Abstract
Effective targeted therapies for small-cell lung cancer (SCLC), the most aggressive form of lung cancer, remain urgently needed. Here we report evidence of preclinical efficacy evoked by targeting the overexpressed cell-cycle checkpoint kinase CHK1 in SCLC. Our studies employed RNAi-mediated attenuation or pharmacologic blockade with the novel second-generation CHK1 inhibitor prexasertib (LY2606368), currently in clinical trials. In SCLC models in vitro and in vivo, LY2606368 exhibited strong single-agent efficacy, augmented the effects of cisplatin or the PARP inhibitor olaparib, and improved the response of platinum-resistant models. Proteomic analysis identified CHK1 and MYC as top predictive biomarkers of LY2606368 sensitivity, suggesting that CHK1 inhibition may be especially effective in SCLC with MYC amplification or MYC protein overexpression. Our findings provide a preclinical proof of concept supporting the initiation of a clinical efficacy trial in patients with platinum-sensitive or platinum-resistant relapsed SCLC.
Introduction
Small-cell lung cancer (SCLC) is the most lethal form of lung cancer. The 5-year overall survival rate is less than 5%, and this has not changed substantially in the past three decades. The standard of care for advanced SCLC is cisplatin-based doublet chemotherapy, which causes an initial tumor regression in most patients. However, chemotherapy resistance rapidly develops (1, 2), and the only approved second-line therapy (topotecan) produces responses in fewer than 20% of patients (reviewed in ref. 3). This aggressive cancer is characterized by the near-ubiquitous inactivation of TP53 and RB1 and, in a subset of SCLC tumors (~16% in patients and 38% in mouse models), mutually exclusive amplification of MYC family members (CMYC, MYCN, and MYCL; refs. 4–9). Over the past few years, identification of EGFR mutations, ALK fusions, and other gene alterations has significantly advanced the therapeutic options for non–small cell lung cancer (NSCLC; refs. 10–12). However, despite a high overall mutation rate in SCLC, there are currently no targeted drugs with proven clinical benefit for this disease. Hence, there is an urgent need to identify new, viable therapeutic targets.
In a previous study, we found that frequent loss of RB1 in SCLC with resulting loss of E2F1 repression is associated with increased expression of several key mediators of DNA damage repair (DDR), including PARP1 (13) and the checkpoint kinase 1 (CHK1) protein (13). PARP is a promising new target for SCLC that is now being tested in clinical trials (13, 14). Studies in other cancer types have shown the efficacy of PARP inhibitors in combination with inhibitors of CHK1 and other DDR proteins (15, 16). CHK1 is an essential serine threonine protein kinase that, in cells with aberrant TP53, becomes the primary mediator of DNA damage–dependent cell-cycle arrest (17–20). Studies in triple-negative breast cancers and head and neck cancers (in which TP53 mutations often occur) have demonstrated that CHK1 inhibitors can augment the effects of DNA-damaging treatments (17–20). More recently, a high-throughput small-molecule drug screen also identified CHK1 as a candidate therapeutic target in SCLC (21).
In this study, we further investigated the therapeutic potential of CHK1 inhibition (through genetic knockdown and pharmacologic inhibition with LY2606368) in a comprehensive panel of SCLC cell lines, syngeneic models, and a genetically engineered mouse model (GEMM), as monotherapy and in combination with cisplatin. The CHK1 inhibitor used in the study, prexasertib (henceforth referred to as LY2606368), is an ATP-competitive protein kinase inhibitor that showed selectivity to CHK1 inhibition over 224 protein kinases tested biochemically (21). LY2606368 also recently completed phase I testing, which demonstrated tolerability as well as single-agent clinical activity in a subset of solid tumors (primarily advanced, metastatic squamous cancers, which are also TP53-mutant cancers; ref. 22). We also sought to determine whether the addition of LY2606368 would enhance the activity of the PARP inhibitor olaparib in SCLC models (13). We show here the efficacy of CHK1 inhibition (alone and with chemotherapy or PARP inhibition) in platinum-sensitive and -resistant SCLC models and apply high-throughput proteomic analysis to identify candidate predictive biomarkers of CHK1 inhibitor response in SCLC.
Materials and Methods
Cell lines and characterization
Cell lines (35 human-derived SCLC, one patient-derived xenograft (PDX)-derived SCLC, three NSCLC, and one large-cell neuroendocrine carcinoma) were provided by Dr. John Minna (The University of Texas Southwestern Medical Center, Dallas, TX) or obtained from ATCC (between the years 2011 and 2015). The GEMM-derived SCLC cell lines were established from mouse tumors with a ROSA26R reporter and conditional deletion of Trp53, p130, and Rb1 (23, 24) and obtained in 2015. Complete cell line information is provided in the Supplementary Materials and Methods and in Supplementary Table S1.
All cell lines were tested and authenticated by short tandem repeat profiling (DNA fingerprinting; ref. 25) within 6 months of the study and routinely tested for Mycoplasma species before any experiments were performed. For details, see Supplementary Materials and Methods.
Chemical compounds
LY2603618 was obtained from Selleckchem. LY2606368 and olaparib were manufactured by MD Anderson’s Institute for Applied Chemical Science. All compounds were dissolved in DMSO for in vitro treatments.
Mice
For the syngeneic mouse model, 6-week-old female athymic nude mice (Envigo) were used. These animals were maintained in accordance with the Institutional Animal Care and Use Committee (IACUC) of The University of Texas MD Anderson Cancer Center (Houston, TX) and the NIH Guide for the Care and Use of Laboratory Animals.
The spontaneous GEMM mice were maintained according to practices prescribed by the NIH at the Stanford Research Animal Facility, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care. The Trp53, Rb1, Rbl2 (p53/Rb/p130) conditional knockout mouse model induced by intratracheal administration of Ad-CMV-Cre for SCLC has been described previously (24, 26). Refer to Supplementary Materials and Methods for details on tumor development and dosing schedule.
Treatment schedule of SCLC in vivo models
Mice with mouse SCLC tumors received one of the following treatments: (i) vehicle; (ii) cisplatin (4 mg/kg daily) once per week; (iii) high-dose LY2606368 (10 mg/kg twice daily) 3 consecutive days per week (weekly dose = 60 mg/kg); (iv) low-dose LY2606368 (16 mg/kg twice daily) once per week (weekly dose = 32 mg/kg); or (v) combination of cisplatin (4 mg/kg daily) and LY2606368 (16 mg/kg twice daily) once per week.
Mice with human SCLC (H69/CR) tumors received one of the following treatments: (i) vehicle; (ii) cisplatin (8 mg/kg) once per week; (iii) LY2606368 (10 mg/kg twice daily) twice per week; or (iv) combination of cisplatin (8 mg/kg) and LY2606368 (10 mg/kg twice daily, days 1 and 2 of each 7-day cycle).
For the PARP inhibitor experiments, mice received one of the following treatments: (i) vehicle; (ii) olaparib (100 mg/kg daily) 5 times per week; (iii) LY2606368 (10 mg/kg twice daily) twice per week; or (iv) combination of olaparib and LY2606368.
Once tumor volume reached 1,500 mm3 or the tumors developed ulceration, mice were euthanized and tumor tissues were isolated. Tumors were snap-frozen in liquid nitrogen for protein isolation or fixed in 4% paraformaldehyde in PBS at 4°C and processed for paraffin histologic analysis. Sections of paraffin-embedded tissues (4 μm) were stained with hematoxylin and eosin.
Development of spontaneous SCLC tumors and dosing schedule
Mice were maintained according to practices prescribed by the NIH at the Stanford Research Animal Facility, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care. The p53/Rb/p130 triple-knockout mouse model for SCLC has been previously described (24, 26). The mice were bred onto a mixed genetic background composed of C57BL/6, 129/SvJ, and 129/SvOla. Tumors were induced in young adult mice by intratracheal instillation of 4 × 107 plaque-forming units of adenovirus-expressing Cre recombinase (Ad-Cre; Baylor College of Medicine, Houston, TX).
Treatment was initiated 4 months after Ad-Cre delivery, at a time when infected mice had developed more than 50 independent lesions (24). Each randomized cohort had 4 males and 3 females. The CHK1 inhibitor, LY2606368(10 mg/kg) was injected subcutaneously at the nape of the neck every 12 hours for three days in a row, for three weeks (i.e., 10 mg/kg, twice daily, days 1–3 of a 7-day cycle, for three cycles). Control mice were injected with vehicle. On the fourth week, mice were injected two more times and euthanized three hours after the last injection. The lungs were fixed in formaldehyde overnight before processing in paraffin and sectioning. Tumor size, number of tumors, and overall tumor burden were measured using ImageJ software.
Calculation of drug parameters
For single-agent analysis, we estimated IC50 values by using the software program drexplorer, which fitted multiple dose–response models and selected the best model using the residual standard error (27). For drug combination analysis, we used MacSynergy II (http://www.uab.edu/medicine/peds/macsynergy) to calculate metrics, including synergistic volume, antagonistic volume, overall volume, and extra kill percentage (28).
In drug combination analysis of cisplatin and LY2606368, the Bliss independence model was used to estimate additive effect (http://onlinelibrary.wiley.com/doi/10.1111/j.1744-7348.1939.tb06990.x/abstract;jsessionid=E87F2639E0C805EA671F6922AFA74C8D.f01t03).
Statistical analysis
We used Spearman rank correlation to assess the association between IC50 values and individual reverse-phase protein analysis (RPPA) protein markers, the ANOVA to assess the association between IC50 values and cMYC status. We used the R software program to perform all statistical analyses (29).
Other methods
The details of other methods, including targeted CHK1 knockdown, cell viability assay, RNA isolation, reverse transcription, PCR, Western blot analysis, RPPA, mice models, tumor engraftment and monitoring, in vivo drug dosing schedule, and immunofluorescence, are given in the Supplementary Materials and Methods.
Results
CHEK1 (CHK1) is overexpressed in SCLC patient tumors
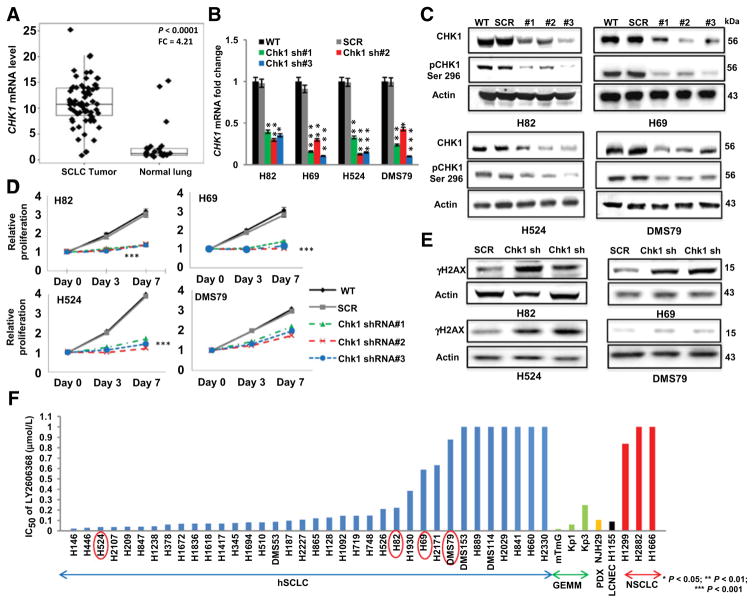
To confirm that CHK1 is overexpressed in human tumors, we analyzed mRNA expression levels in 68 SCLC tumors. Compared with normal lung specimens (n = 26), SCLC specimens had 4.21-fold higher CHK1 expression (P < 0.0001; Fig. 1A). We then performed mRNA profiling of 11 human-derived and 3 GEMM-derived SCLC cell lines and 3 NSCLC cell lines as an independent confirmation of CHK1 gene expression in SCLC. Consistent with our previous proteomic analysis (13), the SCLC cell lines had a median of 4.5-fold higher (range, 2- to 7-fold) CHK1 expression than NSCLC cell lines (P = 0.001; Supplementary Fig. S1A and S1B). Furthermore, expression levels of CHK1 mRNA and CHK1 protein were highly correlated in the SCLC cell lines (n = 54; r = 0.605; P < 0.0001; Supplementary Fig. S1C).
Figure 1.
CHK1 is overexpressed in SCLC, and CHK1 inhibition abrogates the proliferation of genotypically and phenotypically distinct SCLC cell lines. A, RNA sequencing analysis showing the gene expression profile of CHK1 in 68 SCLC and 26 normal lung tissue samples. SCLC tumors had significantly higher (P < 0.0001) CHK1 gene expression levels than normal lung tissue, with a fold change (FC) of 4.21. P <0.0001 was obtained by ANOVA. B, shRNA-mediated knockdown of CHK1 in a subset of human SCLC cell lines. Three separate shRNAs (sh#1, sh#2, and sh#3) targeting CHK1 were applied to each of the cell lines H82, H69, H524, and DMS79. Parental cells (no shRNA; WT) and scramble shRNA cells (SCR) were used as controls in each case. The knockdown efficiency was measured by quantitative real-time PCR analysis of the target gene. GAPDH was used as the reference in this analysis. Data represent mean ± SEM (error bars) of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. C, Western blot analysis confirming the knockdown efficiency of total and phosphorylated (Ser296) CHK1 in total protein lysates from the parental (WT), scramble (SCR), and CHK1-knockdown cell lines. Actin was used as the loading control. D, Proliferation assay results, including parental (WT), scramble (SCR), and CHK1-knockdown cell lines, demonstrating the effect of CHK1 inhibition on the proliferation of SCLC cells. Data represent the mean ± SEM (error bars) of three independent experiments. ***, P < 0.001. E, Western blot analysis showing elevated phospho-H2AX (γH2AX) levels in the total protein lysates of the scramble (SCR) SCLC cell line and two shRNA-transfected SCLC cell lines. The CHK1-knockdown cells showed higher levels of H2AX than the scramble control cells. Actin was used as the loading control. F, Cell viability in response to treatment with LY2606368 in a panel of human SCLC (hSCLC) cell lines (blue bars), GEMM-derived SCLC cell lines (green bars), a PDX-derived cell line (yellow bar), a large-cell neuroendocrine carcinoma (LCNEC) cell line (black bar), and NSCLC cell lines (red bars). The genetic profile information and mean half-maximal inhibitory concentration (IC50) values for each cell line are presented in Supplementary Table S1 and Supplementary Table S3.
CHEK1 (CHK1) knockdown is sufficient to inhibit SCLC cell viability
Given the overexpression of CHK1 in SCLC cell lines and tumors, we first tested whether CHK1 knockdown was sufficient to affect SCLC growth. CHK1 was knocked down using three independent shRNAs in four SCLC cell lines. The efficiency of the knockdown was confirmed by mRNA (Fig. 1B) and immunoblot analysis of total and phospho-CHK1 (Ser296; Fig. 1C). The targeted knockdown of CHK1 significantly abrogated cell proliferation in human SCLC cell lines (P < 0.05; Fig. 1D).
Next, we aimed to assess the effect of targeted CHK1 knockdown on DNA damage in the hSCLC cell lines. Analysis of CHK1 knockdown using the two most efficient shRNA constructs for each cell line detected higher levels of phospho-H2AX (γH2AX) in the CHK1-knockdown cells compared with control cells transduced with scrambled shRNA (Fig. 1E). These results demonstrate that reducing expression of CHK1 promotes DNA double-strand breaks in SCLC cells in vitro.
Human and murine SCLC cells are highly sensitive to CHK1 inhibition
We then investigated the potency of a selective second-generation CHK1 inhibitor, LY2606368 (30), in a comprehensive panel of SCLC cell lines, including 35 that were human-derived, three that were murine-derived, and one that was derived from a PDX (13). These included models with characteristic genetic alterations such as loss of TP53 (100% of samples tested) and RB1 (75% of samples tested) and MYC family amplifications (in about 30% of samples; Supplementary Table S1). We also included three NSCLC cell lines and one large-cell neuroendocrine carcinoma cell line, derived from a high-grade neuroendocrine cancer with a molecular profile similar to that of SCLC (13).
Most of the SCLC cell lines were sensitive to CHK1 inhibition at a concentration (300 nmol/L) achievable at the clinically recommended phase II dose (based on patient pharmacokinetic testing from the recent phase 1 study; ref. 22). Specifically, the antiproliferation half-maximal inhibitory concentration (IC50) of LY2606368 was <100 nmol/L in half of the human SCLC cell lines, two of the three murine SCLC cell lines, and the PDX-derived and large-cell neuroendocrine carcinoma cell lines. In contrast, most NSCLC cell lines were resistant to LY2606368 (IC50 > 0.8 μmol/L; Fig. 1F). Similar to what we observed with LY2606368, human SCLC cells were sensitive to a second CHK1 inhibitor, LY2603618, in a subset of SCLC cell lines (n = 11; 50% of cells had IC50 <6 μmol/L; Supplementary Fig. S1D). Similar to LY2606368, NSCLC cell lines were found to be resistant to the second CHK1 inhibitor, LY2603618 (IC50 >10 μmol/L). Thus, single-agent CHK1 inhibition has activity in a large majority of SCLC cell lines.
LY2606368 potentiates the cytotoxic effect of cisplatin and leads to DNA double-strand breakage and apoptosis
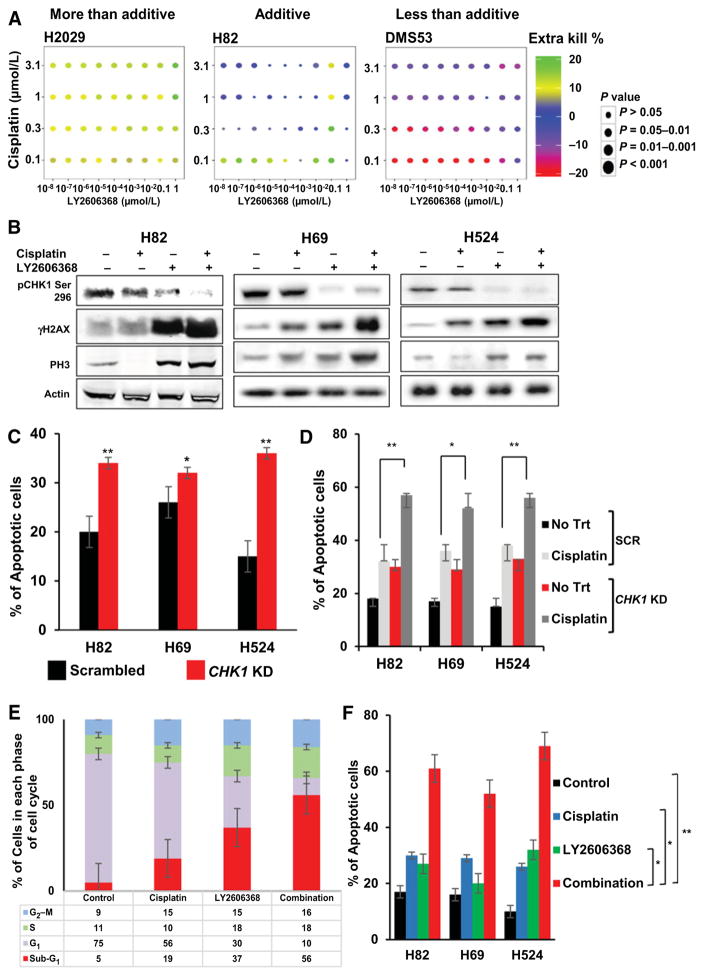
On the basis of prior data suggesting that CHK1 targeting can potentiate the effect of DNA-damaging chemotherapies, we next investigated whether CHK1 inhibition enhances the activity of the most commonly used chemotherapeutic agent for SCLC, cisplatin, in a panel of 35 human SCLC lines. Cell lines were treated with multiple doses of cisplatin 24 hours before treatment with LY2606368, and cell viability was measured 120 hours after treatment with LY2606368. Even at a non–growth-inhibitory dose (1 nmol/L), LY2606368 potentiated the cytotoxic effect of cisplatin in majority of the SCLC lines (Fig. 2A; Supplementary Fig. S2A and S2B). The combination of cisplatin and LY2606368 was also synergistic at higher, more clinically relevant doses of cisplatin (range, 0.1–10 μmol/L) when combined with LY2606368 (Supplementary Fig. S2C).
Figure 2.
LY2606368 enhances the cytotoxic potential of cisplatin in SCLC cell lines. A, Matrix dot plots showing the percentage of greater inhibition of viability (extra kill) achieved by the combination of cisplatin and LY2606368 at various doses compared with that achieved with either drug alone in representative SCLC cell lines. The colors of the dots represent the percentages of efficacy of the combinations, and the sizes of the dots indicate the statistical significance. B, Western blot analysis of total protein lysates collected 24 hours after treatment with LY2606368, revealing elevation of phospho-H2AX (γH2AX) and phospho-histone 3 (PH3) expression in three human SCLC lines—H82, H69, and H524—treated with 10 nmol/L LY2606368 with or without 0.1 μmol/L cisplatin. Actin was used as the loading control. C, Results of a flow cytometry assay using Annexin V–propidium iodide staining, showing apoptosis in cell lines H82, H69, and H524 transfected with shRNA against CHK1 (CHK1 KD) or with scrambled shRNA. Data are derived from two independent experiments conducted in triplicate (error bars, SEM). The P values were calculated using the Student t test. *, P < 0.05; **, P < 0.01. D, Apoptosis in H82, H69, and H524 cells treated with shRNAs targeting CHK1 (CHK1 KD) or scramble shRNA (SCR), treated with 0.1 μmol/L cisplatin. Apoptosis was evaluated using flow cytometry with Annexin V–propidium iodide staining after 48 hours. Data are derived from two independent experiments conducted in triplicate (error bars, SEM). The P values were calculated using the Student t test. *, P < 0.05; **, P < 0.01. E, Cell-cycle analysis in SCLC cell lines (n = 12) treated with cisplatin (0.1 μmol/L), LY2606368 (10 nmol/L), or both. Cell-cycle analysis was done by propidium iodide/RNAse flow cytometry 24 hours after treatment with LY2606368; the data shown represent averages of the two independent experiments conducted in triplicate in 12 cell lines. Error bars, SEM. F, Apoptosis in H82, H69, and H524 cells treated with cisplatin (0.1 μmol/L), LY2606368 (10 nmol/L), or both. Apoptosis was evaluated by a flow cytometry assay with Annexin V–propidium iodide staining 48 hours after treatment with LY2606368. Data are derived from two independent experiments conducted in triplicate. Error bars, SEM. The P values were calculated using the Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In SCLC cell lines H82, H69, and H524, treatment with LY2606368 (24 hours) induced DNA double-strand breaks, as indicated by elevated γH2AX levels (Fig. 2B). There was less induction of γH2AX in H69 cells compared with the other cell lines in response to shRNA-mediated targeted knockdown of CHK1 (Fig. 1E) and single-agent LY2606368 treatment, which is consistent with this cell line being relatively less sensitive to LY2606368. Single-agent cisplatin induced a modest increase in γH2AX levels that was potentiated in presence of LY2606368. However, γH2AX levels appreciably increased when a subtherapeutic dose of cisplatin (0.1 μmol/L) was combined with LY2606368 (10 nmol/L), and this increase was greater than that caused by either drug alone (Fig. 2B). We also observed increased expression of the mitotic marker phospho-histone H3 (PH3) following treatment with LY2606368, with or without cisplatin, in the SCLC cell lines (Fig. 2B). The results indicate that single-agent CHK1 targeting induces DNA damage without abrogating the mitotic potential of the cells. Hence, the cells undergo cell cycle with damaged DNA, which subsequently leads to apoptosis.
An apoptosis assay using Annexin V–propidium iodide staining showed that CHK1 knockdown induced apoptosis in all three cell lines tested (Fig. 2C). This was further enhanced by the addition of a subtherapeutic dose of cisplatin (0.1 μmol/L), as evidenced by increased apoptosis in these cells 48 hours after treatment with cisplatin (Fig. 2D).
Given the pivotal role of CHK1 in the G2–M phase of the cell cycle, we then investigated the effect of pharmacologic inhibition of CHK1 (with or without cisplatin) on the cell-cycle progression. Cell-cycle analysis by flow cytometry in SCLC cell lines (n = 12) showed increased accumulation of cells at the sub-G1 phase following treatment with single-agent LY2606368 (10 nmol/L) for 24 hours. Moreover, CHK1 targeting significantly potentiated the effect of subtherapeutic doses of cisplatin (0.1 μmol/L) in 80% of SCLC cell lines (P < 0.001; Fig. 2E). Furthermore, non–growth-inhibitory doses of LY2606368 (10 nmol/L) in combination with cisplatin (0.1 μmol/L) for 48 hours caused greater apoptosis in SCLC cells than did cisplatin alone (P < 0.05; Fig. 2F).
Taken together, these results suggest that single-agent targeting of CHK1 can increase DNA damage, transient cell-cycle arrest, and eventually apoptosis in most SCLC cell lines tested and potentiates the effect of cisplatin, the most common chemotherapeutic drug used in the treatment of SCLC.
CHK1 inhibition, alone or in combination with cisplatin, causes significant tumor regression in a syngeneic model of SCLC
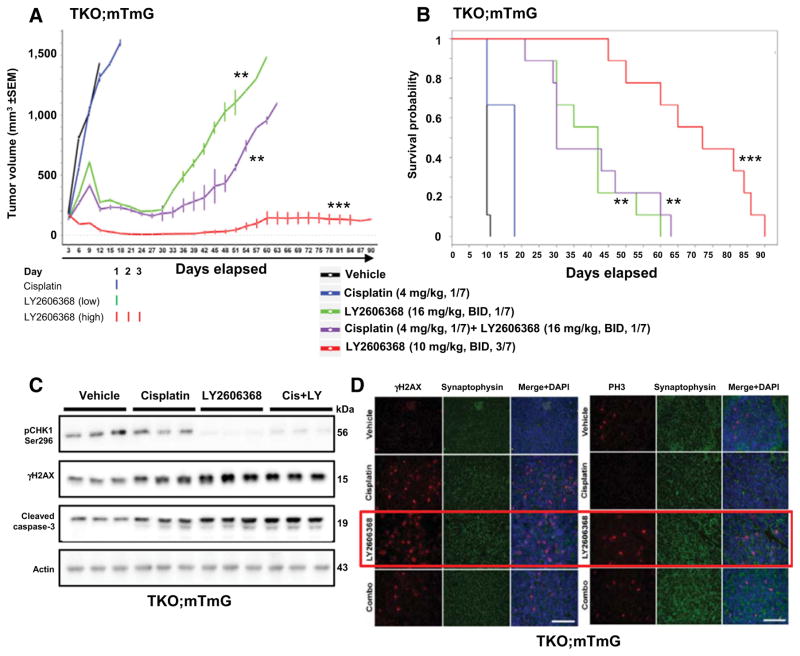
We next investigated the antitumor efficacy of LY2606368 as a single agent and in combination with cisplatin in a syngeneic flank tumor model generated from subcutaneous injection of cells (TKO.mTmG) derived from a GEMM with conditional loss of Trp53, p130, and Rb1 in the neuroendocrine cells of the lung (23, 24). These mice develop tumors that closely resemble human SCLC (Supplementary Fig. S3A; ref. 26). The TKO.mTmG cells expressed high levels of CHK1 and were sensitive to LY2606368 in vitro (IC50 < 30 nmol/L). Tumor-bearing mice (n = 9 per group) were treated with LY2606368, cisplatin, or vehicle.
Within 1 week of the start of treatment with LY2606368, remarkable tumor regression was observed (Fig. 3A; Supplementary Fig. S3B). Of the 9 mice treated with LY2606368 (10 mg/kg, twice daily, days 1–3 of a 7-day cycle; i.e., 60 mg/kg per week), 6 had a complete response (100% reduction) and the other 3 had >75% reduction in tumor volume. The tumor–control ratio at day 12 was 0.02 (P < 0.001). Mice treated with a lower dose of LY2606368 (16 mg/kg, twice daily, day 1 of a 7-day cycle; i.e., 32 mg/kg per week) had stable disease for up to 30 days, with a tumor–control ratio of 0.16 (P < 0.001; Fig. 3A; Supplementary Fig. S3B). In contrast, all vehicle-treated mice (n = 9) experienced rapid tumor progression and were removed from the experiment because of excessive tumor volume within two weeks (Fig. 3A and B; Supplementary Fig. S3B), and mice treated with cisplatin alone (4 mg/kg per week) died owing to tumor burden within the first three weeks (Fig. 3A and B; Supplementary Fig. S3B).
Figure 3.
LY2606368, with or without cisplatin, causes significant tumor regression and increases survival in a syngeneic model of SCLC. A, Tumor volume changes [means ± SEM (error bars)] in mice treated with vehicle (control), LY2606368 [high dose: 10 mg/kg, twice daily (BID), days 1–3 of each 7-day cycle; low dose: 16 mg/kg, twice daily, day 1 of each 7-day cycle), cisplatin (4 mg/kg, once per week), or LY2606368 (low dose) plus cisplatin (n = 9 mice per group). The P values were calculated using the Student t test. B, Survival of mice treated with vehicle (control), LY2606368 (high or low dose), cisplatin, or LY2606368 + cisplatin. The P value was established by the Mantel–Cox test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. C, Western blot analysis of phospho-CHK1 (Ser296), total CHK1, phospho-H2AX (γH2AX), and cleaved caspase-3 in lysates of resected tumors at treatment day 3 from the vehicle, cisplatin, LY2606368 (16 mg/kg, twice daily, 1/7), and combination [cisplatin (4 mg/kg, 1/7) + LY2606368 (16 mg/kg, twice daily, 1/7)] treatment groups. Three animals were included per group. Actin was used as the loading control. D, Immunofluorescence of paraffin-embedded tissue sections from resected TKO; mTmG tumors in treatment groups vehicle, cisplatin (4 mg/kg, 1/7), LY2606368 (10 mg/kg, twice daily, 3/7), and combination [cisplatin (4 mg/kg, 1/7)+ LY2606368 (16 mg/kg, twice daily, 1/7)] probed for γH2AX and phospho-histone H3 (PH3). Synaptophysin was used for cytoplasmic staining control and DAPI was used for nuclear staining control.
Consistent with these findings, overall survival rates of the LY2606368-treated groups were significantly higher than those of the vehicle- and cisplatin-treated groups (P < 0.001; Fig. 3B; Supplementary Table S2). There was no weight loss in any of the treatment groups throughout the course of this experiment (up to 90 days; Supplementary Fig. S3C). In summary, treatment with single-agent LY2606368 (high dose) caused significant tumor regression in a murine model of SCLC, and this was sustainable for up to 90 days.
In the group of mice treated with the combination of cisplatin and LY2606368, we used a lower dose of LY2606368 (32 mg/kg per week), given the risk of overlapping toxicities with cisplatin. This combination was well tolerated and produced significantly more tumor regression than cisplatin alone (P < 0.001; Fig. 3A) but was less potent than the higher dose of single-agent LY2606368.
To determine the effect of treatment with LY2606368 on DNA damage and apoptosis, we then performed immunofluorescence and immunoblot analysis of tumors from the vehicle and LY2606368 (high dose) treatment groups following 3 days of treatment. Consistent with the remarkable tumor regressions, we found that tumors from the LY2606368 (single agent) and combination-treated animals showed reduced levels of phospho-CHK1 (Ser296) and higher levels of γH2AX (marker of DNA damage), PH3 (a marker of mitosis), and cleaved caspase-3 (apoptosis marker) compared with vehicle-treated animals (Fig. 3C). Immunofluorescence further confirmed elevated γH2AX (Fig. 3D, left) and PH3 (Fig. 3D, right) in the tumors of mice treated with LY2606368, either as a single agent or combined with cisplatin. Together, these findings indicate that treatment with LY2606368 significantly augments the DNA damage and apoptosis caused by cisplatin in SCLC cells in this allograft model.
LY2606368 reduces tumor growth in mice with aggressive spontaneous SCLC
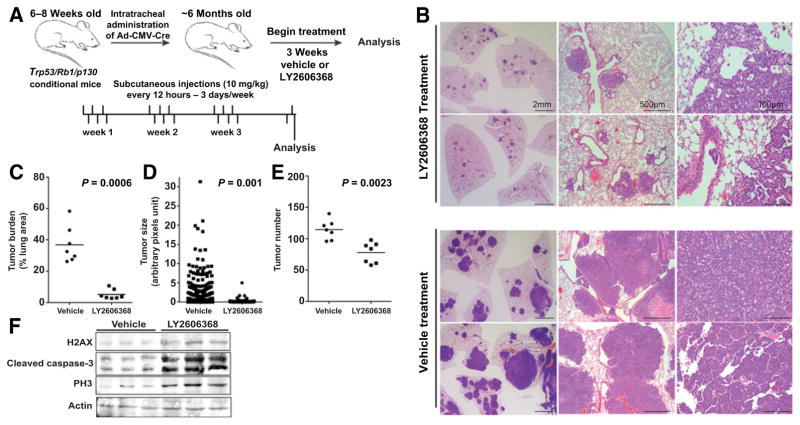
To investigate the effects of LY2606368 in SCLC tumors growing in the endogenous lung microenvironment, we used a GEMM of SCLC, in which conditional deletion of Trp53, Rb1, and p130 tumor-suppressor genes in adult neuroendocrine lung epithelial cells leads to the development of tumors that closely resemble human SCLC (24, 26). On the basis of prior experience with these models, we treated Trp53/Rb1/p130–knockout tumor-bearing mice (n = 7 per group) with LY2606368 (10 mg/kg, twice daily, days 1–3 of a 7-day cycle; i.e., 60 mg/kg per week for 3 weeks) or vehicle control starting four months after administration with Ad-CMV-Cre, when all mice had more than 50 SCLC lesions growing in their lungs (24). Tumor burden was quantified after three weeks of treatment (Fig. 4A). Upon sectioning, tumors in LY2606368-treated mice occupied a significantly smaller fraction of the total lung area (Fig. 4B and C) and were smaller (Fig. 4D) and fewer in number (Fig. 4E; P < 0.001 for all) compared with tumors in vehicle-treated mice. As expected, LY2606368 treatment also appreciably increased markers of DNA damage and apoptosis (γH2AX, PH3, and cleaved caspase-3) compared with vehicle-treated animals (Fig. 4F).
Figure 4.
LY2606368 causes tumor regression in a spontaneous genetically engineered mouse model of SCLC. A, Schematic representation of the treatment schedule in Trp53/Rb1/p130-knockout mice that develop SCLC. Mice (n = 7 per group) were treated with vehicle or LY2606368 (10 mg/kg, twice daily, days 1–3 of each 7-day cycle). B, Representative images (hematoxylin and eosin staining) of tumor burden in Trp53/Rb1/p130-knockout mice treated with vehicle control or LY2606368. Scale bars are indicated. C, Quantification of tumor burden as a percentage of lung area in Trp53/Rb1/p130-knockout mice treated with vehicle control or LY2606368 (n = 7 mice per group, one random lung section quantified per mouse). Per Mann–Whitney test, P = 0.006. D, Quantification of tumor size in Trp53/Rb1/p130-knockout mice treated with vehicle control or LY2606368 (n = 7 mice per group, one random lung section quantified per mouse). Per Mann–Whitney t test, P = 0.001. E, Quantification of tumors in Trp53/Rb1/p130-knockout mice treated with vehicle control or LY2606368 (n = 7 mice per group, one random lung section quantified per mouse). Per Mann–Whitney t test, P = 0.0023. *, P < 0.05; **, P < 0.01; ***, P < 0.001. F, Immunoblot analysis of phospho-H2AX (H2AX), cleaved caspase-3, and PH3 in lysates of resected tumors at treatment day 7 from the vehicle, LY2606368 treatment groups. Three animals were included per group. Actin was used as the loading control.
Platinum-resistant models of SCLC are sensitive to CHK1 inhibitor LY2606368
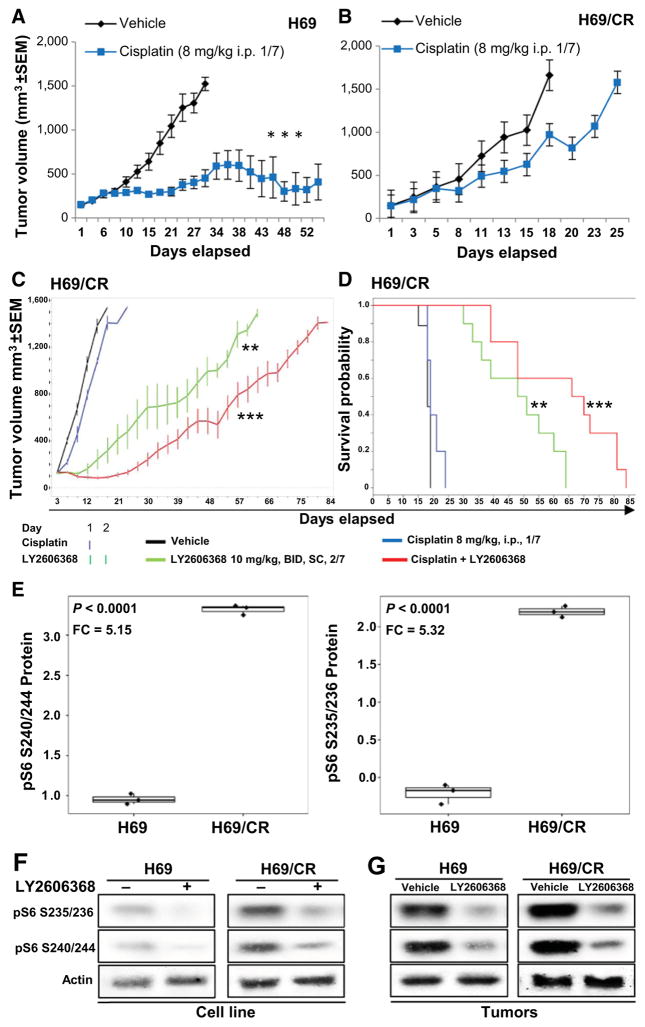
Although most SCLC initially responds to platinum-based therapy, nearly all cases become resistant to platinum-based therapy within a few months, at which point the disease typically does not respond to further therapy. Given the clinical need for active drugs for platinum-resistant SCLC, we next investigated the efficacy of CHK1 inhibition in a SCLC model of acquired chemoresistance in vitro and in vivo. For these experiments, we used human SCLC H69 cells (chemo-sensitive; cisplatin IC50 < 1 μmol/L) that had been previously treated with increasing doses of cisplatin until cells developed resistance to cisplatin [H69/CR (cisplatin resistant); cisplatin IC50 > 10 μmol/L; Supplementary Fig. S4A; ref. 31)]. Notably, treatment with single-agent LY2606368 significantly decreased cell viability and combined synergistically with cisplatin in both H69 (combination index = 0.22; fraction affected = 0.5) and H69/CR (combination index = 0.02; fraction affected = 0.5) models in vitro (Supplementary Fig. S4B and S4C), thus demonstrating that LY2606368 appreciably augments the cytotoxic effect of cisplatin even in a platinum-resistant human SCLC model in vitro.
After confirming that the chemoresistant phenotype was maintained in xenograft models (Fig. 5A and B), we investigated the effect of LY2606368 (with or without cisplatin) in the H69/CR xenograft mice. After two weeks of treatment, vehicle-treated mice had rapid tumor progression and died of tumor burden by day 15 (Fig. 5C and D). Similarly and as expected, cisplatin-treated mice (8 mg/kg, once per week) also had rapid disease progression with all mice dying due to tumor burden by day 21 (Fig. 5C and D). In contrast, single-agent LY2606368 (10 mg/kg, 2/7 = 40 mg/week) delayed tumor growth relative to cisplatin alone (Fig. 5C). This effect on tumor growth and survival was even greater when LY2606368 was combined with cisplatin, even though single-agent cisplatin had no significant activity. Specifically, the tumor–control ratio at day 15 was 0.88 for cisplatin (P < 0.01), 0.2 for single-agent LY2606368 (P < 0.001), and 0.06 for the combination group (P < 0.001). The survival rate of the LY2606368 + cisplatin-treated mice was also higher than vehicle-treated and single agent-treated mice (P = 0.02; Fig. 5D). The combination doses were well tolerated with no observed toxicity. In this model, we used a lower dose of LY2606368 (40 mg/kg/week) than in the spontaneous or syngeneic model (60 mg/kg/week) to avoid toxicity with a higher dose of cisplatin (8 mg/kg/week). To determine the effect of LY2606368 treatment on DNA damage and apoptosis in platinum-resistant models, we compared pre-versus post-LY2606368–treated H69/CR cell lines by Western blot analysis. We observed an increased in markers of DNA damage (γH2AX, RAD51) and apoptosis (cleaved PARP and cleaved caspase-3) 24 hours after treatment (Supplementary Fig. S4D). These in vitro findings were confirmed by RPPA in LY2606368-treated H69/CR xenograft tumors. Similar to the in vitro findings, we observed increased expression of γH2AX (P < 0.0001, FC = 3.66) and other DNA repair or apoptosis proteins in LY2606368-treated animals (Supplementary Fig. S4E).
Figure 5.
Therapeutic targeting of CHK1 overcomes acquired cisplatin resistance in SCLC. A and B, Tumor volume changes (mean ± SEM; error bars) in mice treated with vehicle (control) or cisplatin in flank xenograft models of parental H69 cells (A), or cisplatin-resistant H69 cells (H69/CR; B). The P values were calculated using the Student t test. C, Tumor volume changes (mean ± SEM) in mice treated with vehicle (control), cisplatin (8 mg/kg once per week), LY2606368 [10 mg/kg, twice daily (BID), twice weekly], or the combination of cisplatin and LY2606368. The P values were calculated using the Student t test. D, Survival of mice treated with vehicle, LY2606368, or cisplatin. The P values were derived by the Mantel–Cox test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. E, Box plots of RPPA markers differentially expressed in H69 and H69/CR cells, as determined by t test. The analysis showed significantly higher expression of pS6 S240/244 and pS6 S235/236 in the cisplatin-resistant cells compared with cisplatin-sensitive cells. FC, fold change. F, Immunoblot analysis of H69 and H69/CR cells before and after treatment with LY2606368 (24 hours; 100 nmol/L). The basal expression level of pS6 S240/244 and pS6 S235/236 was lower in nontreated H69 cells than in nontreated H69/CR cells, and this was further abrogated by treatment with LY2606368. Actin was used as a loading control. G, Western blot analysis of tumor lysates from H69 and H69/CR tumors with or without treatment with LY2606368. The basal expression level of pS6 S240/244 and pS6 S235/236 was lower in H69 vehicle tumors compared with H69/CR vehicle tumors, and this was further abrogated by treatment with LY2606368. Actin was used as a loading control.
To better understand signaling pathways that may mediate platinum resistance in H69/CR cells and how these pathways are affected by LY2606368, we compared the protein expression profile of H69 (parental) and H69/CR using a RPPA. Of the 195 total or phosphorylated proteins quantified, pS6 S240/244 (P < 0.0001; FC = 5.15) and pS6 S235/236 (P < 0.0001; FC = 5.32) were the top two markers expressed at higher levels in the cisplatin-resistant H69/CR model than in the H69 (parental) model (Fig. 5F). This is in agreement with our earlier observations, in which we have shown that phosphorylation of the mTOR pathway and pS6 (a downstream target) is associated with resistance to DDR inhibitors targeting PARP (32) and that a strong positive correlation exists between PARP inhibitor sensitivity and cisplatin sensitivity (33).
Given this observation, we then investigated the effect of CHK1 inhibition on phospho-pS6 levels in the platinum-sensitive and platinum-resistant models. Western blot analysis of H69 and H69/CR cells confirmed higher protein expression of pS6 S240/244 and pS6 S235/236 prior to treatment in H69/CR cells than in H69 cells, and this was significantly abrogated 24 hours after treatment with LY2606368 (Fig. 5G). Immunoblot analysis of lysates of tumors from the mice treated with LY2606368 (single agent) or vehicle collected on day 3 of treatment (from the animal study as shown in Fig. 5C and D) also showed reduced levels of pS6 S235/236 and pS6 S240/244 in tumor lysates from mice treated with LY2606368 (Fig. 5H). We also observed significant increase in the basal expression of total p90RSK and phospho p90RSK_Thr359 in both H69/CR cell line and tumors (Supplementary Fig. S4F). Given the role of p90RSK as a regulator of the cell cycle and mTOR pathway and that p90RSK is a secondary target of LY2606368, we investigated the effect of LY2606368 on p90RSK in H69/CR models and found that treatment inhibited phospho p90RSK_Thr359 in vivo (Supplementary Fig. S4G) and in vitro (Supplementary Fig. S4H).
Taken together, these results suggest that the antitumor effect of LY2606368 in the H69/CR model may be mediated through the regulation of the p90RSK/mTOR pathway.
CHK1 and PARP inhibitor combination decrease viability and cause tumor regression in SCLC models
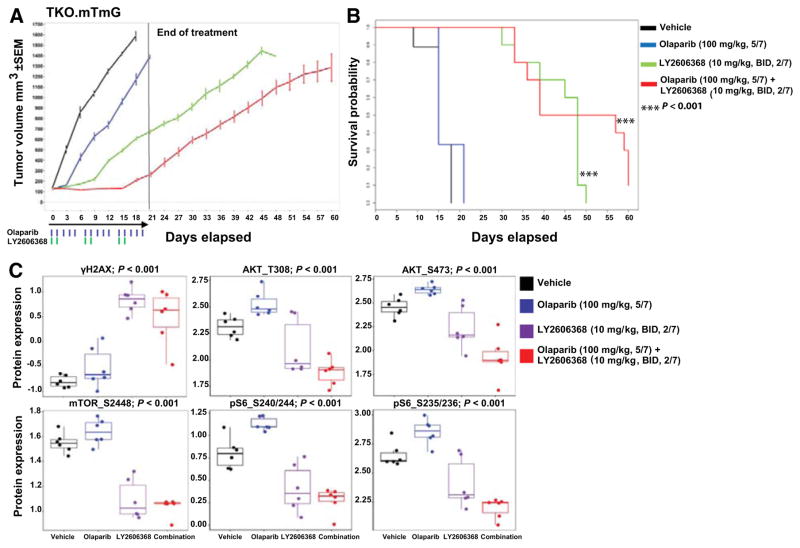
We have previously shown that another important DNA repair protein, PARP1, is also overexpressed in SCLC, and that PARP inhibition is active in SCLC models (13, 33). However, some SCLC cells have primary resistance to single-agent PARP inhibition in vitro (Supplementary Fig. S5A). We predicted that combinations of PARP inhibitors and CHK1 inhibitors, by preventing DNA repair while simultaneously abrogating the G2 cell-cycle checkpoint, may enhance the effect of PARP inhibition. To test this, we treated nine cell lines with LY2606368, olaparib (a PARP inhibitor approved by the FDA for the treatment of ovarian cancer), or both. We found that the addition of LY2606368 to olaparib decreased proliferation in SCLC cell lines with primary resistance to PARP targeting (Supplementary Fig. S5B).
To further investigate the combination treatment in vivo, we used the syngeneic GEMM-derived triple-knockout SCLC model (TKO.mTmG; ref. 26). Tumor-bearing mice were treated with olaparib (100 mg/kg, five times weekly), LY2606368 (10 mg/kg, twice daily, days 1 and 2 of a 7-day cycle), or the combination of the drugs for three cycles and then followed off treatment. Mice treated with olaparib (100 mg/kg, days 1–5 of a 7-day cycle, intraperitoneally; n = 10) had progressive disease with no tumor regression and no survival benefit as compared with vehicle (Fig. 6A and B). Given the potential for overlapping hematologic toxicities between CHK1 and PARP inhibitors, a moderately reduced dose of LY2606368 was used for this experiment (compared with the higher single-agent dose of LY2606368 used in the earlier experiments using this model).
Figure 6.
CHK1 targeting augments the effect of PARP inhibitors in SCLC models. A, Tumor volume changes (mean ± SEM) in mice treated with vehicle (control), LY2606368 [10 mg/kg, twice daily (BID), days 1 and 2 of each 7-day cycle], olaparib (100 mg/kg, five times weekly), or LY2606368 + olaparib (n = 10 mice per group). The P values were calculated using the Student t test. B, Survival of mice treated with vehicle (control), LY2606368, olaparib, or LY2606368 + olaparib. The P value was established by the Mantel–Cox test. C, RPPA analysis corresponding to the indicated AKT/mTOR pathway and DNA damage markers in SCLC in vivo tumor samples treated with vehicle, olaparib (100 mg/kg, 5/7), LY2606368 (10 mg/kg, twice daily, 2/7), or combination and collected at the end of the first treatment cycle (false discovery rate < 0.05).
Animals were treated for 21 days and then monitored off treatment. All 10 mice treated with LY2606368 (10 mg/kg, twice daily, days 1 and 2 of each 7-day cycle) had delayed tumor progression and survived significantly longer than those treated with vehicle or single-agent olaparib (Fig. 6A and B). Mice (n = 10) treated with the combination of olaparib and LY2606368 had the greatest delay in tumor growth of all the arms and survived three times longer than vehicle- or olaparib-treated mice (Fig. 6A and B). Specifically, the tumor–control ratio at day 18 was 0.73 for single-agent olaparib (P = 0.01), 0.42 for single-agent LY2606368 (P < 0.001), and 0.16 for LY2606368 + olaparib (P < 0.001), demonstrating the efficacy of the combination. On the basis of these results, we conclude that even with a relatively short duration of treatment, the addition of LY2606368 improves response to olaparib and prolongs survival in this mouse model. The single-agent and combination doses were well tolerated with no observed toxicity.
Interestingly, using RPPA, we found that phosphorylation (activation) of proteins in the AKT/mTOR pathway AKT_T308, mTOR_S2448, pS6_S240/244, and pS6_S235/236 (P < 0.001) were increased in olaparib-treated groups (Fig. 6C), consistent with prior findings by our group in other SCLC models that mTOR pathway activation is an adaptive response promoting PARP inhibitor resistance (32). However, LY2606368 treatment as a single agent and in combination with olaparib abrogated the activation of AKT/mTOR pathway. Thus, LY2606368 may potentiate the antitumor response of olaparib by abrogating the activation of the AKT/mTOR pathway.
cMYC overexpression predicts sensitivity to CHK1 inhibition in SCLC
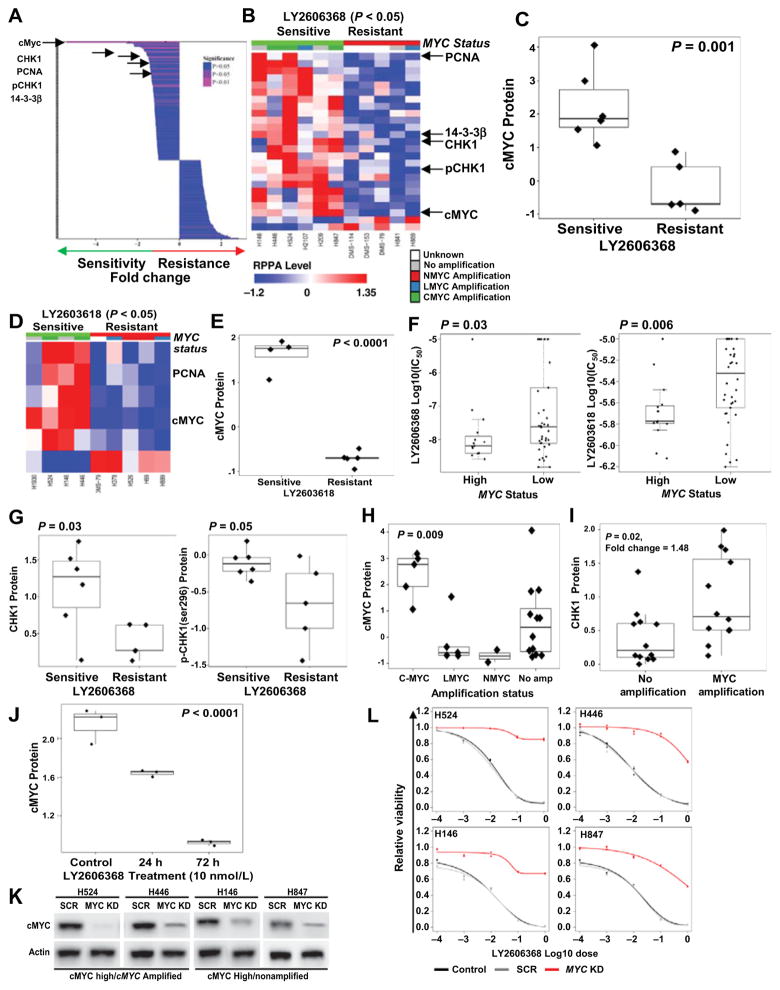
Although LY2606368 inhibited SCLC growth in most SCLC models, we identified a subset of SCLC cell lines with relatively greater resistance to CHK1 inhibition. Proteomic profiles (by RPPA) of the SCLC cell lines most sensitive to LY2606368 (IC50 < 100 nmol/L; n = 6) and those that were the least sensitive (IC50 > 1 μmol/L; n = 5) were compared by t test. This analysis revealed significantly increased expression of cMYC protein (P = 0.002; FC = 5.39) as one of the top five markers associated with LY2606368 sensitivity (Figures 7A, 7B, and 7C). This observation was further strengthened by the finding that cMYC expression was also the top marker associated with sensitivity to a second CHK1 inhibitor, LY2603618 (P < 0.0001; Figs. 7D and E). In the full panel of cell lines in which we had tested LY2606368, Spearman correlation between RPPA markers and IC50 across all human SCLC cell lines (n = 29; as a continuous variable) also identified cMYC expression as a top marker of sensitivity to LY2606368 (P = 0.08) and LY2603618 (P = 0.009; Figures S6A and S6B).
Figure 7.
cMYC expression predicts sensitivity to CHK1 inhibition in SCLC. A, Spearman correlation of differential expression of 195 RPPA markers and dichotomized half-maximal inhibitory concentration (IC50; most resistant cells, n = 5; most sensitive cells, n = 6; from Supplementary Table S1) of LY2606368. The top markers of sensitivity are marked with arrows. B, Heatmap of RPPA markers that were significantly correlated with sensitivity to LY2606368 in human SCLC cell lines (P < 0.05). In the top index, cell lines with relatively low LY2606368 and LY2603618 IC50 values are marked in green and those with relatively high IC50 values are marked in red. The cell lines are grouped according to their MYC status and marked as having no MYC amplification (gray), MYCN amplification (red), MYCL amplification (blue), MYC amplification (green), or unknown (white). The overexpressed markers are shown in red, and markers with reduced expression are shown in blue. C, Box plot of cMYC protein correlated with dichotomized IC50 of LY2606368 as determined by t test. The top markers of sensitivity to CHK1 inhibitors were total cMYC, CHK1, pCHK1, PCNA, and 14-3-3β. D, Heatmap of RPPA markers that were significantly correlated with sensitivity to LY2603618 in human SCLC cell lines (P < 0.05). In the top index, cell lines with relatively low LY2603618 IC50 values are marked in green and those with relatively high IC50 values are marked in red. The color indexing was similar to that of Fig. 7B. E, Box plot of cMYC protein correlated with dichotomized IC50 of LY2603618 as determined by t test. F, Comparison (t test) of the IC50 of two CHK1 inhibitors, LY2606368 and LY2603618, with MYC mRNA expression, divided into MYC-high and MYC-low groups on the basis of bimodal distribution of MYC mRNA in 63 human SCLC cell lines. The IC50 values of the CHK1 inhibitors LY2606368 (P = 0.03) and LY2603618 (P = 0.006) are lower in the MYC-high group. t test analysis also shows higher level of CHK1 and pCHK1 (Ser296) in cell lines sensitive to LY2606368 (Fig. 7G). H, cMYC protein expression across different MYC amplification subtypes, determined by ANOVA, demonstrating high expression of cMYC in both MYC-amplified and nonamplified cells, with the highest expression in the MYC-amplified subset (P = 0.009). I, Comparison (t test) of MYC-amplified (MYC, MYCL, and MYCN) and nonamplified SCLC cells revealing greater expression of the target, CHK1, in the MYC-amplified cells. J, RPPA analysis comparing cMYC expression between nontreated and cells treated with LY2606368 for 24 and 72 hours shows decreased expression of cMYC after CHK1 inhibitor treatment. K, Western blot analysis confirming shRNA-mediated knockdown of MYC in four human SCLC cell lines: H446 and H524 (high cMYC expression and MYC-amplified) and H146 and H847 (high cMYC expression and not MYC-amplified). L, Cell viability assay, including scramble (SCR) and MYC-knockdown (KD) cell lines, demonstrating the effect of MYC inhibition on the viability of SCLC cells after treatment with LY2606368. Data represent the mean ± SEM (error bars) of three independent experiments.
These findings were further validated using MYC mRNA expression data from cell lines treated with LY2606368 and LY2603618 in a recently published National Cancer Institute drug screen (34). Specifically, response of 63 cell lines to the CHK1 inhibitors was compared between MYC-high and MYC-low cell lines (determined by bimodal distribution of MYC mRNA) (35, 36). Consistent with our proteomic findings, MYC-high cells were significantly more sensitive to both LY2606368 (P = 0.03) and LY2603618 (P = 0.006; Fig. 7F).
Increased expression of CHK1 (P < 0.03; FC = 1.66), and phospho-CHK1_S296 (P < 0.05; FC = 1.47) were also among the top markers of LY2606368 sensitivity (Fig. 7A, B, and G). PCNA and 14-3-3β, two important proteins implicated in cell-cycle regulation (37, 38), were also associated with sensitivity in both dichotomized (P < 0.05; Fig. 7A and B; and Supplementary Fig. S7A) and continuous (P < 0.05; Supplementary Figs. S6A and S7B) IC50 correlations of LY2606368 and LY2603618 (Fig. 7D).
Although cMYC and CHK1 protein levels were elevated in MYC-amplified SCLC cell lines (P = 0.009 for cMYC and P = 0.02 for CHK1), a substantial subset of cell lines without MYC amplification also had levels of cMYC and CHK1 similar to those of MYC-amplified lines (Fig. 7H and I; Supplementary Table S1). Using a bimodal analysis (36), we found elevated MYC expression in 35.9%, as compared with MYC amplification in only 18% cell lines (Supplementary Fig. S8A). Similarly, in SCLC patient tumors (n = 81), MYC overexpression (mRNA) was almost 3 times more common than MYC amplifications (19% of tumors with over-expression as compared with 7% with amplification; Supplementary Fig. S8B). This finding suggests that MYC expression levels may identify a substantially larger population of SCLC patients who may benefit from CHK1 targeting as compared with MYC amplification alone.
Interestingly, LY2606368 treatment reduced total levels of cMYC protein in sensitive cell lines after 24 and 72 hours of LY2606368 treatment (P < 0.0001; Fig. 7J), while there was no significant change in cMYC protein (P = 0.6) or MYC mRNA posttreatment in LY2606368-resistant models (data not shown). To investigate the role of high cMYC expression in promoting sensitivity to CHK1 inhibition, we next performed siRNA-mediated knockdown of MYC in cell lines sensitive to LY2606368 (Fig. 7K). Knockdown of MYC in four SCLC cell lines—H524 and H446 (high cMYC expression and MYC-amplified) and H146 and H847 (high cMYC expression and not MYC-amplified)—induced resistance to CHK1 inhibition in these cells (Fig. 7L). To further test the functional role of MYC expression in LY2606368 sensitivity, we overexpressed MYC in three SCLC cell lines (H889, H2029, H2330) with low MYC expression and higher IC50 to LY2606368 (>1 μmol/L; Supplementary Fig. S8C). Overexpression of MYC sensitized all three cell lines to LY2606368 (P < 0.05; Supplementary Fig. S8D). Thus, cMYC expression level is directly linked to CHK1 inhibitor sensitivity in SCLC models.
Discussion
Given the critical role of CHK1 in G2 checkpoint regulation, especially in cancer types in which G1 regulation is impaired owing to TP53 loss (17–19, 39), in the current study we investigated the role of CHK1 targeting in SCLC using a large number of molecularly characterized cell lines and animal models. We confirmed that CHK1 is overexpressed in SCLC patient tumors and studied the effect of targeted CHK1 knockdown and pharmacologic inhibition in vitro and in vivo. We found that a second-generation CHK1 inhibitor, LY2606368, had remarkable single-agent activity, enhanced the cytotoxic potential of clinically relevant dose of cisplatin (including in platinum-resistant models), and, when combined with the FDA-approved PARP inhibitor olaparib, led to tumor regression and prolonged survival. Given the clinical importance of predictive biomarkers for targeted therapeutics, we then identified candidate protein biomarkers of CHK1 inhibitor response, and here, we propose for the first time that cMYC protein overexpression is a predictor of CHK1 inhibitor sensitivity in SCLC models. Together, these data indicate that CHK1 is a promising therapeutic target for SCLC, especially in the subset of tumors with cMYC protein overexpression.
The current study shows that genetic knockdown of CHK1 is sufficient to decrease viability and increase apoptosis in SCLC cells, a finding that is in agreement with a previous study showing that siRNA knockdown of CHK1 caused >50% growth inhibition in neuroblastoma cells, another high-grade neuroendocrine cancer (32). Although a prior study showed that CHK1 targeting could enhance the cytotoxicity of cisplatin in a small number of SCLC cell lines (40), the current study significantly expands upon this work and reports additional novel findings. First, by leveraging a large panel of molecularly profiled cell lines, we identify for the first time biomarker-defined SCLC subsets with enhanced vulnerability to CHK1 inhibition. Furthermore, we used LY2606368, the first clinically viable CHK1 inhibitor to date, which has recently completed phase I testing (22) and is now being tested in ongoing clinical trials for multiple cancer types. Finally, the current study shows for the first time the remarkable effect of single-agent CHK1 targeting in a spontaneous GEMM model and syngeneic in vivo model, as well as the effects of CHK1–PARP combinations in SCLC. In this study, we have used multiple doses of LY2606368 (60 mg/kg/week for single agent and 40 mg/kg/week for combination with PARP inhibitor) to achieve optimal responses with the single-agent treatment and limit toxicities that were anticipated with combination therapy. The monotherapy dose of 60 mg/kg/week led to a durable response is translatable to patients based on recent phase I clinical trial results (22).
Although platinum-based chemotherapy is the mainstay of treatment for patients with SCLC, the disease eventually becomes resistant to cisplatin and carboplatin (often within a matter of months), at which point other available chemotherapies such as topotecan have limited benefit. Therefore, identifying targeted therapies with activity in platinum-resistant SCLC represents an important goal for the field and one with immediate clinical implications. In contrast to previous findings that platinum resistance predicts resistance to PARP inhibitors (another DDR inhibitor) in SCLC and other cancer types (33), we found that H69/CR cells (a model of acquired platinum resistance; ref. 31) were sensitive to LY2606368 both in vitro and in vivo. Moreover, treatment with LY2606368 significantly enhanced the cytotoxic potential of cisplatin in this model. Activity of LY2606368 in the setting of platinum resistance may be due to its downregulation of phospho-pS6, a downstream target of AKT/mTOR pathways that we found to be overexpressed in H69/CR cells and that we previously reported is associated with PARP inhibitor resistance (32). Given that the mTOR inhibitor everolimus was recently approved by the FDA for the treatment of lung neuroendocrine tumors, this finding may have important implications beyond the current study in terms of the role of this pathway in drug resistance in SCLC.
Our group previously identified PARP as a novel therapeutic target for SCLC, and several clinical trials with PARP inhibitors are ongoing (13). As with other targeted drugs, understanding combinations that can enhance PARP inhibitor efficacy or delay resistance will be important to maximize the potential therapeutic impact of PARP inhibitors. Prior studies have demonstrated that inhibitors of PARP1 interact with CHK1 inhibitors to cause DNA damage and cell death in various cancers (15, 41), but the combination of PARP inhibition with CHK1 inhibition has not yet been explored in SCLC. Using a panel of nine cell lines and a syngeneic xenograft model, we found that the combination of LY2606368 with the FDA-approved PARP inhibitor olaparib was synergistic (in 40%) or additive (in 60%) in SCLC cell lines, including those with primary resistance to single-agent olaparib. In vivo, the combination of LY2606368 with olaparib resulted in significantly greater tumor regression and prolonged survival compared with either single-agent alone. RPPA analysis demonstrated that PARP inhibition and platinum resistance are both associated with activation the mTOR pathway, but that, LY2606368 downregulates AKT/mTOR pathway activation. We propose that CHK1 targeting reverses PARP inhibitor resistance, at least in part, through modulation of the AKT/mTOR pathway. Further studies are needed to fully define the pathway by which PARP1 and CHK1 inhibitors interact to lead to cell death in SCLC.
The identification of molecularly defined subsets of lung cancer patients with different sensitivities to a targeted therapy has substantial implications for clinical development. We found that high cMYC protein expression was the top marker of sensitivity to LY2606368 and LY2603618. cMYC protein overexpression was observed in both MYC-amplified and nonamplified cell lines. This finding is consistent with SCLC patient tumor profiles published by George and colleagues (5), in which MYC over-expression (mRNA) was observed three times more often than MYC amplification in patient tumors. This suggests that cMYC overexpression (which can be determined by IHC) has the potential to identify an even larger subset of SCLC patients, beyond those with MYC amplification alone, who may benefit from CHK1 targeting. Our finding is consistent with previous studies in which CHK1 inhibitors (UCN-01 or SB-218078) were particularly active in MYC-driven lymphoma and pancreatic cancers with an inactivated TP53 background (42). Other studies have shown that MYC overexpression increases CHK1 expression, thereby enhancing the DNA repair capacity (43). This finding was confirmed in our SCLC models where the MYC mRNA expression correlated with higher protein expression of CHK1 (P = 0.01) in SCLC cell lines thus showing enhanced DNA repair capacity in these models. Cancers overexpressing MYC become “addicted” to CHK1, and concurrent inactivation of TP53 makes cells completely dependent on CHK1 for genomic integrity (43, 44). Thus, CHK1 inhibition may be especially effective in MYC-driven cancer cells such as SCLC, pushing them to DNA damage and cell death. Taken together, these data suggest that in SCLC, higher cMYC expression may lead to increased CHK1 (target) expression and promote therapeutic vulnerability to CHK1 inhibitors, but further investigation of the mechanism through which cMYC regulates CHK1 is warranted.
There is an urgent clinical need for effective therapeutic targets for SCLC. In the current study, we provide compelling preclinical evidence of significant efficacy of a CHK1 inhibitor as monotherapy and in combination with chemotherapy or olaparib in SCLC models, including those with acquired cisplatin resistance. Given that platinum resistance ultimately develops in nearly all patients with SCLC and is associated with resistance to other available chemotherapies, the finding that LY2606368 has activity in a platinum-resistant model has important clinical implications. Combination therapies may inhibit development of drug resistance to PARP inhibitors, which are now in clinical trials for the treatment of SCLC, and in the current study, we show for the first time that targeting CHK1 may help overcome de novo resistance to PARP inhibitors in this disease. Finally, we describe a candidate biomarker, MYC overexpression, which predicts sensitivity to CHK1 targeting and could be translated into the clinic to identify a biomarker-defined patient population that may get relatively greater benefit from CHK1 inhibitors. These findings support the further investigation of CHK1 targeting and related biomarkers in SCLC. Toward this goal, the first clinical trial of LY2606368 for patients with SCLC (NCT02735980) will explore the translational application of these preclinical findings.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Emily Roarty, Department of Thoracic Head and Neck Medical Oncology, The University of Texas MD Anderson Cancer Center for reviewing and editing the manuscript.
Grant Support
This work was supported by NIH (R01CA207295 to L.A. Byers), The University of Texas MD Anderson Cancer Center Small Cell Lung Cancer Working Group and Abell Hangar Foundation Distinguished Professor Endowment to B.S. Glisson, Sidney Kimmel Scholar Award to L.A. Byers, by Free to Breathe (formerly National Lung Cancer Research Partnership), North Carolina Lung Cancer Partnership (L.A. Byers), the LUNGevity Foundation (L.A. Byers), by Uniting Against Lung Cancer (L.A. Byers), R. Lee Clark Fellow Award, made possible by the Jeanne F. Shelby Scholarship Fund, to L.A. Byers, MD Anderson Physician Scientist Award to L.A. Byers, National Cancer Institute Cancer Clinical Investigator Team Leadership Award (P30 CA016672 to L.A. Byers), the generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program, and The Susan G. Komen Foundation award to H. Piwnica-Worms. LY2606368 and olaparib provided by The Pharmaceutical Chemistry Facility at MD Anderson, supported by the NIH/National Cancer Institute (P30CA016672); flow cytometry studies done at The Flow Cytometry and Cellular Imaging Core Facility, North Campus, The University of Texas MD Anderson Cancer Center, funded by NCI Cancer Center Support Grant (P30CA016672).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
D.S. Shames reports receiving ownership interest (including patents) in Genentech Inc. and UT Southwestern. J. Heymach is a consultant/advisory board member for AstraZeneca and Lilly. L.A. Byers is a consultant/advisory board member for AstraZeneca. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: T. Sen, C.A. Stewart, J. Minna, J. Sage, D.L. Gibbons, J. Heymach, L.A. Byers
Development of methodology: T. Sen, C.A. Stewart, L.A. Byers
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): T. Sen, C.A. Stewart, S. Cristea, A. Valliani, D.S. Shames, A. Redwood, Y. Fan, J. Minna, D.L. Gibbons, H. Piwnica-Worms, J. Wang, L.A. Byers
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): T. Sen, P. Tong, S. Cristea, A. Valliani, D.S. Shames, L. Li, J. Sage, D.L. Gibbons, J. Wang, L.A. Byers
Writing, review, and/or revision of the manuscript: T. Sen, C.A. Stewart, S. Cristea, A. Valliani, D.S. Shames, A. Redwood, L. Li, B.S. Glisson, J. Minna, J. Sage, D.L. Gibbons, H. Piwnica-Worms, J. Heymach, L.A. Byers
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): T. Sen
Study supervision: T. Sen, J. Wang, L.A. Byers
References
- 1.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–44. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 2.William WN, Jr, Glisson BS. Novel strategies for the treatment of small-cell lung carcinoma. Nat Rev Clin Oncol. 2011;8:611–9. doi: 10.1038/nrclinonc.2011.90. [DOI] [PubMed] [Google Scholar]
- 3.Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer. 2015;121:664–72. doi: 10.1002/cncr.29098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–8. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 5.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–6. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–43. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 9.Jahchan NS, Lim JS, Bola B, Morris K, Seitz G, Tran KQ, et al. Identification and targeting of long-term tumor-propagating cells in small cell lung cancer. Cell Rep. 2016;16:644–56. doi: 10.1016/j.celrep.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–74. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 13.Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2:798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunn PA, Jr, Minna JD, Augustyn A, Gazdar AF, Ouadah Y, Krasnow MA, et al. Small cell lung cancer: can recent advances in biology and molecular biology be translated into improved outcomes? J Thorac Oncol. 2016;11:453–74. doi: 10.1016/j.jtho.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth L, Cruickshanks N, Ridder T, Dai Y, Grant S, Dent P. PARP and CHK inhibitors interact to cause DNA damage and cell death in mammary carcinoma cells. Cancer Biol Ther. 2013;14:458–65. doi: 10.4161/cbt.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karnak D, Engelke CG, Parsels LA, Kausar T, Wei D, Robertson JR, et al. Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res. 2014;20:5085–96. doi: 10.1158/1078-0432.CCR-14-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadhikar MA, Sciuto MR, Alves MV, Pickering CR, Osman AA, Neskey DM, et al. Chk1/2 inhibition overcomes the cisplatin resistance of head and neck cancer cells secondary to the loss of functional p53. Mol Cancer Ther. 2013;12:1860–73. doi: 10.1158/1535-7163.MCT-13-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122:1541–52. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–23. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–22. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong D, Infante J, Janku F, Jones S, Nguyen LM, Burris H, et al. Phase I study of LY2606368, a checkpoint kinase 1 inhibitor, in patients with advanced cancer. J Clin Oncol. 2016 doi: 10.1200/JCO.2015.64.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahchan NS, Dudley JT, Mazur PK, Flores N, Yang D, Palmerton A, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–77. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010;70:3877–83. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Nat Acad Sci U S A. 2001;98:8012–7. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol. 2015;10:553–64. doi: 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong P, Coombes KR, Johnson FM, Byers LA, Diao L, Liu DD, et al. drexplorer: A tool to explore dose-response relationships and drug-drug interactions. Bioinformatics. 2015;31:1692–4. doi: 10.1093/bioinformatics/btv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prichard MN, Shipman C. A three-dimensional model to analyze drug-drug interactions. Antiviral Res. 1990;14:181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 29.R Development Core Team. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 30.King C, Diaz HB, McNeely S, Barnard D, Dempsey J, Blosser W, et al. LY2606368 causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol Cancer Ther. 2015;14:2004–13. doi: 10.1158/1535-7163.MCT-14-1037. [DOI] [PubMed] [Google Scholar]
- 31.Twentyman PR, Wright KA, Mistry P, Kelland LR, Murrer BA. Sensitivity to novel platinum compounds of panels of human lung cancer cell lines with acquired and inherent resistance to cisplatin. Cancer Res. 1992;52:5674–80. [PubMed] [Google Scholar]
- 32.Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Stewart CA, et al. Activation of the PI3K/mTOR pathway following PARP inhibition in small cell lung cancer. PLoS ONE. 2016;11:e0152584. doi: 10.1371/journal.pone.0152584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardnell RJ, Feng Y, Diao L, Fan YH, Masrorpour F, Wang J, et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res. 2013;19:6322–8. doi: 10.1158/1078-0432.CCR-13-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, et al. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J Nat Cancer Inst. 2016;108:djw122. doi: 10.1093/jnci/djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong P, Chen Y, Su X, Coombes KR. SIBER: systematic identification of bimodally expressed genes using RNAseq data. Bioinformatics. 2013;29:605–13. doi: 10.1093/bioinformatics/bts713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Wen S, Symmans WF, Pusztai L, Coombes KR. The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform. 2009;7:199–216. doi: 10.4137/cin.s2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermeking H. The 14-3-3 cancer connection. Nat Rev Cancer. 2003;3:931–43. doi: 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- 38.Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochem Soc Trans. 2009;37:605–13. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- 39.Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol Oncol. 2011;5:368–73. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson R, Meuth M, Woll P, Zhu Y, Danson S. Treatment with the Chk1 inhibitor Go6976 enhances cisplatin cytotoxicity in SCLC cells. Int J Oncol. 2012;40:194–202. doi: 10.3892/ijo.2011.1187. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell C, Park M, Eulitt P, Yang C, Yacoub A, Dent P. Poly(ADP-ribose) polymerase 1 modulates the lethality of CHK1 inhibitors in carcinoma cells. Mol Pharmacol. 2010;78:909–17. doi: 10.1124/mol.110.067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–5. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoglund A, Nilsson LM, Muralidharan SV, Hasvold LA, Merta P, Rudelius M, et al. Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells. Clin Cancer Res. 2011;17:7067–79. doi: 10.1158/1078-0432.CCR-11-1198. [DOI] [PubMed] [Google Scholar]
- 44.Wang WJ, Wu SP, Liu JB, Shi YS, Huang X, Zhang QB, et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013;73:1219–31. doi: 10.1158/0008-5472.CAN-12-1408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.