells by 58%, and the miR-137 inhibitor increased luciferase expression activity by 25% (Fig. 1B). miR-137 decreased luciferase activity in these cells in a dose-dependent manner and reached a ceiling effect at 150–200 nmol/L (Fig. 1E).
Fig. 1.
miR-137 regulates the expression of DAT in SK-N-SH cells. A The binding site of miR-137 in the 3′UTR of SLC6A3 mRNA. The mutated bases of the miR-137 binding site are shown in red. The sequence alignments of the seed regions of the binding sites for miR-137 are shown for the indicated species. The conserved sequences are shown in gray. B Effects of miR-137/anti-miR-137 on the activity of the luciferase reporter construct with the SLC6A3 3′UTR in SK-N-SH cells, which were transfected with different constructs for 5 h, followed by transient transfection with miR-137/anti-miR-137 (100 nmol/L) for 48 h. DAT mRNA levels were assessed by quantitative PCR (C), and protein levels were analyzed by Western blot (D) in SK-N-SH cells transfected with miR-137, anti-miR-137, or ctl-miR (100 nmol/L) for 72 h (representative result from three independent experiments). E Effects of different concentrations of miR-137 or anti-miRNA on the luciferase activity in SK-N-SH cells. Different concentrations of miR-137 or anti-miRNA were transfected for 48 h. F SK-N-SH cells were transfected with wild-type or mutated constructs for 5 h, followed by transient transfection with miR-137/anti-miR-137 (100 nmol/L) for 48 h. *P < 0.05; **P < 0.01; ***P < 0.001, NS, not significant; WT, wild-type; Hsa, Homo sapiens; Ptr, Pan troglodytes; Mml, Macaca mulatta; Cpo, Cavia porcellus.
We then investigated the effects of miR-137 on the mRNA (Fig. 1C) and protein (Fig. 1D) expression levels of the DAT in SK-N-SH cells. The results showed that the mRNA levels of their DAT genes underwent the same changes in luciferase expression activity. miR-137 reduced the mRNA levels of the DAT genes by 55%, and the miR-137 inhibitor increased them by 38% (Fig. 1C). These results showed that miR-137 influenced the expression of DATs in SK-N-SH cells.
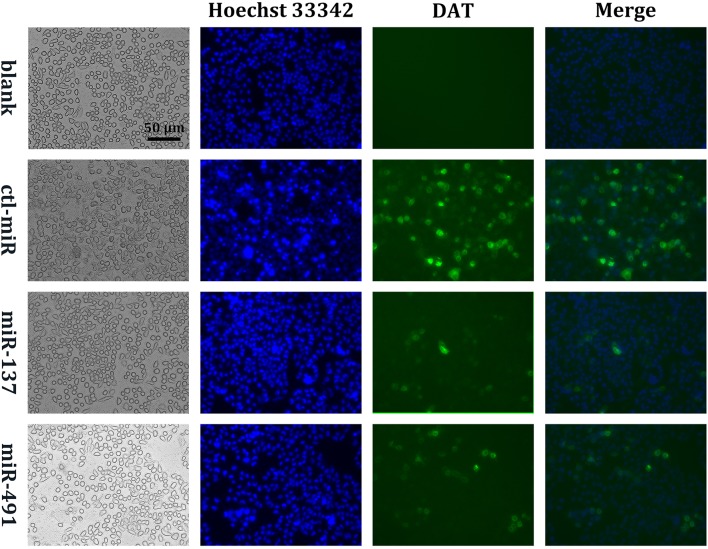
Next, we tested whether the effect of miR-137 on SLC6A3 expression in SK-N-SH cells occurs via the predicted active site. We generated a point mutation in the predicted miR-137 seed region using overlap extension PCR [26], inserted the mutated SLC6A3 mRNA 3′UTR downstream of the psiCHECK2 reporter gene, and transfected the mutated plasmid and miRNA into SK-N-SH cells. The results showed that the regulatory effects of miR-137 on the reporter genes disappeared after mutation of its seed region (Fig. 1A, F). We inserted the DAT gene protein coding sequence (CDS) and 3′UTR downstream of the green fluorescent protein (GFP) gene in pcDNA3.1 and constructed a fusion expression plasmid: pcDNA3.1-GFP-DAT-CDS-3′UTR. We then transfected HepG2 cells with the plasmid and miRNA together and detected GFP expression under a fluorescence inverted microscope (Olympus, Tokyo, Japan) after 48 h of incubation. The reason for choosing HepG2 cells was that their DAT expression was relatively low. The results showed that miR-137 significantly reduced the fusion protein expression of GFP and the DAT in HepG2 cells (Fig. 4). These results demonstrated that the predicted site was indeed the site of action of miR-137.
Fig. 4.
Fluorescence microscopic imaging of the DAT. After being treated with miRNAs for 72 h, DAT-overexpressing HepG2 cells were incubated with 10 μg/mL Hoechst 33342 at 37 °C for 15 min. HepG2 cells were visualized and photographed using a fluorescence microscope with a 20× objective. ‘Blank’ indicates that cells did not overexpress DAT (representative result from three independent experiments; scale bar 50 μm).
miR-491 Regulates DAT Transcription by Targeting the VNTR Seed Region
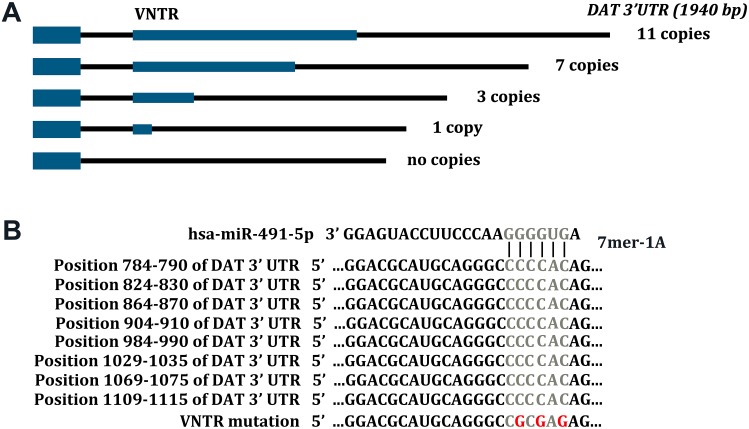
The VNTR sequence is an important functional site of the SLC6A3 3′UTR, and its copy-number usually ranges from 3 to 11 [11]. Previous studies have shown that a change in copy-number is closely linked with many diseases, such as idiopathic epilepsy, ADHD, alcohol and cocaine dependence, susceptibility to Parkinson’s disease, and resistance to nicotine dependence [6, 11, 14, 16, 17]. The seed region of miR-491 in VNTR is highly conserved between humans and rhesus monkeys (Fig. 2A, B).
Fig. 2.
Schematic diagram of VNTRs and miR-491 seed region in SLC6A3 mRNA. A Human SLC6A3 mRNA harbors 3–11 VNTRs within its 3′UTR. The relative positions of the VNTRs are indicated in blue. Position 1 is the first nucleotide following the termination codon. B Sequence alignment of the seed region in the SLC6A3 3′UTR for miR-491.
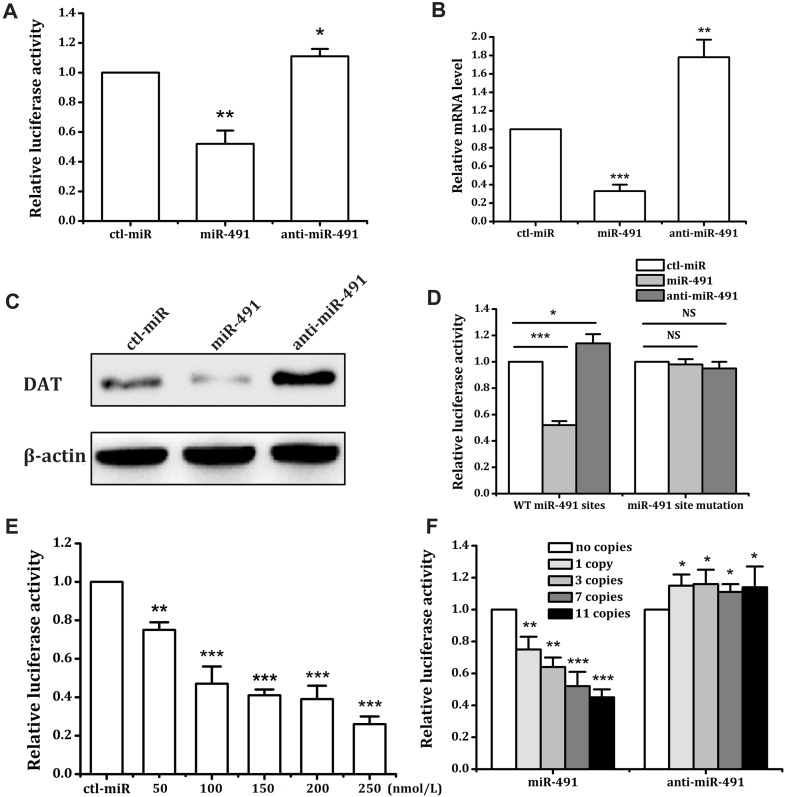
Our bioinformatics analysis showed that the miR-491 seed region exists on the VNTR sequence of the 3′UTR of SLC6A3 mRNA. miR-491 reduced the luciferase activity of SK-N-SH cells by 48%, and the inhibitor increased luciferase activity by 11% (Fig. 3A), acting in a dose-dependent manner (Fig. 3E). We inserted synthetic DNA fragments (Invitrogen) with different VNTR copy-numbers downstream of luciferase reporter genes, constructed numerous reporter plasmids with various VNTR copy-numbers, and then transferred the reporter plasmids and miRNA into SK-N-SH cells by transient transfection (Fig. 2A). The cells were tested after 24 h of culture [24]. Our results showed that miR-491 decreased the luciferase expression in SK-N-SH cells with plasmid transfection copy numbers of 1, 3, 7, and 11 by 25%, 36%, 48%, and 55%, respectively. The miR-491 inhibitor increased the luciferase expression activity in these cells with plasmid transfection copy numbers of 1, 3, 7, and 11 by 15%, 16%, 11%, and 14%, respectively (Fig. 3F). We investigated the effects of miR-491 on mRNA (Fig. 3B) and protein (Fig. 3C) expression levels in SK-N-SH cells. The results showed that miR-491 significantly decreased the SLC6A3 mRNA level by 67%, and the inhibitor significantly increased the mRNA level by 78% (Fig. 3B). These results showed that miR-491 may influence the DAT expression in SK-N-SH cells similarly to miR-137.
Fig. 3.
miR-491 modulates DAT expression at the post-transcriptional level through 3′UTR VNTRs. A Effects of miR-491/anti-miR-491 on the activity of the luciferase reporter construct with the SLC6A3 3′UTR in SK-N-SH cells. miR-491 or anti-miRNA (150 nmol/L) was transfected for 48 h. DAT mRNA levels were assessed by quantitative PCR (B), and protein levels were analyzed by Western blot (C) in HepG2 cells transfected with miRNA, anti-miRNA, or ctl-miR (150 nmol/L) for 72 h (representative result from three independent experiments). D The SLC6A3 3′UTR (7 VNTRs) with mutation of the seed region of miR-491. SK-N-SH cells were transfected with different constructs for 5 h, followed by transient transfection with miR-491/anti-miR-491 (150 nmol/L) for 48 h. E Different concentrations of the miR-491 were transfected for 48 h. The effect of ctl-miR on luciferase activity was defined as 1 for each concentration. F Effects of miR-491/anti-miR-491 (150 nmol/L) on the activity of the luciferase reporter constructs with different VNTRs. *P < 0.05; **P < 0.01; ***P < 0.001 vs ctl-miR, NS not significant, WT wild-type.
We then tested whether miR-491, like miR-137, induces the expression of SK-N-SH cell DAT genes through the predicted active site. We synthesized a DNA fragment with a point mutation in the predicted miR-491 seed region, inserted 7 VNTRs of the miR-491 seed region mutation downstream of the psiCHECK2 reporter gene, and transfected the mutated plasmid and miRNA into SK-N-SH cells. The results showed that the regulatory effects of miR-491 on reporter genes disappeared after the mutation of its seed region (Fig. 3D). Similar to the miR-137 experiment, we transfected HepG2 cells with the fusion plasmid pcDNA3.1-GFP-DAT-CDS-3′UTR and miRNA together. We then observed GFP expression under a fluorescence inverted microscope after 48 h of incubation. The results showed that miR-491 significantly reduced the expression of the GFP-DAT fusion protein in HepG2 cells (Fig. 4). Using the predicted active site and overexpression fusion protein, we demonstrated that the predicted site is indeed the binding site of miR-491.
Altogether, our results revealed that miR-491 can regulate DAT expression post-transcriptionally by targeting the VNTR sequence. Moreover, the regulatory effects of miR-491 on DAT expression were closely linked with the VNTR copy number in the SLC6A3 3′UTR.
Effects of miR-137 and miR-491 on Dopamine Transport
The function of the DAT is to transport DA in the synaptic cleft back into the synaptic interior so that the neurotransmitter is prevented from continuously inducing its effects. To determine whether miR-137 and miR-491 influence the transport of extracellular DA in neurons by regulating DAT expression, after miRNA transfection, we placed neurons in 5 μg/mL DA solution that was prepared with MEM, followed by incubation for 5 h. After washing with PBS, the cells were lysed and intracellular DA levels were assessed using a DA ELISA kit. Our results confirmed that miR-137 and miR-491 significantly reduced neuronal DA uptake by 36% and 53%, respectively (Fig. 5A, B).
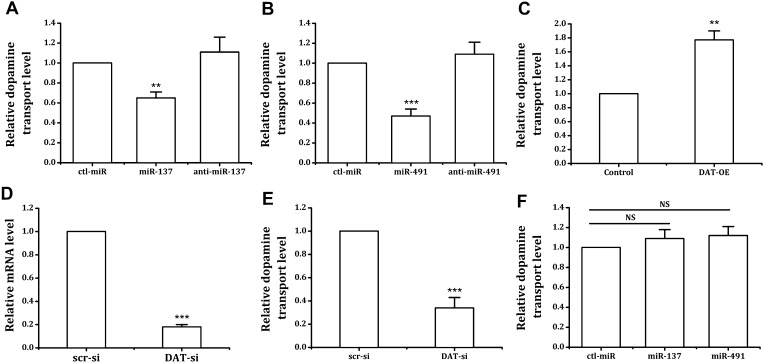
Fig. 5.
miR-137 and miR-491 regulate DAT-dependent DA uptake into nerve cells. A, B The effects of miR-137 (A) and miR-491 (B) on DA transport in nerve cells. SK-N-SH cells were incubated with 5 μg/mL DA for 5 h after treatment with miR-137/anti-miR-137 (100 nmol/L) (A) or miR-491/anti-miR-491 (150 nmol/L) (B) for 72 h. C After transfection of the DAT-OE plasmid, DA in SK-N-SH cells increased. After transfection with SLC6A3 siRNA, the levels of SLC6A3 mRNA (D) and DA (E) significantly decreased. F SLC6A3 silencing prevented the effects of miR-137 and miR-491 on DA transport. *P < 0.05; **P < 0.01; ***P < 0.001 vs ctl-miR, NS, not significant; DAT-OE, DAT overexpression,
To further verify that the effects of miRNA on neuronal DA transport are mediated by the DAT, we conducted gene silencing and overexpression experiments. There was a significant positive correlation between the DA transport and the expression of DATs in SK-N-SH cells (Fig. 5C–F). After the neurons were transfected with siRNA using RNAiMAX for 36 h and the SLC6A3 gene was silenced, the neurons were transfected with miRNA and incubated with 5 μg/mL DA. The test results of intracellular DA levels confirmed that after SLC6A3 gene silencing, the regulatory effects of miR-137, miR-491, and their inhibitors on neuronal DA transport were lost (Fig. 5F). Conversely, DAT overexpression significantly increased the DA uptake of SK-N-SH cells (Fig. 5C). These results showed that miR-137 and miR-491 affected DA uptake in cells by regulating DAT expression.
Discussion
DAT, a member of the Na+/Cl−-dependent transporter gene family, is a membrane protein located at the terminals of dopaminergic neurons [1, 2]. Most mesotelencephalic DA neurons in the human brain express high levels of the DAT throughout their entire somatodendritic and axonal domains, whereas a smaller subpopulation of mesencephalic DA cells and all hypothalamic DA cell groups express little or no DAT [6, 27]. The DAT transports DA in the synaptic cleft through the presynaptic membrane for reuse after its physiological effects have taken place [8, 9]. The DAT can stop the transfer of information between cells, plays a crucial role in controlling DA balance, and its regulatory mechanism has been the focus of much research on drug development for the treatment of addiction and depression [6, 8–10, 28, 29]. Studies of DAT-knockout mice have reported that DA synthesis increases two-fold, but total DA levels are reduced by 95%, thus causing a significant change in the neurochemical process [30]. This shows that the DAT plays a crucial role in both terminating DA signaling and refilling DA vesicles [29, 30]. The DAT plays a critical role in addiction to psychoactive drugs, including opiates, amphetamines, and cocaine, and the pathogenesis of Parkinson’s disease and other nervous system diseases [1, 5, 31]. Research has shown that the fine-tuning of DA balance can be achieved by carefully manipulating the DA channel. Multiple mechanisms can affect DAT activity and the distribution of its expression within cells [1, 4, 28, 29, 32–34]. Rapidly controlled signaling pathways that are associated with DAT expression include the activation of G-protein-coupled receptors and intracellular second-messenger systems, thus influencing protein–protein interactions. Multiple regulatory modes can lead to changes in DAT function, which is considered to be an underlying cause of major changes in DA transport and behaviors that are modulated by DA [29, 35–37].
Cocaine can modulate the expression of the pitx3 gene and subsequently the expression of DA receptors, DATs, and tyrosine hydroxylase via miR-133b, suggesting that miRNAs may play an important role in embryogenesis and drug addiction [38]. The regulatory effect of miRNA and the stability of the DAT gene appear to be inextricably linked, and miRNAs play an important role in regulating DAT expression and function. The multiple effects of miR-137 have been extensively investigated. It is a brain-enriched miRNA with a critical role in regulating nervous system development and mediating synaptic plasticity [39–41]. Furthermore, miR-137 is also a candidate gene for schizophrenia susceptibility [40, 41], tumor growth, and the inhibition of invasion [42–45]. Our results indicated that miR-137 affects the expression of luciferase through the SLC6A3 3′UTR, thereby significantly reducing the expression levels of neuronal DAT mRNA and proteins and reducing neuronal DA transport. After SLC6A3 silencing, the effect of miR-137 on DA transport in neurons disappeared. When the DAT was overexpressed, however, miR-137 had a stronger effect on DA transport. Therefore, the DAT appears to be a direct target of miR-137.
The SLC6A3 gene comprises 15 exons that span 60 kb on chromosome 5p15.32. The 40-bp VNTR polymorphism is located in exon 15 of the SLC6A3 gene [1]. The VNTR of the SLC6A3 3′-UTR consists of a 40-bp repeat with 3–13 copies, and the 9- and 10-repeat alleles are the most frequently found in the human population. This polymorphism has been linked to various disease phenotypes that involve disturbances in the regulation of dopaminergic neurotransmission, such as alcoholism, cocaine abuse, working memory, Alzheimer’s disease, ADHD, and emotional behavior [11, 12, 14, 17, 46–51]. Research has shown that changes in the number of VNTR copies are closely related to DAT expression. For example, the expression level of the 9-repeat allele is lower than the 10-repeat allele [11]. MIR-491 is a tumor-suppressor gene. Two mature microRNAs, miR-491-3p and miR-491-5p, are encoded by MIR-491 [52] and both mature forms are involved in the modulation of crucial cancer hallmarks: proliferation, invasion, and stem-cell propagation [52–54]. Targeted intervention in the miR-491-related signaling pathway can enhance the efficacy of chemotherapy against cancers [52, 53]. In the present study, we found that the seed region of miR-491 was located on the VNTR sequence in the 3′UTR of SLC6A3 mRNA, and the regulatory effect of miR-491 on DAT expression depended on the VNTR copy-number in the 3′UTR of the DAT gene mRNA. miR-491 regulated the expression and function of the DAT through its active site on the VNTR sequence. Altogether, we demonstrated that miR-491 can regulate DAT expression post-transcriptionally by targeting the VNTR sequence, and miRNA disturbances may be involved in the pathophysiology of a variety of diseases.
The VNTR of SLC6A3 mRNA is highly conserved in humans and rhesus monkeys. Since the origin of VNTR, the regulation of the DA signaling system in primates may differ from that in other species. With the variation of VNTR copy number, the number in the miR-491 seed region in the SLC6A3 mRNA 3′UTR and the ability of miR-491 to regulate SLC6A3 expression was correspondingly changed. This mechanism may be a unique means of DAT regulation in humans and rhesus monkeys. Therefore, clarification of the action of miR-491 in regulating the expression and function of the DAT by targeting the seed region of VNTR is important.
The seed region of miR-491 is located in the VNTR fragment of the DAT mRNA 3′UTR, and the effect of miR-491 on DAT expression is dependent on the VNTR copy-number. The seed region of miR-137 is located in the DAT mRNA 3′UTR fragment outside the VNTR sequence, and the effect of miR-137 on DAT expression and function does not depend on the VNTR copy-number. After transfection of anti-miR-137 and anti-miR-491, the endogenous miR-137 and miR-491 were inhibited and the negative regulation of the endogenous miRNAs on its target gene DAT was released, and therefore the levels of mRNA and protein were up-regulated.
However, we were unable to find an effective experimental animal model to demonstrate that miR-137 and miR-491 affect DA function and animal behavior through post-transcriptional regulation of the DAT because of the limitation associated with the binding-site homology of the two miRNAs. We hope to be able to use central neurons or myocardial cells with comparatively higher levels of DAT expression and suitable animal models to further study the effects of miR-137 and miR-491 on the expression and function of the DAT and the relationship between miRNA disturbances and DAT-related diseases within the scope of medical research ethics.
In summary, our results demonstrated that miRNAs have important regulatory effects on the expression and function of the DAT, a key element in the DA signaling pathway. miRNA interventions may be considered a practical therapeutic strategy for diseases associated with DAT dysfunction. These findings extend our understanding of the DAT and the DA signaling system.
Acknowledgments
This work was supported by grants from the National Postdoctoral Science Foundation, China (2014M552219), the Natural Science Foundation of Guangdong Province, China (2015A030313889, 2015A030401013, 2014A030313709, and 2014A030313710), the Science and Technology Planning Project of Shenzhen Municipality, China (ZDSYS201504301045406, JCYJ20150403110829621, JCYJ20150403091443301, JCYJ20140415162542975, JCYJ20140415162338855, JCYJ20140828163634004, and JCYJ20120616144352139), and the Health and Family Planning Commission Project of Shenzhen Municipality, China (201401026).
Contributor Information
Lin Lu, Email: linlu@bjmu.edu.cn.
Yun Chen, Email: chenyunpku@sina.cn.
References
- 1.Pramod AB, Foster J, Carvelli L, Henry LK. SLC6 transporters: structure, function, regulation, disease association and therapeutics. Mol Aspects Med. 2013;34:197–219. doi: 10.1016/j.mam.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hitri A, Hurd YL, Wyatt RJ, Deutsch SI. Molecular, functional and biochemical characteristics of the dopamine transporter: regional differences and clinical relevance. Clin Neuropharmacol. 1994;17:1–22. doi: 10.1097/00002826-199402000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Iversen L. Neurotransmitter transporters and their impact on the development of psychopharmacology. Br J Pharmacol. 2006;147(Suppl 1):S82–S88. doi: 10.1038/sj.bjp.0706428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikolaus S, Antke C, Hautzel H, Mueller HW. Pharmacological treatment with L-DOPA may reduce striatal dopamine transporter binding in in vivo imaging studies. Nuklearmedizin. 2016;55:21–28. doi: 10.3413/Nukmed-0764-15-08. [DOI] [PubMed] [Google Scholar]
- 5.Wise RA. Dopamine, learning and motivation. Nat Rev Neurosci. 2004;5:483–494. doi: 10.1038/nrn1406. [DOI] [PubMed] [Google Scholar]
- 6.McHugh PC, Buckley DA. The structure and function of the dopamine transporter and its role in CNS diseases. Vitam Horm. 2015;98:339–369. doi: 10.1016/bs.vh.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Otani S, Bai J, Blot K. Dopaminergic modulation of synaptic plasticity in rat prefrontal neurons. Neurosci Bull. 2015;31:183–190. doi: 10.1007/s12264-014-1507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block ER, Nuttle J, Balcita-Pedicino JJ, Caltagarone J, Watkins SC, Sesack SR, et al. Brain region-specific trafficking of the dopamine transporter. J Neurosci. 2015;35:12845–12858. doi: 10.1523/JNEUROSCI.1391-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng MH, Bahar I. Molecular mechanism of dopamine transport by human dopamine transporter. Structure. 2015;23:2171–2181. doi: 10.1016/j.str.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mergy MA, Gowrishankar R, Davis GL, Jessen TN, Wright J, Stanwood GD, et al. Genetic targeting of the amphetamine and methylphenidate-sensitive dopamine transporter: on the path to an animal model of attention-deficit hyperactivity disorder. Neurochem Int. 2014;73:56–70. doi: 10.1016/j.neuint.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sery O, Paclt I, Drtilkova I, Theiner P, Kopeckova M, Zvolsky P, et al. A 40-bp VNTR polymorphism in the 3′-untranslated region of DAT1/SLC6A3 is associated with ADHD but not with alcoholism. Behav Brain Funct. 2015;11:21. doi: 10.1186/s12993-015-0066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ettinger U, Merten N, Kambeitz J. Meta-analysis of the association of the SLC6A3 3′-UTR VNTR with cognition. Neurosci Biobehav Rev. 2016;60:72–81. doi: 10.1016/j.neubiorev.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 13.Sano A, Kondoh K, Kakimoto Y, Kondo I. A 40-nucleotide repeat polymorphism in the human dopamine transporter gene. Hum Genet. 1993;91:405–406. doi: 10.1007/BF00217369. [DOI] [PubMed] [Google Scholar]
- 14.VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats element in DAT1 regulates in vitro dopamine transporter density. BMC Genet. 2005;6:55. doi: 10.1186/1471-2156-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandenbergh DJ, Persico AM, Hawkins AL, Griffin CA, Li X, Jabs EW, et al. Human dopamine transporter gene (DAT1) maps to chromosome 5p15.3 and displays a VNTR. Genomics. 1992;14:1104–1106. doi: 10.1016/S0888-7543(05)80138-7. [DOI] [PubMed] [Google Scholar]
- 16.Jeong SH, Choi KS, Lee KY, Kim EJ, Kim YS, Joo EJ. Association between the dopamine transporter gene (DAT1) and attention deficit hyperactivity disorder-related traits in healthy adults. Psychiatr Genet. 2015;25:119–126. doi: 10.1097/YPG.0000000000000086. [DOI] [PubMed] [Google Scholar]
- 17.Sambataro F, Podell JE, Murty VP, Das S, Kolachana B, Goldberg TE, et al. A variable number of tandem repeats in the 3′-untranslated region of the dopamine transporter modulates striatal function during working memory updating across the adult age span. Eur J Neurosci. 2015;42:1912–1918. doi: 10.1111/ejn.12956. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Zhou S, Ding F, Gu X. Non-coding RNAs as emerging regulators of neural injury responses and regeneration. Neurosci Bull. 2016;32:253–264. doi: 10.1007/s12264-016-0028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan J. Path from schizophrenia genomics to biology: gene regulation and perturbation in neurons derived from induced pluripotent stem cells and genome editing. Neurosci Bull. 2015;31:113–127. doi: 10.1007/s12264-014-1488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, et al. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33:1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Zhang J, Du Y, Jia X, Yang F, Si S, et al. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. 2015;243:523–532. doi: 10.1016/j.atherosclerosis.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 26.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 27.Ciliax BJ, Drash GW, Staley JK, Haber S, Mobley CJ, Miller GW, et al. Immunocytochemical localization of the dopamine transporter in human brain. J Comp Neurol. 1999;409:38–56. doi: 10.1002/(SICI)1096-9861(19990621)409:1<38::AID-CNE4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 28.Rothman RB, Baumann MH, Prisinzano TE, Newman AH. Dopamine transport inhibitors based on GBR12909 and benztropine as potential medications to treat cocaine addiction. Biochem Pharmacol. 2008;75:2–16. doi: 10.1016/j.bcp.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Olivares J, Torres-Salazar D, Owens WA, Baust T, Siderovski DP, Amara SG, et al. Inhibition of dopamine transporter activity by G protein betagamma subunits. PLoS ONE. 2013;8:e59788. doi: 10.1371/journal.pone.0059788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones SR, Gainetdinov RR, Jaber M, Giros B, Wightman RM, Caron MG. Profound neuronal plasticity in response to inactivation of the dopamine transporter. Proc Natl Acad Sci USA. 1998;95:4029–4034. doi: 10.1073/pnas.95.7.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blum K, Oscar-Berman M, Barh D, Giordano J, Gold M. Dopamine genetics and function in food and substance abuse. J Genet Syndr Gene Ther. 2013 doi: 10.4172/2157-7412.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baik JH. Dopamine signaling in reward-related behaviors. Front Neural Circuits. 2013;7:152. doi: 10.3389/fncir.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velazquez-Sanchez C, Garcia-Verdugo JM, Murga J, Canales JJ. The atypical dopamine transport inhibitor, JHW 007, prevents amphetamine-induced sensitization and synaptic reorganization within the nucleus accumbens. Prog Neuropsychopharmacol Biol Psychiatry. 2013;44:73–80. doi: 10.1016/j.pnpbp.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan RA, Foster JD. Mechanisms of dopamine transporter regulation in normal and disease states. Trends Pharmacol Sci. 2013;34:489–496. doi: 10.1016/j.tips.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksen J, Jø rgensen TN, Gether U. Regulation of dopamine transporter function by protein-protein interactions: new discoveries and methodological challenges. J Neurochem. 2010;113:27–41. doi: 10.1111/j.1471-4159.2010.06599.x. [DOI] [PubMed] [Google Scholar]
- 36.Sager JJ, Torres GE. Proteins interacting with monoamine transporters: current state and future challenges. Biochemistry. 2011;50:7295–7310. doi: 10.1021/bi200405c. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Terry DS, Shi L, Quick M, Weinstein H, Blanchard SC, et al. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barreto-Valer K, Lopez-Bellido R. Macho Sanchez-Simon F, Rodriguez RE. Modulation by cocaine of dopamine receptors through miRNA-133b in zebrafish embryos. PLoS ONE. 2012;7:e52701. doi: 10.1371/journal.pone.0052701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strazisar M, Cammaerts S, van der Ven K, Forero DA, Lenaerts AS, Nordin A, et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol Psychiatry. 2015;20:472–481. doi: 10.1038/mp.2014.53. [DOI] [PubMed] [Google Scholar]
- 40.Olde Loohuis NF, Nadif Kasri N, Glennon JC, van Bokhoven H, Hebert SS, Kaplan BB, et al. The schizophrenia risk gene MIR137 acts as a hippocampal gene network node orchestrating the expression of genes relevant to nervous system development and function. Prog Neuropsychopharmacol Biol Psychiatry. 2016 doi: 10.1016/j.pnpbp.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma G, Yin J, Fu J, Luo X, Zhou H, Tao H, et al. Association of a miRNA-137 polymorphism with schizophrenia in a Southern Chinese Han population. Biomed Res Int. 2014;2014:751267. doi: 10.1155/2014/751267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen H, Wang L, Ge X, Jiang CF, Shi ZM, Li DM, et al. MicroRNA-137 inhibits tumor growth and sensitizes chemosensitivity to paclitaxel and cisplatin in lung cancer. Oncotarget. 2016;7:20728–20742. doi: 10.18632/oncotarget.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neault M, Mallette FA, Richard S. miR-137 modulates a tumor suppressor network-inducing senescence in pancreatic cancer cells. Cell Rep. 2016;14:1966–1978. doi: 10.1016/j.celrep.2016.01.068. [DOI] [PubMed] [Google Scholar]
- 44.Liang ML, Hsieh TH, Ng KH, Tsai YN, Tsai CF, Chao ME, et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget. 2016;7:19723–19737. doi: 10.18632/oncotarget.7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun J, Zheng G, Gu Z, Guo Z. MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J Neurooncol. 2015;122:481–489. doi: 10.1007/s11060-015-1753-x. [DOI] [PubMed] [Google Scholar]
- 46.Ivashchenko DV, Shuvalov SA, Chuprova NA, Kibitov AO. The association of polymorphisms in DAT (40 bp VNTR, C > T 3′UTR) and DBH (-1021 C/T) genes with the severe complications of alcohol withdrawal state. Psychiatr Genet. 2015;25:268–269. doi: 10.1097/YPG.0000000000000101. [DOI] [PubMed] [Google Scholar]
- 47.Stolf AR, Szobot CM, Halpern R, Akutagava-Martins GC, Muller D, Guimaraes LS, et al. Crack cocaine users show differences in genotype frequencies of the 3′UTR variable number of tandem repeats of the dopamine transporter gene (DAT1/SLC6A3) Neuropsychobiology. 2014;70:44–51. doi: 10.1159/000365992. [DOI] [PubMed] [Google Scholar]
- 48.Feher A, Juhasz A, Pakaski M, Kalman J, Janka Z. Association between the 9 repeat allele of the dopamine transporter 40 bp variable tandem repeat polymorphism and Alzheimer’s disease. Psychiatry Res. 2014;220:730–731. doi: 10.1016/j.psychres.2014.07.060. [DOI] [PubMed] [Google Scholar]
- 49.Giana G, Romano E, Porfirio MC, D’Ambrosio R, Giovinazzo S, Troianiello M, et al. Detection of auto-antibodies to DAT in the serum: interactions with DAT genotype and psycho-stimulant therapy for ADHD. J Neuroimmunol. 2015;278:212–222. doi: 10.1016/j.jneuroim.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Brown BK, Murrell J, Karne H, Anand A. The effects of DAT1 genotype on fMRI activation in an emotional go/no-go task. Brain Imaging Behav. 2016 doi: 10.1007/s11682-016-9516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kasparbauer AM, Rujescu D, Riedel M, Pogarell O, Costa A, Meindl T, et al. Methylphenidate effects on brain activity as a function of SLC6A3 genotype and striatal dopamine transporter availability. Neuropsychopharmacology. 2015;40:736–745. doi: 10.1038/npp.2014.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Liu Y, Granberg KJ, Wang Q, Moore LM, Ji P, et al. Two mature products of MIR-491 coordinate to suppress key cancer hallmarks in glioblastoma. Oncogene. 2015;34:1619–1628. doi: 10.1038/onc.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng G, Jia X, Peng C, Deng Y, Yin J, Zhang Z, et al. The miR-491-3p/mTORC2/FOXO1 regulatory loop modulates chemo-sensitivity in human tongue cancer. Oncotarget. 2015;6:6931–6943. doi: 10.18632/oncotarget.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore LM, Zhang W. MIR-491: CDKN2A tumor suppressor co-pilot. Oncoscience. 2015;2:825–826. doi: 10.18632/oncoscience.201. [DOI] [PMC free article] [PubMed] [Google Scholar]