Abstract
Cytokinesis, the terminal event in the canonical cell cycle, physically separates daughter cells following mitosis. For cleavage to occur in many eukaryotes, a cytokinetic ring must assemble and constrict between divided genomes. Although dozens of different molecules localize to and participate within the cytokinetic ring, the core machinery comprises linear actin filaments. Accordingly, formins, which nucleate and elongate F-actin (filamentous actin) for the cytokinetic ring, are required for cytokinesis in diverse species. In the present article, we discuss specific modes of formin-based actin regulation during cell division and highlight emerging mechanisms and questions on this topic.
Keywords: actin, cytokinesis, cytoskeleton, formin
Cytokinesis is an actomyosin-based process in many eukaryotes
To complete daughter cell separation following mitosis, many well-studied organisms utilize a contractile-based apparatus termed the CR (cytokinetic ring). This structure, which is structurally and functionally based on F-actin (filamentous actin) and non-muscle myosin II, enables constriction at the division site during cytokinesis. In metazoans, cytokinetic constriction manifests as an ingressing cleavage furrow, which narrows into a thread-like midbody that later becomes resolved during cellular abscission. In fungi, such as yeasts, the constricting CR also guides new cell wall deposition at the division plane [1]. Given that this structure can both organize and drive cleavage of the cellular cytoplasm, proper CR functioning represents a conserved determinant of cell division integrity.
In multiple species, formins localize to the future division site, where they assemble F-actin for CR formation. Although it is clear that both the localization and activity of cytokinetic formins is controlled to ensure proper CR performance, our understanding of how this regulation occurs and which formin activities are specifically affected by various signalling inputs has only begun to take shape. In the present mini-review, we introduce formin molecules in more detail and then discuss recent insights into formin function and regulation during cytokinesis.
Formin domains and mechanisms of action
The formin family of F-actin assembly factors are defined by FH (formin homology) domains. The FH1 and FH2 domains comprise a characteristic core responsible for multiple formin activities (Figure 1). Dimeric FH2 domains [2] are sufficient to nucleate F-actin assembly in vitro [3,4], but in vivo may also interact with cofactors such as budding yeast Bud6, which enhances nucleation efficiency [5]. Whereas FH2 domains remain processively associated to F-actin barbed ends with high affinity [4,6,7], rapid F-actin elongation also requires FH1 domain binding to the actin monomer-binding protein profilin [3,8,9], which increases local G-actin (globular actin) concentrations and orients monomers on to the FH2-associated barbed end [8,10,11].
Figure 1. Multiple domains contribute to formin function and regulation.
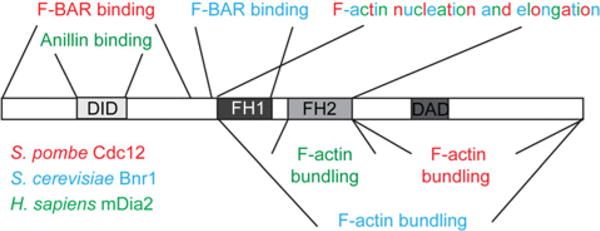
Formins contain signature FH domains and often contain DID and DAD motifs that can interact to autoinhibit nucleation and elongation activities. S. pombe Cdc12 (red), S. cerevisiae Bnr1 (blue) and Homo sapiens mDia2 (green) catalyse F-actin nucleation and elongation via their FH1–FH2 core, and each also contains an F-actin bundling domain within its C-terminus. In addition, cytokinetic formins participate in important protein–protein interactions at the CR, and the binding domains responsible for these interactions reside within these proteins’ N-terminal halves.
How are the actin assembly properties of the formin FH1–FH2 core regulated? A common mode of formin regulation involves two additional formin domains, the N-terminal DID (diaphanous inhibitory domain) and the C-terminal DAD (diaphanous autoregulatory domain) (Figure 1). DID–DAD cis interactions can establish an autoinhibitory conformation that precludes F-actin assembly by the FH1–FH2 core [12]. Formins autoregulated by DID–DAD interactions must be freed from autoinhibition for full activity. Interaction with Rho proteins, via Rho-binding domains on formins, commonly relieves DID– DAD autoinhibitory interactions partially [12], although some formins, including Saccharomyces cerevisiae Bni1 and mammalian FHOD1, are freed from autoinhibition by phosphorylation [13,14].
Beyond nucleation and elongation, formins modify the actin cytoskeleton through additional activities, such as F-actin bundling and severing. Intriguingly, some formins, including mammalian formins FRL1 and mDia2 and plant formin FH8, perform these activities via their FH1–FH2 domains, whereas others, including mammalian formins INF2 and FRL2, possess distal domains responsible for these functions [12] (Figure 1). In the future, it will be important to clarify the conservation of these additional activities among eukaryotic formins and the extent to which they influence formin-mediated actin assembly.
Cytokinetic formin function and regulation in eukaryotes
Formins participate in cytokinesis in a variety of eukaryotes, ranging from single-celled yeast to multicellular organisms. In S. cerevisiae, two formins, Bni1 and Bnr1, localize to the bud neck and together are required for assembly of an actin-based CR [15,16]. A strain lacking both formins exhibits a high degree of multinucleation, whereas those lacking either individually undergo relatively efficient cytokinesis [16]. Although F-actin is required for budding yeast cytokinesis [17], myosin-independent cytokinesis can occur in certain strain backgrounds by a mechanism relying on septins and abnormal cell wall deposition [18]. Formins affect myosin-independent cytokinesis in S. cerevisiae because Bni1-mediated actin assembly influences septin ring formation [19], and Bnr1 and Bni1 interact with the chitin synthase activator Hof1 [20], an F-BAR protein whose homologue Cdc15 in fission yeast Schizosaccharomyces pombe likewise participates in a formin interaction during cytokinesis [21] (Figure 1). Rho binding and phosphorylation affect cytokinetic formin targeting and activation in S. cerevisiae. Specifically, Bnr1- and Bni1-dependent actin assembly is influenced by Rho1, Rho3 and Rho4 GTPases [15,22], and phosphorylation of Bni1 by Prk1 facilitates Bni1 activation to relieve autoinhibition [14].
In metazoans, formins are central players in cytokinesis, although the range of their activities towards actin and their mechanisms of regulation are less characterized. Both Caenorhabditis elegans and Drosophila melanogaster require formins CYK-1 and Diaphanous respectively for early embryonic divisions, owing to their essential roles in building the cytokinetic machinery [23,24]. In human cells, formin mDia2 localizes to the division site and nucleates F-actin for CR formation [25]. Furthermore, mDia2, similar to budding yeast Bnr1 [26], bundles F-actin in vitro [27] (Figure 1), hinting that cytokinetic formins may generally cross-link F-actin to aid in CR formation. At the conclusion of cytokinesis, mDia2 is primed for degradation by ubiquitination [28], providing a turnover mechanism that may be pertinent to CR disassembly.
Despite the studies described above, our understanding of how cytokinetic formins function and are regulated is incomplete. To provide a more comprehensive view, our laboratories are interested in defining mechanisms controlling the sole fission yeast cytokinetic formin, Cdc12. Although it is the least abundant protein that has been measured at the fission yeast CR [29], Cdc12 is essential for S. pombe cell division. In its absence, cells undergo mitosis, but never form a CR, and cdc12-null cells die as multinucleates [30]. As the timing and relative order of events has been well described for S. pombe cytokinesis and a catalogue of molecular participants has been established [31], S. pombe provides a useful genetically tractable system in which to investigate formin contributions to cell division. As discussed below, many elements of Cdc12 function and regulation remain to be clarified.
Localization of S. pombe Cdc12 during cell division
During interphase, S. pombe Cdc12 forms spot-like structures (Figure 2), which traverse the cell on actin and microtubule networks [32]. Spots serve as reservoirs for Cdc12 and other CR components during interphase [33], including F-BAR protein Cdc15. Cdc12 spots disassemble in wild-type cells as the CR is assembled at mitotic onset [33,34]. Medial nodes established by anillin-like Mid1 in late G2-phase organize two modules that recruit and concentrate Cdc12 medially [35]; one is based on myosin and IQGAP Rng2 and the other on F-BAR Cdc15. Whereas mDia2 interacts with anillin [36], Cdc15 is the sole node protein known to bind Cdc12 directly [21] (Figure 1), but another must exist because Cdc12 is still recruited medially in the absence of Cdc15 [35]. Upon formin activation, nodes coalesce into a complete ring structure [37] via a search–capture–pull–release mechanism involving myosin and Cdc12-nucleated actin filaments from opposite nodes [38]. In the absence of nodes, medial Cdc12 is more broadly distributed, and CR assembly is delayed as a result [39]. Intriguingly, pre-existing actin cables are also incorporated into assembling CRs [40], as are non-medial F-actin nucleated by small subpopulations of Cdc12 scattered throughout the cell [41]. F-actin polymerization remains dynamic at the CR throughout cytokinesis [40]. Fittingly, Cdc12 stays at the CR through its constriction, although Cdc12 also disassembles with the constricting CR [30]. Whether and how Cdc12 activities are modulated throughout cytokinesis has not been extensively investigated.
Figure 2. Sid2-mediated phosphorylation of S. pombe Cdc12 during cytokinesis relieves formin multimerization and facilitates SIN-dependent cytokinesis.
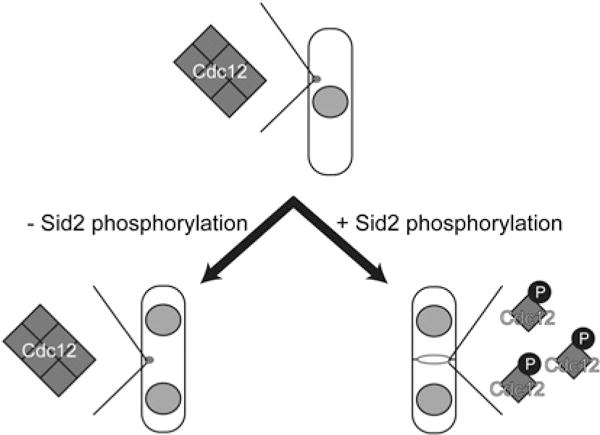
During interphase, Cdc12 forms spot-like structures, which disassemble before CR assembly. Multimerization through a C-terminal domain facilitates Cdc12 spot formation during interphase. Upon SIN activation, Sid2-mediated phosphorylation reverses Cdc12 multimerization, ensuring the efficient formin de-clustering required for SIN-dependent CR assembly and maintenance.
Regulation of Cdc12 activities during cytokinesis
When compared with the catalytic activity of cytokinetic formins in other species, the Cdc12 FH1–FH2 core is a more efficient F-actin nucleator [42]. However, how Cdc12-mediated F-actin nucleation is activated in vivo is presently unknown. Like other formins, Cdc12-mediated F-actin elongation is regulated by profilin [8], which binds the proline-rich FH1 domain [30]. Molecularly, profilin ‘gates’ Cdc12 capping activity, freeing the barbed end for polymerization upon binding to the FH1 domain [8,34]. Because the FH1–FH2 core of Cdc12 essentially caps F-actin when not bound to profilin [8,34,43], cells expressing mutant Cdc12 incapable of binding profilin exhibit abnormal CR and actin dynamics [34]. Notably, profilin influences the competition between Cdc12 and S. pombe capping proteins for F-actin barbed end binding. Whereas barbed ends elongate when bound to Cdc12–profilin complexes, barbed ends bound to capping proteins do not [43]. Presumably, any modulation of this competition would influence CR structure during cytokinesis. Another CR factor, tropomyosin, can either stimulate or inhibit Cdc12-mediated actin assembly in vitro depending on the context. During Cdc12-mediated F-actin elongation, tropomyosin enhances filament length by slightly increasing the elongation rate and by enabling F-actin annealing. However, this annealing activity may ultimately ‘trap’ Cdc12 between actin filaments and/or promote Cdc12 dissociation from barbed ends [44]. How these complex effects on Cdc12-mediated actin assembly are co-ordinated in vivo is important to clarify in future studies.
Does the central cell cycle machinery influence Cdc12 activity during cytokinesis? Because Cdc12 hyperactivity results in cell lethality [8], it seemed plausible that Cdc12 function is temporally and spatially fine-tuned by cell cycle signals. The common formin DID–DAD autoinhibitory interaction does not appear to operate, or, at least, not to have a significant effect in the case of Cdc12 [34]. However, overexpression of a Cdc12 fragment lacking the C-terminus induces cytokinesis in interphase [45]. This finding indicated that the Cdc12 C-terminus has intrinsic inhibitory activity and also suggested that a mechanism restraining Cdc12 activity normally operates during interphase. Yet, how cell cycle cues affect the ability of the C-terminus to inhibit Cdc12 activity or how the C-terminus influences Cdc12 function remained unclear.
We found that Cdc12 is phosphorylated in a cell-cycle-dependent manner, with hyperphosphorylation occurring immediately before cytokinesis [46]. Using a candidate kinase approach, we identified the SIN (septation initiation network), a conserved kinase cascade related to the Hippo pathway [47], as a key participant in Cdc12 phosphorylation during cell division [46]. Specifically, Sid2, the terminal kinase in the SIN cascade, phosphorylates Cdc12 on four residues located outside the characteristic formin domains. Eliminating the four Sid2 phosphosites on Cdc12 caused persistent Cdc12 clustering in spots (Figure 2). CR assembly can occur in the absence of Mid1 or nodes through a SIN-dependent pathway [48,49]. However, hyper-clustering of the non-phosphorylatable Cdc12 mutant precluded Mid1-independent CR assembly (Figure 2), validating loss of a critical SIN input. We found further that Sid2 phosphorylation facilitates formin de-clustering during cytokinesis by inhibiting multimerization of a novel C-terminal domain, explaining why the Cdc12 C-terminus is inhibitory to its function when overproduced [46]. Nonetheless, oligomerization through this domain is important earlier in CR assembly for Cdc12-mediated F-actin bundling (Figure 1). Therefore SIN-mediated phosphoregulation of Cdc12 functions as an oligomeric switch, which temporally distinguishes and allows for Cdc12-dependent F-actin bundling during CR assembly and Cdc12 de-clustering later during cytokinesis [46].
Concluding points
In summary, cytokinetic formins are critical for cell division in multiple species, and these formins are subject to precise cell cycle control. Focusing on S. pombe Cdc12, our laboratory and others have uncovered several new aspects of cytokinetic formin regulation, which provide insights into the process of cell division, and may inform our understanding of formin function in other contexts and in other organisms.
Multiple domains, including a newly identified C-terminal multimerization domain, contribute to Cdc12 function and regulation during cytokinesis (Figure 1). Separation of the Cdc12 multimerization domain, which permits F-actin bundling, from its FH1–FH2 core presents an opportunity for differential regulation of distinct facets of Cdc12-mediated actin assembly. In the future, it will be important to assess whether the Cdc12 C-terminus affects nucleation and elongation activities of the FH1–FH2 core in addition to directly conferring F-actin bundling capacity. What role does the Cdc12 N-terminus play in S. pombe cytokinetic formin regulation? It is very likely that the Cdc12 N-terminus mediates important protein–protein interactions at the CR. The Cdc12 N-terminus is sufficient for CR targeting during cytokinesis [34] and has been demonstrated to bind the F-BAR protein Cdc15 [21] (Figure 1), providing a possible link between F-actin and the division site cortex. Determining whether Cdc15 or yet unidentified N-terminal interactors influence Cdc12 during cytokinesis should clarify the extent to which this region participates in Cdc12 regulation. Although the Cdc12 DID and DAD motifs appear to be unimportant [34], it remains possible that Cdc12 is autoinhibited in a non-canonical fashion, and further investigation of Cdc12 self-interactions could be informative.
It seems likely that other master cell cycle regulators besides the SIN participate in S. pombe Cdc12 regulation, because a mutant lacking Sid2-targeted phosphosites is still phosphorylated, but to a lesser degree than the wild-type protein, as cells enter mitosis. Identification of kinases responsible and analysis of their effects on Cdc12 activity, protein–protein interactions and targeting is important for assessing how these phosphorylation events control S. pombe cytokinesis. An interesting possibility is that both SIN-independent and SIN-dependent Cdc12 phosphorylations co-operate in full activation of Cdc12 during cytokinesis. Is Cdc12 targeted by post-translational modifications other than phosphorylation? Because S. pombe cells are highly sensitive to changes in Cdc12 protein levels [8], it is possible that Cdc12, like mDia2 [28], is actively targeted for degradation by ubiquitination. If so, this modification could facilitate Cdc12 turnover within the CR during CR disassembly and aid in Cdc12 clearance from the division site. In summary, a more comprehensive understanding of the roles that post-translational modifications serve in cytokinetic formin regulation should enhance our understanding of the molecular cues guiding eukaryotic cell division.
Acknowledgments
We thank members of the Gould and Kovar laboratories for helpful discussions.
Funding
Our work is supported by the National Institutes of Health [grant numbers T32-CA119925 (to K.A.B.), GM079265 (to D.R.K.) and GM101035 (to K.L.G.)].
Abbreviations used
- CR
cytokinetic ring
- DAD
diaphanous autoregulatory domain
- DID
diaphanous inhibitory domain
- F-actin
filamentous actin
- FH
formin homology
- G-actin
globular actin
- SIN
septation initiation network
References
- 1.Balasubramanian MK, Srinivasan R, Huang Y, Ng KH. Comparing contractile apparatus-driven cytokinesis mechanisms across kingdoms. Cytoskeleton. 2012;69:942–956. doi: 10.1002/cm.21082. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 3.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 4.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 5.Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 11.Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 13.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBOJ. 2008;27:618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Neo SP, Cai M. Regulation of the yeast formin Bni1p by the actin-regulating kinase Prk1p. Traffic. 2009;10:528–535. doi: 10.1111/j.1600-0854.2009.00893.x. [DOI] [PubMed] [Google Scholar]
- 15.Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol. 2002;12:1864–1870. doi: 10.1016/s0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- 16.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBOJ. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolliday N, Pitcher M, Li R. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol Biol Cell. 2003;14:798–809. doi: 10.1091/mbc.E02-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p: implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- 21.Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Y, Pruyne D, Bretscher A. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J Cell Biol. 2003;161:1081–1092. doi: 10.1083/jcb.200212040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Severson AF, Baillie DL, Bowerman B. A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- 24.Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 27.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 28.DeWard AD, Alberts AS. Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J Biol Chem. 2009;284:20061–20069. doi: 10.1074/jbc.M109.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 30.Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- 33.Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 39.Saha S, Pollard TD. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Mol Biol Cell. 2012;23:3982–3992. doi: 10.1091/mbc.E12-07-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 41.Huang J, Huang Y, Yu H, Subramanian D, Padmanabhan A, Thadani R, Tao Y, Tang X, Wedlich-Soldner R, Balasubramanian MK. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J Cell Biol. 2012;199:831–847. doi: 10.1083/jcb.201209044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yonetani A, Chang F. Regulation of cytokinesis by the formin cdc12p. Curr Biol. 2010;20:561–566. doi: 10.1016/j.cub.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bohnert KA, Grzegorzewska AP, Willet AH, Vander Kooi CW, Kovar DR, Gould KL. SIN-dependent phosphoinhibition of formin multimerization controls fission yeast cytokinesis. Genes Dev. 2013;27:2164–2177. doi: 10.1101/gad.224154.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hergovich A, Hemmings BA. Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin Cell Dev Biol. 2012;23:794–802. doi: 10.1016/j.semcdb.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]


