Abstract
Cytokinesis represents the final stage in the cell cycle, in which two daughter cells, each with their complement of the duplicated genome, physically separate. At the core of this process sits highly conserved machinery responsible for specifying the plane of division, building a contractile apparatus and ultimately cleaving cells in two. Although the ‘parts list’ of contributing proteins has been well described, mechanisms by which these parts are spatially and temporally regulated are only beginning to be understood. With advancements in biochemical and proteomic analyses, recent work has uncovered multiple new roles for post-translational modifications in the regulation of cytokinesis. Here, we review these latest findings and interpret our current understanding of cytokinesis in light of relevant modifications.
Modifications as a means to regulate cytokinesis
In a variety of organisms and cell types, cytokinesis follows nuclear division and directs the cleavage of the cellular cytoplasm. Although this process ultimately manifests itself with the physical separation of two daughter cells, myriads of preliminary events set the necessary framework to ensure the accurate execution of this concluding stage in the cell cycle. Cytokinesis initially requires the selection of an appropriate division site. Following this, a contractile apparatus assembles at the division site and undergoes constriction. Daughter cells fully separate as cleavage completes during abscission, and each daughter cell then initiates its division cycle anew (Figure 1). Actin filaments combined with nonmuscle myosin II provide the actomyosin core of the cytokinetic machinery in many organisms. Additionally, dozens of accessory proteins localize to the site of division and dynamically impact cytokinesis. Given the conservation of many of these factors among eukaryotes, regulatory mechanisms that mediate this process in one organism probably mirror those used by others (reviewed in [1,2]).
Figure 1.
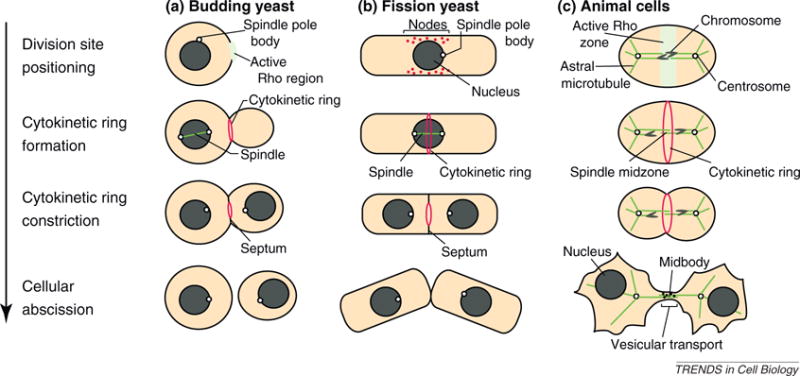
Stages of eukaryotic cytokinesis. Schematic representations of the process of cytokinesis in (a) the budding yeast S. cerevisiae, (b) the fission yeast S. pombe and (c) animal cells. Examples of cells at individual stages of cytokinesis are presented, with progression through the cell cycle oriented downward. In budding yeast and animal cells, the cytokinetic apparatus is positioned and assembled from an active Rho region. In wild type fission yeast, cytokinetic ring assembly instead initiates from node-like structures containing formin and myosin II, although initial ring assembly can occur via node-independent mechanisms in mutants lacking these structures. Following the constriction of the cytokinetic apparatus in budding and fission yeasts, new cell wall material is deposited at the division site to form a septum, which is subsequently cleaved to allow for cell separation (reviewed in [1,2]). In animal cells, vesicular transport to the midbody, the microtubule-based remnant of the anaphase spindle midzone, likewise promotes abscission via its effects on membrane composition at the division site as well as its delivery of important cleavage factors (reviewed in [52]).
The post-translational modification of proteins serves as one means by which protein activity, localization and interactions acquire temporal and spatial specificity. Although roles for modifications in general cell cycle control have been known for some time (reviewed in [3,4]), their varied contributions to cytokinesis have only recently gained appreciation on a broader spectrum, most notably because of improved techniques in targeted and genome-wide proteomics. In this review, we highlight recent advances in the field of cytokinesis, with a specific focus on regulatory post-translational modifications (Figure 2). In addition to addressing the consequences of phosphorylation and ubiquitination in individual stages of cytokinesis, we present smaller discussions of other modifications emerging as modulators of cytokinesis with the goal of underscoring not only the diversity in signaling achieved through these mechanisms but also their potential for interplay.
Figure 2.
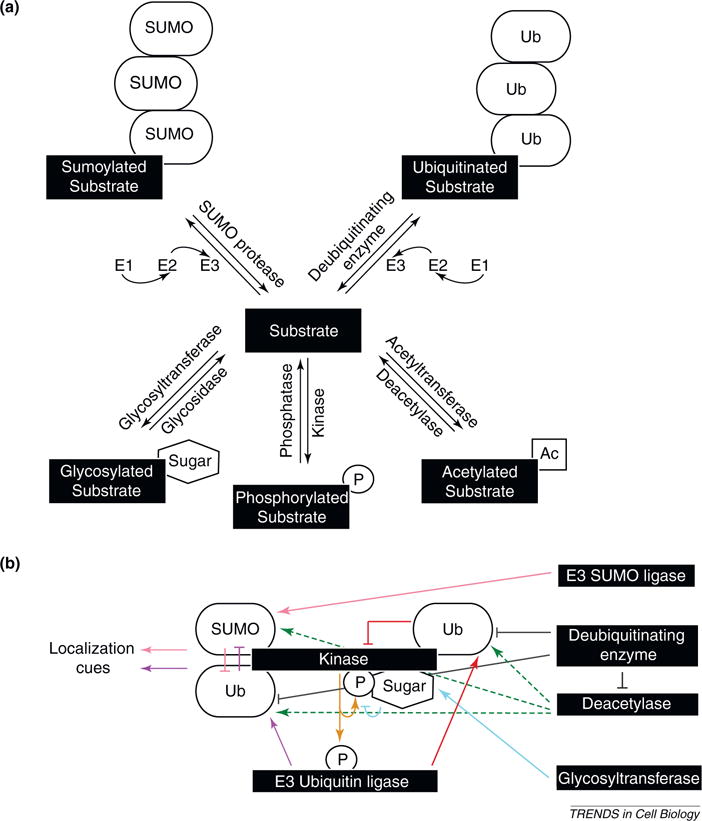
Regulatory post-translational modifications guide cytokinesis. (a) Enzymes responsible for catalyzing or reversing the post-translational modifications of interest are indicated. Ubiquitination and sumoylation are unique in that they require a cascade of E1 activating, E2 conjugating and E3 ligase enzymes and can result in mono-, multi- or polymodifications. (b) Proteins can be influenced by different modifications simultaneously. The schematic broadly illustrates potential for crosstalk based on known modifications of cytokinesis regulators. Protein kinases phosphorylate a variety of proteins during cytokinesis, including themselves and E3 ubiquitin ligases (yellow lines). The phosphorylation of some E3 ubiquitin ligases then drives their activation, leading to the degradation of key cytokinesis factors, including many of the kinases initially responsible for this phosphorylation (red lines) [99,100]. However, not all ubiquitination causes degradation. In fact, E3 ligases can also mediate regulatory ubiquitination, which commonly affects protein localization in direct and indirect ways (purple lines) [70,81]. Modification by SUMO has been reported to similarly regulate protein localization (pink lines) [94] and, on a global scale, ubiquitination and sumoylation could potentially compete for the same sites on proteins (purple and pink lines), given that they both target lysines. Deubiquitinating enzymes, which remove ubiquitin from proteins, presumably influence this balance (gray lines). In addition to ubiquitin and SUMO, acetyl groups are also attached to lysine residues. Deacetylase enzymes could thereby free potential lysines for either ubiquitination or sumoylation (green lines, which are dashed because the effect is indirect). Crosstalk between acetylation and ubiquitination is probably complicated by interactions among enzymes involved in these modifications, with a deubiquitinating enzyme inhibiting deacetylase activity in one reported case [98]. Thus, if relevant deubiquitinating enzymes are indeed active, they might remove ubiquitin from key substrates, inactivate relevant deacetylases and allow for the accumulation of acetyl groups on lysines previously modified by ubiquitin. Additionally, just as ubiquitination, sumoylation and acetylation can compete for sites, recent findings suggest that glycosylation can occur on residues identical to or flanking phosphorylation sites, and this can in turn affect the phosphorylation state of glycosylated proteins, many of which are kinases (blue lines) [90]. Accordingly, crosstalk during cytokinesis is multifaceted, with singular events often acting both upstream and downstream of numerous others. Ub, ubiquitin; SUMO, small ubiquitin-like modifier; P, phosphate group; Ac, acetyl group.
Phosphorylation
The covalent attachment of phosphate groups to proteins by kinases is a widespread post-translational modification (Figure 2a). This modification can exert positive, negative and even cooperative effects depending on the context (Figure 2b). The reversal of phosphorylation by phosphatases contributes an additional level of control to this modification. Although the description below focuses on symmetric cell divisions, phosphorylation contributes to the regulation of asymmetric divisions as well (Box 1).
Box 1. Phosphoregulation of asymmetric cell divisions.
Asymmetric cell divisions result in daughter cells with unique molecular identities. As part of this process, distinct fate determinants differentially segregate to opposite halves of dividing cells early in the division process. Neuroblasts of D. melanogaster have emerged as leading models for the study of asymmetric cell division (reviewed in [101]). In this organism, the Par and Pins complexes initially define the apical cortex. Atypical protein kinase C (aPKC), which is a member of the Par complex, is activated following Aurora A phosphorylation of another Par complex component, namely Par6 [102]. Active aPKC in turn blocks basal determinants from the apical side via both direct phosphorylation and indirect mechanisms [101,103]. Partner of Inscruteable (Pins), the namesake of the Pins complex, meanwhile binds proteins that can attach to microtubules at the apical cortex, thereby establishing proper spindle alignment and spindle-based furrow positioning [101]. Aurora A phosphorylates Pins in its central linker region. The loss of this phosphorylation impedes the recruitment of proteins involved in microtubule attachment to the apical cortex and severely disrupts spindle orientation [104]. Interestingly, the site phosphorylated by Aurora A in Drosophila is conserved in the mammalian Pins ortholog, and this site is phosphorylated by the aPKC of the mammalian Par complex to likewise affect protein binding and spindle orientation in epithelial cells [105]. Therefore, the phosphorylation of Pins orthologs might represent a conserved means for guiding spindle orientation and division plane positioning during asymmetric cell divisions in various organisms. Even so, recent evidence from Drosophila and C. elegans suggests that spindle-independent pathways based on unequal myosin partitioning also drive asymmetric division plane positioning [106,107]. In Drosophila, the required polarization of myosin depends on apical Pins [106]. Therefore, it will be interesting to assess whether the direct phosphorylation of Pins or other polarity factors contributes to the spindle-independent placement of the cleavage furrow.
Positioning and assembly of the contractile apparatus
In the fission yeast Schizosaccharomyces pombe, two separate but integrated mechanisms, both of which possess ties to upstream kinase regulation, are involved in the assembly of a contractile apparatus (Figure 3a), known as a cytokinetic ring because of its shape. The first mechanism, known as the search, capture, pull and release model, is based on the observation that formin Cdc12 and type II myosin Myo2 localize to a broad series of medial nodes at the onset of mitosis, where they respectively mediate F-actin nucleation and actin filament condensation into a ring structure [5,6]. The recruitment of both Cdc12 and Myo2 into nodes depends on the anillin-related protein Mid1. Nuclear export via phosphorylation by the polo-like kinase Plo1 concentrates Mid1 medially in early mitosis (Figure 3a, left), thereby coupling the nucleus to division plane positioning in this organism [7,8]. Cues from distal cell tips, including the inhibitory phosphorylation of the Mid1-binding protein kinase Cdr2 by the DYRK-family kinase Pom1, meanwhile reinforce medial node distribution to promote the assembly of the cytokinetic ring in the middle of the cell [9–11]. De novo ring assembly from node-like bands of myosin II has also been reported in animal cells [12,13]. Whether this represents a conserved assembly mechanism and whether phosphorylation plays a role in this process in animal cells have yet to be fully addressed.
Figure 3.
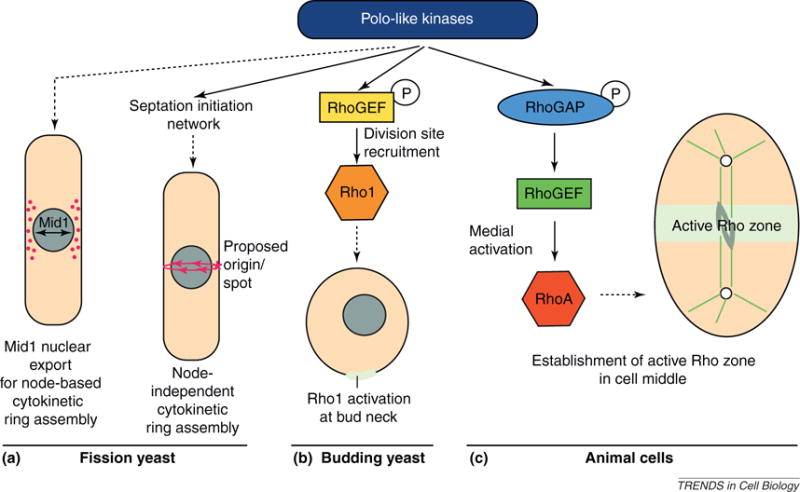
Upstream polo kinase function in cytokinetic ring assembly. Polo-like kinases execute a conserved role in regulating the assembly of the cytokinetic apparatus. In the fission yeast S. pombe, the polo-like kinase Plo1 promotes the accumulation of the anillin-like protein Mid1 in medial nodes [7,8], to which it subsequently recruits cytokinesis factors such as formin and myosin II [5,6] (a, left). However, cytokinesis can occur independently of both Mid1 and nodes through Plo1-mediated activation of the septation initiation network [14,16], and analysis of F-actin in the cytokinetic ring suggests that this might occur through a spot-leading cable model [15] (a, right). In the budding yeast S. cerevisiae (b) and animal cells (c), signaling through polo-like kinases establishes active Rho zones, which are needed for the positioning and assembly of the cytokinetic apparatus. In budding yeast (b), the polo-like kinase Cdc5 phosphorylates (‘P’) RhoGEFs, thereby targeting them to the bud neck and activating Rho1 in this area [25,26]. In animals (c), the polo-like kinase Plk1 instead phosphorylates RhoGAP, which subsequently recruits RhoGEF medially and promotes RhoA activation in the middle of the cell [27,28]. In the figure, solid arrows denote direct activation and/or recruitment, whereas dashed arrows indicate the cellular implications of relevant upstream signaling.
Nonetheless, in the absence of detectable nodes and Midi, the formation of a cytokinetic ring still occurs in S. pombe, albeit with reduced speed and efficiency [14]. The spot/leading cable mechanism of ring assembly might account for this fact, because electron microscopy suggests that actin filaments arise from a single aster and spread around the circumference of the cell to form the cytokinetic ring [15] (Figure 3a, right). A conserved signaling network, known as the septation initiation network in fission yeast, has been implicated in Mid1-independent ring assembly [16]. The septation initiation network, which is orthologous to the mitotic exit network of the budding yeast Saccharomyces cerevisiae, consists of a GTPase-regulated kinase cascade that is initially triggered through polo-like kinase. The most downstream kinase, known as Dbf2 in S. cerevisiae and Sid2 in S. pombe, phosphorylates Cdc14 family phosphatases to control their cytoplasmic accumulation [17,18], which is significant given the role of Cdc14 phosphatases in reversing phosphorylations catalyzed by M phase cyclin-dependent kinases (CDKs). Although signaling through some kinases, including polo, advances cytokinesis, other kinases, such as M phase CDKs, instead inhibit cytokinesis. Indeed, the dephosphorylation of several microtubule-binding CDK substrates, including protein regulator of cytokinesis 1 (PRC1), facilitates the formation of the spindle midzone [19], and the reduction of CDK activity in mitotically arrested mammalian cells causes cytokinesis [20]. In S. pombe, the Cdc14 family phosphatase Clp1, which is activated via the autodephosphorylation of CDK phosphosites [21], dephosphorylates the cytokinetic F-BAR scaffold Cdc15 [22]. The activation of the septation initiation network controls Cdc15 hypophosphorylation [16], which is logical given the role of Sid2-mediated phosphorylation in Clp1 regulation [18]. The dephosphorylation of Cdc15 subsequently promotes Cdc15 scaffolding activity and cytokinetic ring assembly [16,23]. The removal of CDK phosphorylations on the cytokinetic IQGAP in the polymorphic fungus Candida albicans likewise promotes ring formation [24], although it is unclear whether this can be triggered via related signaling pathways. Currently, it is also unknown if the S. pombe septation initiation network or orthologous pathways in other organisms mediate the direct phosphorylation of ring components other than Cdc14 phosphatases to control ring assembly.
In both models of S. pombe ring assembly, signaling begins through polo-like kinase (Figure 3a). In budding yeast (Figure 3b) and animal cells (Figure 3c), polo also influences small Rho GTPases, Rho1 and RhoA, respectively, to control the early stages of cytokinesis and cytokinetic ring assembly. Rho proteins become activated through the loading of GTP by guanine nucleotide exchange factors (GEFs) and inactivated following GTP hydrolysis mediated by GTPase-activating proteins (GAPs). In budding yeast, the polo-like kinase Cdc5 phosphorylates Rho1 GEFs to target them to the bud neck [25]. Rho1 GEFs subsequently bind and activate Rho1 at this site (Figure 3b) [25,26]. In animal cells, polo signaling likewise controls RhoA function, with polo-like kinase Plk1 phosphorylating the RhoGAP Cyk4/MgcRacGAP [27,28]. Cyk4 associates with the kinesin MKLP1 to form centralspindlin, a microtubule-bundling complex that stably accumulates at the spindle midzone once CDK phosphosites on MKLP1 are reversed and once Aurora B phosphorylates MKLP1 to locally impede the 14-3-3 inhibition of central-spindlin in this region [29,30]. Plk1-mediated phosphorylation of Cyk4 then primes centralspindlin for the recruitment of the RhoGEF Ect2 to the midzone and the subsequent medial activation of RhoA [27,28] (Figure 3c). Rho GTPase flux achieved through active RhoGAP meanwhile limits the lateral spread of active Rho zones such that they are maintained medially [31]. RhoGEF and GAP activities are themselves modulated through phosphorylation by diverse kinases such as Aurora B, CDKs and polo-like kinases [32–36]. The protein phosphatase PP2A can reverse such phosphorylation [32], consistent with protein kinases and phosphatases operating in concert to dictate the timing of cytokinesis in various organisms.
Constriction of the contractile apparatus
Although the exact mechanism guiding the constriction of the cytokinetic ring has not been fully defined, the general assumption is that ring constriction proceeds similarly to the constriction of muscle sarcomeres, with antiparallel F-actin sliding on myosin II [1]. Therefore, regulatory inputs impacting myosin II profoundly shape this step of cytokinesis. Like other type II myosins, those involved in cytokinesis possess both heavy and light chains. In animal cells, regulatory light chain phosphorylation at two sites, threonine 18 and serine 19, enhances myosin II ATPase activity as well as myosin II filament formation [37]. Consistent with a role for light chain phosphorylation in cytokinesis, diphosphorylated myosin II promotes total cellular contractility [38,39], and cancer cells that fail cytokinesis exhibit reduced myosin light chain phosphorylation [40]. Although light chain phosphorylation was also previously thought to act as the predominant determinant of the cortical localization of myosin II during cytokinesis [37], more recent research has suggested that factors independent of phosphorylation can control this targeting and thereby impact initial ring assembly [41,42].
A variety of kinases, including two Rho-effector kinases, ROCK and citron kinase, as well as a calmodulin-effector kinase, namely myosin light chain kinase, contribute to myosin II diphosphorylation [37]. Scaffolding through the septin SEPT2 is reported to enable many of these kinases to assemble together with myosin II for robust phosphorylation [43]. The dissociation of ROCK and citron kinases from myosin II has been associated with cleavage furrow regression in Chinese hamster cells [43]. Correspondingly, the silencing of ROCK signaling has been posited to cause the late cytokinesis failure observed during the polyploidization of human megakaryocytes [44], and the knock-down of citron kinase in HeLa cells blocks cytokinesis [45]. The effect of ROCK on myosin II function during cytokinesis is complicated by its phosphorylation of myosin light chain phosphatase. Such phosphorylation indirectly heightens myosin light chain phosphorylation by opposing the binding of myosin light chain phosphatase to myosin II and by also inhibiting the activity of the phosphatase [37]. Unlike Rho-effector kinases, myosin light chain kinase becomes activated by calcium-calmodulin binding. In sea urchin eggs, the forced release of calcium in early metaphase triggers premature cortical contraction, suggesting a prominent role for myosin light chain kinase in cytokinetic contractility in this organism [46]. Consistent with transient calcium-calmodulin-myosin light chain kinase inter-actions inducing localized cellular contraction, the maximal equatorial binding of calcium-calmodulin to myosin light chain kinase occurs just prior to ring constriction during cytokinesis in rat kidney cells [47]. Although all of these kinases can shape myosin activation during cytokinesis, more research is required to detail their relative timings and contributions in different organisms and cell types.
In addition to activating roles for myosin phosphorylation, some identified phosphorylation events instead confer inhibitory cues for cytokinesis. For example, the S. pombe PAK-related kinase Pak1/Orb2 phosphorylates the myosin II light chain Rlc1 to inhibit the constriction of the cytokinetic ring [48], and the phosphorylation of the myosin II heavy chain in the slime mold Dictyostelium actually prevents the formation of myosin II filaments that are responsible for myosin II function in this organism [49]. The involvement of heavy chain phosphorylation in myosin II regulation during cytokinesis has furthermore been documented in fission yeast, where such phosphorylation contrarily stimulates constriction through an unknown mechanism [50]. Clearly, myosin II function in various organisms is controlled through a complicated array of phosphorylation events, although the exact locations and effects of these phosphosites might differ among species.
Separation of daughter cells
Just as the initiation of cytokinetic ring assembly requires concentrated, medial RhoA activation in animal cells, the completion of cytokinesis only occurs as active RhoA declines at the cleavage furrow. Protein kinase C ε (PKCε) promotes this local decrease in active RhoA following the priming phosphorylation of PKCε, 14-3-3 binding and full activation [51]. However, the relevant PKCε substrate mediating this effect has yet to be identified.
Concomitant with such Rho inhibition, the local lipid composition at the furrow must be dynamically refashioned for cytokinesis to complete [52]. Lipid kinases perform crucial roles in this task and thereby deserve recognition as key players in molecular modifications during cytokinesis. Despite being broadly distributed on the plasma membrane earlier in mitosis, the phosphoinositide phosphatidyl-4,5-bisphosphate (PtdIns(4,5)P2) concentrates in the cytokinetic furrows of fission yeast and mammalian cells [52–55]. The recruitment of relevant phosphoinositide kinases to the furrow is thought to be largely responsible for such accumulation [52,53]. Significantly, the loss of PtdIns(4,5)P2 from mammalian furrows results in late cytokinesis failure, probably because of an inability to recruit post-furrowing factors such as septins and ezrin/radixin/moesin proteins and related defects in linking the cytokinetic apparatus to the plasma membrane [52–54]. Unlike PtdIns(4,5)P2, another phosphoinositide, namely phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3), is undetectable at the cleavage furrow but instead localizes to the poles of dividing Dictyostelium cells. The experimental disruption of kinases controlling PtdIns(3,4,5)P3 destroys this localization pattern and prevents the completion of cytokinesis following ingression [56], likewise underscoring the significance of spatial control in lipid kinase function.
During abscission, additional regulatory factors tied to phosphoinositide signaling are shuttled to the furrow. Indeed, the disruption of cellular trafficking, especially of two Rab GTPases, Rab11 and Rab35, affects the final steps of cytokinesis in higher eukaryotes [52,55,57,58]. In the fruit fly Drosophila melanogaster, the kinase Four wheel drive catalyzes the formation of phosphatidylinositol-4-phosphate (PtdIns(4)P). The subsequent incorporation of PtdIns(4)P into Golgi-derived vesicles promotes the trafficking of Rab11 to the midbody, the microtubule remnant of the anaphase spindle midzone that links the two daughter cells [59]. It will be interesting to examine whether lipid kinase function also directs Rab11 trafficking in mammals given the role of Rab11 in mammalian abscission [57,58]. The disruption of Rab35-based cycling meanwhile perturbs the cellular distribution of PtdIns(4,5)P2, probably by affecting the transport of relevant kinases to the furrow [52,55]. Therefore, lipid kinases act both upstream and downstream of Rab GTPase-mediated endocytosis at the cleavage furrow. Moreover, phosphoinositides at the midbody can also directly bind factors needed for abscission. For example, phosphatidylinositol-3-phosphate (PtdIns(3)P) recruits the centrosomal protein FYVE-CENT to the midbody. Here, FYVE-CENT binds CHMP4B [60], which is an ESCRT-III component, and this interaction is significant given the proposed function of ESCRT-III in membrane bending and scission [61]. Given that lipid kinase activity directs secretory machinery and transport in yeasts as well [62], these enzymes could potentially affect cell separation in multiple organisms through similar pathways.
In addition to cues that trigger abscission, chromatin persisting in the plane of division has been reported to delay the completion of cytokinesis. In both budding yeast [63,64] and human cells [65], Aurora B kinase activity safeguards cells from dividing through unsegregated chromosomes. In these organisms, Aurora B transfers with the rest of the chromosomal passenger complex to the spindle midzone in anaphase following the reversal of CDK-mediated phosphorylation on the passenger protein INCENP [66,67]. Thus, Aurora B is nicely positioned at this stage of the cell cycle to detect chromatin in the cleavage plane. Indeed, acetylated chromatin stalled in the vicinity of the central spindle activates Aurora B in budding yeast [63]. Active Aurora B then initiates a NoCut checkpoint by targeting the abscission inhibitors Boi1 and Boi2 to the bud neck, where they prevent chromosome cutting by the cytokinetic apparatus [64]. In HeLa cells, chromosome bridges likewise prolong Aurora B activity at the midbody, and this stabilizes ingressed furrows and protects against tetraploidization caused by furrow regression [65]. Although the mechanism by which such stabilization occurs is currently unclear, it has been suggested that Aurora B phosphorylation of the centralspindlin kinesin MKLP1 could be involved [65]. Aurora B-mediated abscission checkpoints do not operate in S. pombe, and the generality of these mechanisms awaits further examples.
Ubiquitination
Ubiquitin is a small, 76-residue protein that is attached to substrates via a cascade of E1 activating, E2 conjugating and E3 ligase enzymes (Figure 2a). Although widely recognized for its role in proteasomal degradation, ubiquitination also primes proteins for various regulatory functions (Figure 2b).
Positioning and assembly of the contractile apparatus
As previously noted, Rho signaling contributes primary cues for cytokinetic ring assembly in animal cells. A crucial upstream regulator of such signaling, via the phosphorylation and clustering of the centralspindlin complex at the midzone [29] and the direct phosphorylation of various Rho GAPs and GEFs [32,34,36], is Aurora B kinase. Once recruited to the midzone, Aurora B establishes a phosphorylation gradient that spatially organizes the informational outputs required for cytokinesis [68]. Although much of the signal originating from midzone-activated Aurora B comes in the form of phosphorylation, the initial recruitment and accumulation of Aurora B at the midzone in animal cells requires ubiquitin modification. Aurora B associates with numerous Bric-a-brac-Tramtrack-Broad (BTB) domain adaptors for the Cullin3 E3 ubiquitin ligase [69,70]. The BTB adapter KLHL21 specifically mediates Aurora B transfer to microtubules at the midzone, and this targeting seems partially enzymatically driven given that Cullin3-KLHL21 complexes ubiquitinate Aurora B [70]. Interestingly, Aurora A kinase is likewise ubiquitinated in human cells. Although such ubiquitination targets Aurora A for degradation at anaphase, this allows for the formation of a mature spindle midzone [71]. Thus, through its destruction ubiquitinated Aurora A paves the way for ubiquitinated Aurora B to exert its cytokinesis functions at the spindle midzone. In this way, the ubiquitination of both Aurora A and Aurora B affect subsequent Aurora B phosphorylation events that guide cytokinesis.
Constriction of the contractile apparatus
One important issue in cytokinesis concerns what happens to ring components as the cleavage apparatus constricts (Figure 4). In fission yeast, the local levels of several ring proteins, including the IQGAP Rng2, the F-BAR scaffold protein Cdc15 and the formin Cdc12, decrease at the cytokinetic ring in relation to the reduction in ring size [72]. Interestingly, the disassembly of cleavage furrow components in definable units has been proposed to contribute to the scalability of this process in animal cells [73]. Given its role in protein degradation, ubiquitination represents one means by which the clearance of proteins from the cleavage furrow could be achieved (Figure 4), and there are several examples of ubiquitin-mediated degradation of cytokinetic proteins. In budding yeast, the ubiquitin-mediated destruction of the cytokinetic IQGAP Iqg1 and the F-BAR scaffold Hof1 fosters the disassembly of the contractile apparatus and promotes actomyosin constriction, respectively [74,75]. Both Iqg1 and Hof1 are phosphoproteins [76], and the phosphorylation of Hof1 has been suggested to affect its degradation [75], indicating potential links between phosphorylation and ubiquitination in such regulation. Anillin and anillin-like proteins are also targeted for degradation by ubiquitin-mediated pathways in animals and fission yeast [77,78], with the destruction of fission yeast anillin-like protein Mid2 impacting septin dynamics at the ring [78]. mDia2, the mammalian formin responsible for cytokinetic actin [79], is also degraded via ubiquitin modification at the completion of the cell cycle [80], although the exact contribution of such ubiquitination to ring constriction is unknown. Thus, protein turnover through ubiquitin pathways contributes to the proper cycling of cleavage furrow components in various organisms (Figure 4), which probably impacts the integrity of constriction.
Figure 4.
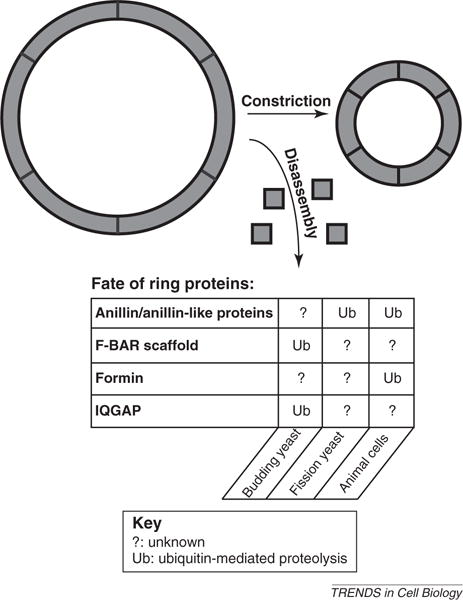
Involvement of ubiquitination in cytokinetic ring disassembly. In various organisms, the contractile apparatus disassembles as it constricts. In animal cells, constriction is proposed to occur through the shortening of contractile units (separated by black lines in the overall ring structure), which thereby couples initial ring size to constriction rate [73]. In the fission yeast S. pombe, the cytokinetic IQGAP Rng2, the F-BAR scaffold Cdc15 and the formin Cdc12 have been shown to decrease in levels within the cytokinetic ring relative to the decrease in overall ring size [72]. Although ubiquitin-mediated degradation (signified by ‘Ub’ in thefigure’s table) has not been linked to the disassembly of these factors in S. pombe, the F-BAR scaffold and IQGAP proteins in the budding yeast S. cerevisiae are targeted for destruction via ubiquitination, and such ubiquitination promotes ring disassembly and constriction in this organism [74,75]. Furthermore, formin and anillin are degraded at the completion of the cell cycle in animal cells [77,80], suggesting a potential link between ubiquitination and ring disassembly in this organism, although the exact contribution of such ubiquitination awaits further analysis. An anillin-like protein Mid2 is likewise degraded in fission yeast via ubiquitin pathways, and the destruction of this protein has been posited to affect the dynamics of septin structures [78]. Therefore, the ubiquitination of conserved ring components is common in various organisms, and such modification probably allows constriction to proceed appropriately.
Ubiquitin signaling also confers a checkpoint function just prior to ring constriction. When chromosomes are not properly attached to the mitotic spindle in fission yeast, the E3 ubiquitin ligase Dma1 blocks the localization of the polo-like kinase Plo1 by ubiquitinating the scaffold of the septation initiation network [81]. This antagonizes the septation initiation network, probably by preventing the upstream Plo1-mediated phosphorylation of factors in this signaling pathway [81]. Thus, Dma1 ubiquitination indirectly curbs Plo1 kinase function and consequently inhibits downstream kinases in the septation initiation network. Dma1 possesses a phosphothreonine-binding FHA domain in addition to a RING finger domain, suggesting that phosphorylation of its binding partners is likely to impact its recruitment and ubiquitin-mediated checkpoint function. Related E3 ubiquitin ligases in humans, known as CHFR and RNF8, are also involved in mitotic checkpoints, although their relation with polo-like kinases and a possible involvement in cytokinesis remain unclear [82,83].
Separation of daughter cells
During the final stages of cytokinesis in animal cells, GFP-ubiquitin accumulates as a dense cluster on the midbody, specifically concentrating on the midbody ring that encircles antiparallel midbody microtubules [84]. Indeed, several proteins capable of binding to ubiquitin, including ESCRT components, localize to the midbody [60,85]. BRUCE, a large, multidomain protein possessing hybrid E2/E3 ubiquitin ligase activity, regulates ubiquitination at the midbody ring and impacts various facets of abscission signaling, including the assembly of the midbody structure and targeting of membrane to the cleavage furrow [84]. Accordingly, BRUCE probably links ubiquitination to these crucial processes. In addition, given that BRUCE shuttles with Rab11-associated endosomes [84], which as previously discussed traffic to the midbody in Drosophila partly through the incorporation of PtdIns(4)P [59],it seems reasonable to speculate that relevant lipid kinases could affect BRUCE function and ubiquitination at the midbody.
Ubiquitin modification of the midbody ring furthermore primes it for degradation. However, this does not occur through the proteasome. Instead, ubiquitin at the midbody ring binds the autophagy factor p62 during abscission, thereby coupling midbody ring clearance following daughter cell separation to these late stages in cytokinesis [86].
Considering the dramatic increase in ubiquitin at the midbody just prior to abscission, it seems reasonable that ubiquitin is participating in regulatory roles that have yet to be described. Currently, whether ubiquitination affects abscission in lower eukaryotes is unclear, although this deserves further analysis given the involvement of ubiquitination in the preceding stages of fungal cytokinesis.
Additional modifications of interest
In addition to phosphorylation and ubiquitination, other modifications dynamically regulate cytokinesis factors (Figure 2). However, the contributions of these additional modifications to the control of cytokinesis have yet to receive as much attention as have phosphorylation and ubiquitination. We highlight emerging roles for three of these modifications, namely glycosylation, sumoylation and acetylation, in brief discussions below, and we also note links that exist among these and other modifications in this process.
Glycosylation
Glycosylation entails the addition of sugar moieties to relevant substrates. In various organisms, including the nematode worm Caenorhabditis elegans [87] and D. melanogaster [88], defects in protein glycosylation have been reported to affect cell separation via the disruption of intracellular transport pathways. Even so, how glycosylation impinges upon other post-translational modifications in cytokinesis remained largely unaddressed until recent investigations of O-linked beta-N-acetylglucosamine (O-GlcNAc). As with general glycosylation defects, the disruption of O-GlcNAc modification impairs cytokinesis [89]. These defects probably result from altered phosphorylation signaling, because glycoproteomic and phosphoproteomic screens of animal proteins that localize to the mitotic spindle and midbody have demonstrated that O-GlcNAc modification affects the phosphorylation status of cytokinesis regulators, including CDK and the chromosomal passenger complex [90]. Sites modified by O-GlcNAc can coincide with or neighbor phosphorylated sites, indicating potential crosstalk [90]. Therefore, although glycosylation probably exerts effects of its own, it also presumably feeds into additional regulatory circuits that complicate the linearity of its signaling. Future efforts to unravel these complicated webs, to define how glycosylation cooperates with or antagonizes nearby modifications and to understand whether responsible glycosyltransferases are themselves regulated should be of considerable interest.
Sumoylation
SUMO, or small ubiquitin-like modifier, is structurally similar to ubiquitin and is attached to substrates through a similar E1, E2 and E3 enzymatic cascade. However, SUMO modification does not promote the degradation of target proteins but rather confers specific regulatory functions. In cytokinesis, two main targets of sumoylation, the septins and the chromosomal passenger complex, have been identified. In budding yeast, septins were the first identified sumoylation targets [91]. These proteins localize to a ring at the S. cerevisiae bud neck and act as scaffolds for proteins involved in various aspects of cytokinesis. The disruption of septin sumoylation impairs septin ring organization and disassembly [91]. Thus, cell cycle-regulated nucleocytoplasmic transport of the SUMO machinery is used to properly control septin sumoylation and desumoylation [92]. In mammalian cells, two members of the chromosomal passenger complex, Aurora B kinase and Borealin, are sumoylated [93,94]. During mitosis, Aurora B mutants lacking SUMO modifications persist at chromosome arms rather than relocating to the spindle midzone, causing cytokinesis failure [94]. In budding yeast, the Survivin ortholog Bir1 is also sumoylated [95], suggesting that sumoylation confers a conserved means of regulating the chromosomal passenger complex. Therefore, it will be interesting to examine how SUMO modification influences phosphate and ubiquitin modifications that control the targeting of the chromosomal passenger complex to the midzone [66,69,70].
Acetylation
Although historically well known as a histone modification, acetylation, or the attachment of acetyl groups to substrates, also occurs on other proteins and affects myriads of cellular processes, including cytokinesis. For instance, increasing evidence suggests that the acetylation of tubulin subunits represents one means by which microtubule properties are controlled at the animal midbody to guarantee proper midbody-based signaling during cytokinesis. The N-acetyltransferase NAT10 transfers from the centrosome to the midbody during late mitosis, where it influences the acetylation of this structure. Reducing NAT10 doubles the amount of time needed to complete cytokinesis and results in an increase in multinucleate cells, indicating a general cytokinesis defect when midbody acetylation is disrupted [96]. In addition, the knockdown of the Arf family GTPase Arl3, which also localizes to the midbody, alters the cellular levels of tubulin acetylation and can impair midbody disassembly [97]. Potential connections between the acetylation and ubiquitination pathways have also been proposed to occur via the histone deacetylase inhibitor CYLD, a deubiquitinating enzyme that localizes to the midbody and controls the rate of cytokinesis through its interactions with HDAC6 at this site [98]. Therefore, although specific mechanisms contributing to midbody acetylation have yet to be fully described, it seems reasonable that this modification controls cytokinesis partly through its modulation of participating microtubules.
Concluding remarks
In summary, cytokinesis factors are subject to a vast array of modifications, and these modifications have been found to operate both individually and interdependently in numerous stages of cytokinesis (Box 2)(Figure 2b). Presumably, thousands of individual modification events have yet to be defined, leaving considerable room for discovery in this field. Future research should not only attempt to identify these events but also focus on defining the upstream controls and downstream consequences of known modifications (Box 3). Despite current shortcomings, our present inventory of post-translational modifications in cytokinesis is impressive. Even so, this knowledge base provides us with even more challenges, because we must now consider how all of these modifications are coordinated to confer robust control (Box 2). Only by doing so will we achieve a full appreciation of the intricate mechanisms by which post-translational modifications guide cytokinesis.
Box 2. Additional crosstalk between modifications during cytokinesis.
In this review, examples of potential and documented interplay between protein modifications during cytokinesis have been provided. However, these examples are by no means exhaustive. For example, the importance of polo-like kinases was highlighted in the discussion of phosphorylation-driven cytokinetic ring assembly (Figure 3). However, polo-like kinases are themselves controlled by post-translational modification. In human cells, the anaphase-promoting complex/cyclosome, an E3 ubiquitin ligase, targets the polo-like kinase Plk1 for proteolysis [100], indicating a direct role for ubiquitination in silencing Plk1 function at the end of cell division. Therefore, direct ubiquitin-dependent mechanisms as well as indirect ones, such as the ubiquitination of polo scaffolds [81], control this kinase family. Interestingly, the anaphase-promoting complex/cyclosome is activated by multiphosphorylation [99]. This is significant given that the anaphase-promoting complex/cyclosome triggers the destruction of animal anillin [77] and the budding yeast IQGAP Iqg1 [74]. Thus, the phosphorylation of the anaphase-promoting complex/cyclosome affects ring constriction via promoting the ubiquitin-mediated degradation of cytokinetic proteins. Furthermore, recognition by another E3 ubiquitin ligase, namely the Skp1-cullin-F-box complex, generally requires substrate phosphorylation [108]. During cytokinesis, the proteolysis of the S. cerevisiae F-BAR protein Hof1 and the S. pombe anillin-related protein Mid2 relies on this complex [75,78]. Both proteins are phosphorylated in vivo [75,109], suggesting added crosstalk occurs through the phosphoregulated targeting of substrates to E3s during ring constriction. Thus, as indicated by these additional examples of crosstalk between phosphorylation and ubiquitination on regulators of cytokinesis, interplay amongst modifications is multifaceted and seemingly limitless (Figure 2b).
In light of such complexity, bioinformatics tools for analyzing networks of post-translational modifications have begun to evolve. The development of PHOSIDA [110], for example, has eased the task of mining published datasets for varied modifications on a specific protein. This tool integrates approximately 70 000 known phosphorylation sites along with the largest databases of protein acetylation and glycosylation sites into one search engine for multiple species. A search of animal septin SEPT2, described in the text as a scaffold for protein kinases that affect myosin phosphorylation [43], identified phosphorylation sites as well as a unique acetylation site. Given that such easily available information could direct future hypotheses pertaining to the scaffolding function of SEPT2 in myosin activation, our understanding of crosstalk between modifications in cytokinesis should be enhanced through the continued improvement of this and similar databases.
Box 3. Outstanding questions.
What are the targets of kinase networks that control the assembly of the cytokinetic apparatus, and what is the contribution of post-translational modifications to the maintenance of Rho zones in relevant organisms?
What is the complete repertoire of cytokinesis factors that must be degraded as the cytokinetic apparatus constricts to ensure that this stage of cytokinesis proceeds appropriately?
What specific role(s) does ubiquitin play at the animal cell midbody during abscission, and does ubiquitination also affect abscission in lower eukaryotes such as yeasts?
How do post-translational modifications modulate checkpoint pathways controlling cytokinesis, and what are the relevant targets involved in this signaling?
How is proper crosstalk of various modifications achieved to ensure coherent informational output during cytokinesis?
Acknowledgments
We apologize to colleagues whose work we did not discuss because of space constraints. We thank members of the Gould laboratory for helpful discussions and critical reading of this manuscript. K.A.B. was supported by National Institute of Health grant T32-CA119925. K.L.G. is an Investigator of the Howard Hughes Medical Institute.
References
- 1.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King RW, et al. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 4.Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- 5.Coffman VC, et al. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vavylonis D, et al. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 7.Almonacid M, et al. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol. 2009;19:961–966. doi: 10.1016/j.cub.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Bahler J, et al. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, et al. Polarity determinants Tea1p Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell. 2007;12:987–996. doi: 10.1016/j.devcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Moseley JB, et al. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Wang YL. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol Biol Cell. 2008;19:318–326. doi: 10.1091/mbc.E07-08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner M, et al. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol. 2007;17:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, et al. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamasaki T, et al. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohl DA, et al. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J Cell Biol. 2009;184:527–539. doi: 10.1083/jcb.200812022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CT, et al. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glotzer M. The 3Ms ofcentral spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf F, et al. ‘The end of the beginning’: cdk1 thresholds and exit from mitosis. Cell Cycle. 2007;6:1408–1411. [PubMed] [Google Scholar]
- 21.Wolfe BA, et al. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev Cell. 2006;11:423–430. doi: 10.1016/j.devcel.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Clifford DM, et al. The Clp1/Cdc14 phosphatasecontributesto the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts-Galbraith RH, et al. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li CR, et al. The IQGAP Iqg1 is a regulatory target of CDK for cytokinesis in Candida albicans. EMBO J. 2008;27:2998–3010. doi: 10.1038/emboj.2008.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida S, et al. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–111. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida S, et al. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfe BA, et al. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009;7:e1000110. doi: 10.1371/journal.pbio.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkard ME, et al. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 2009;7:e1000111. doi: 10.1371/journal.pbio.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douglas ME, et al. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr Biol. 2010;20:927–933. doi: 10.1016/j.cub.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishima M, et al. Cell cycle regulation of central spindle assembly. Nature. 2004;430:908–913. doi: 10.1038/nature02767. [DOI] [PubMed] [Google Scholar]
- 31.Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nat Cell Biol. 2009;11:71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toure A, et al. Phosphoregulation of MgcRacGAP in mitosis involves Aurora B and Cdk1 protein kinases and the PP2A phosphatase. FEBS Lett. 2008;582:1182–1188. doi: 10.1016/j.febslet.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 33.Asiedu M, et al. Phosphorylation of MyoGEF on Thr-574 by Plk1 promotes MyoGEF localization to the central spindle. J Biol Chem. 2008;283:28392–28400. doi: 10.1074/jbc.M801801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birkenfeld J, et al. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosario CO, et al. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA. 2010;107:6888–6893. doi: 10.1073/pnas.0910941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minoshima Y, et al. Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell. 2003;4:549–560. doi: 10.1016/s1534-5807(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 37.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Mizutani T, et al. Diphosphorylation of the myosin regulatory light chain enhances the tension acting on stress fibers in fibroblasts. J Cell Physiol. 2006;209:726–731. doi: 10.1002/jcp.20773. [DOI] [PubMed] [Google Scholar]
- 39.Mizutani T, et al. Regulation of cellular contractile force in response to mechanical stretch by diphosphorylation of myosin regulatory light chain via RhoA signaling cascade. Cell Motil Cytoskeleton. 2009;66:389–397. doi: 10.1002/cm.20378. [DOI] [PubMed] [Google Scholar]
- 40.Wu Q, et al. Deficiency in myosin light-chain phosphorylation causes cytokinesis failure and multipolarity in cancer cells. Oncogene. 2010;29:4183–4193. doi: 10.1038/onc.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uehara R, et al. Determinants of myosin II cortical localization during cytokinesis. Curr Biol. 2010;20:1080–1085. doi: 10.1016/j.cub.2010.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beach JR, Egelhoff TT. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J Biol Chem. 2009;284:27377–27383. doi: 10.1074/jbc.M109.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joo E, et al. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev Cell. 2007;13:677–690. doi: 10.1016/j.devcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Lordier L, et al. Megakaryocyte endomitosis is a failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling. Blood. 2008;112:3164–3174. doi: 10.1182/blood-2008-03-144956. [DOI] [PubMed] [Google Scholar]
- 45.Gruneberg U, et al. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucero A, et al. A global, myosin light chain kinase-dependent increase in myosin II contractility accompanies the metaphase-anaphase transition in sea urchin eggs. Mol Biol Cell. 2006;17:4093–4104. doi: 10.1091/mbc.E06-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chew TL, et al. A fluorescent resonant energy transfer-based biosensor reveals transient and regional myosin light chain kinase activation in lamella and cleavage furrows. J Cell Biol. 2002;156:543–553. doi: 10.1083/jcb.200110161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loo TH, Balasubramanian M. Schizosaccharomyces pombe Pak-related protein Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J Cell Biol. 2008;183:785–793. doi: 10.1083/jcb.200806127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosgraaf L, van Haastert PJ. The regulation of myosin II in Dictyostelium. Eur J Cell Biol. 2006;85:969–979. doi: 10.1016/j.ejcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Sladewski TE, et al. Regulation of fission yeast myosin-II function and contractile ring dynamics by regulatory light-chain and heavy-chain phosphorylation. Mol Biol Cell. 2009;20:3941–3952. doi: 10.1091/mbc.E09-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saurin AT, et al. The regulated assembly of a PKCepsilon complex controls the completion of cytokinesis. Nat Cell Biol. 2008;10:891–901. doi: 10.1038/ncb1749. [DOI] [PubMed] [Google Scholar]
- 52.Echard A. Membrane traffic and polarization of lipid domains during cytokinesis. Biochem Soc Trans. 2008;36:395–399. doi: 10.1042/BST0360395. [DOI] [PubMed] [Google Scholar]
- 53.Emoto K, et al. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J Biol Chem. 2005;280:37901–37907. doi: 10.1074/jbc.M504282200. [DOI] [PubMed] [Google Scholar]
- 54.Field SJ, et al. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr Biol. 2005;15:1407–1412. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 55.Kouranti I, et al. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 56.Janetopoulos C, et al. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Wilson GM, et al. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fielding AB, et al. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polevoy G, et al. Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J Cell Biol. 2009;187:847–858. doi: 10.1083/jcb.200908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sagona AP, et al. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat Cell Biol. 2010;12:362–371. doi: 10.1038/ncb2036. [DOI] [PubMed] [Google Scholar]
- 61.Hanson PI, et al. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yakir-Tamang L, Gerst JE. A phosphatidylinositol-transfer protein and phosphatidylinositol-4-phosphate 5-kinase control Cdc42 to regulate the actin cytoskeleton and secretory pathway in yeast. Mol Biol Cell. 2009;20:3583–3597. doi: 10.1091/mbc.E08-10-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mendoza M, et al. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat Cell Biol. 2009;11:477–483. doi: 10.1038/ncb1855. [DOI] [PubMed] [Google Scholar]
- 64.Norden C, et al. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 65.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 66.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 67.Hummer S, Mayer TU. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr Biol. 2009;19:607–612. doi: 10.1016/j.cub.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 68.Fuller BG, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sumara I, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 70.Maerki S, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Floyd S, et al. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr Biol. 2008;18:1649–1658. doi: 10.1016/j.cub.2008.09.058. [DOI] [PubMed] [Google Scholar]
- 72.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 73.Carvalho A, et al. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 2009;137:926–937. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 74.Tully GH, et al. The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol Biol Cell. 2009;20:1201–1212. doi: 10.1091/mbc.E08-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blondel M, et al. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Albuquerque CP, et al. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 78.Tasto JJ, et al. An anillin homologue Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe S, et al. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeWard AD, Alberts AS. Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J Biol Chem. 2009;284:20061–20069. doi: 10.1074/jbc.M109.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson AE, Gould KL. Dma1 ubiquitinates the SIN scaffold Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J. 2011;30:341–354. doi: 10.1038/emboj.2010.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 83.Tuttle RL, et al. Defective in mitotic arrest 1/ring finger 8 is a checkpoint protein that antagonizes the human mitotic exit network. Mol Cancer Res. 2007;5:1304–1311. doi: 10.1158/1541-7786.MCR-07-0388. [DOI] [PubMed] [Google Scholar]
- 84.Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132:832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 85.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 86.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 87.Wang H, et al. The terminal phase of cytokinesis in the Caenorhabditis elegans early embryo requires protein glycosylation. Mol Biol Cell. 2005;16:4202–4213. doi: 10.1091/mbc.E05-05-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang L, Ten Hagen KG. Dissecting the biological role of mucin-type O-glycosylation using RNA interference in Drosophila cell culture. J Biol Chem. 2010;285:34477–34484. doi: 10.1074/jbc.M110.133561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slawson C, et al. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 90.Wang Z, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Makhnevych T, et al. The role of karyopherins in the regulated sumoylation of septins. J Cell Biol. 2007;177:39–49. doi: 10.1083/jcb.200608066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klein UR, et al. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell. 2009;20:410–418. doi: 10.1091/mbc.E08-05-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fernandez-Miranda G, et al. SUMOylation modulates the function of Aurora-B kinase. J Cell Sci. 2010;123:2823–2833. doi: 10.1242/jcs.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montpetit B, et al. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen Q, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315:1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 97.Zhou C, et al. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell. 2006;17:2476–2487. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wickstrom SA, et al. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kraft C, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chia W, et al. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wirtz-Peitz F, et al. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Atwood SX, Prehoda KE. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnston CA, et al. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hao Y, et al. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Curr Biol. 2010;20:1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cabernard C, et al. A spindle-independent cleavage furrow positioning pathway. Nature. 2010;467:91–94. doi: 10.1038/nature09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ou G, et al. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science. 2010;330:677–680. doi: 10.1126/science.1196112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ang XL, Wade Harper J. SCF-mediated protein degradation and cell cycle control. Oncogene. 2005;24:2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- 109.Wilson-Grady JT, et al. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 110.Gnad F, et al. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]


