Abstract
The differentiation of multipotent stem cells toward a pancreatic lineage provides us with an alternative cell-based therapeutic approach to type 1 diabetes and enables us to study pancreas development. The current study aims to study the effect of growth factors such as activin A or nicotinamide, alone and in combinations with the transcription factor, PDX1 (pancreatic and duodenal homeobox-1), on human amnion epithelial cells (hAECs) toward a pancreatic lineage. Ectopic expression of Pdx1 followed by treatment of hAECs with nicotinamide for 4 days resulted in strong induction of pancreatic endoderm and pancreatic progenitor genes, including NKX6.1 and NEUROD1. Pancreatic lineage cells expressing PDX1, SOX17, and RFX6 are derived from Pdx1-transduced hAECs treated with activin A or nicotinamide, but not cells treated with activin A or nicotinamide alone. Our study provides a novel culture protocol for generating pancreas-committed cells from hAECs and reveals an interplay between Pdx1 and activin A/nicotinamide signaling in early pancreatic fate determination.
Keywords: : amnion, pancreas, activin A, nicotinamide, PDX1, qPCR
Introduction
With the global prevalence of diabetes estimated at 9% of all individuals above 18 years of age, the World Health Organization estimates that diabetes will be the 7th leading cause of death by the year 2030 (WHO Diabetes factsheet). Of this, 70,000 cases of type 1 diabetes are newly diagnosed every year (Guariguata, 2011). The identification of cellular or tissue replacements for the pancreas is thus the need of the hour. While the Edmonton protocol was a breakthrough toward this (Shapiro et al., 2000), the associated long-term difficulties, namely lack of an adequate number of donors, need for lifelong immunosuppression, and loss of graft function over time, require us to look for alternative methods (Ryan et al., 2005; Shapiro et al., 2006).
Multipotent cells from various sources such as embryonic stem (ES) cells, postpartum tissue, and even adult tissue such as bipotent cells from the hepatic duct have been explored as cellular therapy options for type 1 diabetes (Bonfanti et al., 2015; D'Amour et al., 2006; Kadam et al., 2010). The amnion derived from the inner cell mass of the blastocyst of the developing embryo can potentially harbor multipotent cells and is therefore an attractive choice to explore. In fact literature already exists for the successful differentiation of amnion-derived cells in culture (Hou et al., 2008; Miki et al., 2005). The current protocol aims to further the knowledge obtained from the differentiation of human amnion epithelial cells (hAECs) down the pancreatic lineage. In particular, we aim to study the effect of morphogens, individually or in combination, at the molecular level.
PDX1 (pancreatic and duodenal homeobox-1) is a master regulator of pancreatic development and is important in determining definitive endoderm (DE) and pancreatic endoderm cell fates (D'Amour et al., 2006; Jennings et al., 2013; Kelly et al., 2011; Ohlsson et al., 1993). PDX1 is also required in adult islets for the proper functioning of the beta cells (Bastidas and Showalter, 2013; Khoo et al., 2012). Ectopic expression of PDX1 is seen to play a critical role in the developmental reprogramming of cells of unrelated tissues such as keratinocytes and amniotic fluid stem (AFS) cells into cells of the pancreas (Kajiyama et al., 2010; Mauda-Havakuk et al., 2011; Tang et al., 2006; Zhou et al., 2013). The current study thus aimed to harness the activation of PDX1 for the successful production of therapeutically relevant β cells.
Activin A, a member of the TGF-β superfamily, has been shown to either maintain the stemness of human ES cells or promote an endodermal fate depending on the time and dosage of exposure (Beattie et al., 2005; D'Amour et al., 2005). However, the effect of this growth factor on the differentiation of other multipotent stem cells, such as hAECs, has not been studied. The present study aims to fill that gap.
Nicotinamide has been used to differentiate hAECs into functional insulin-producing cells in the presence or absence of serum in the culture media (Hou et al., 2008; Miki et al., 2005). The suggested mechanism of action is the simultaneous proliferation and differentiation of epithelial cells by its action as a PARP (Poly-ADP Ribose synthetase) inhibitor (Otonkoski et al., 1993). However, the duration of the culture period is at least 2 weeks long. This study uses previously established protocols for pancreatic differentiation using nicotinamide by itself or in combination with other growth stimuli with the aim to reduce the duration of the differentiation process. This may have important implications in a clinical setting where timely intervention is needed.
Materials and Methods
Dulbecco's modified Eagle's medium-Low glucose (DMEM-LG), fetal bovine serum (FBS), 100 × Insulin, transferrin, selenite (ITS) Liquid Media Supplement, epidermal growth factor (EGF), and Nicotinamide were all obtained from Sigma-Aldrich®; Nonessential amino acids (NEAA) was obtained from GE Healthcare Life Sciences; 100 × Penicillin–Streptomycin was obtained from Gibco™; and Activin A from R&D systems.
Culture of cells
Uncultured (p0) hAECs were kindly provided by Dr. Sean Murphy (WFIRM). Cells were cultured up to passage 2 (p2) in complete medium (DMEM-LG supplemented with 10% FBS, 1% NEAA, 1% ITS, 10 ng/mL EGF, and 1% penicillin–streptomycin), under standard cell culture conditions (5% CO2/37°C/humidified). Cells were cultured without EGF at passage 2.
Differentiation of p2 hAECs
p2 hAECs were seeded at a density of 10,000 cells/cm2 in six-well plates and allowed to adhere overnight under standard culture conditions.
Adenoviral transduction
Adenovirus expressing mouse Pdx1 (mPdx1) was a gift from Christopher Newgard and Sarah Ferber at Duke University. GFP adenoviral vector was constructed using the pAdTrack-CMV plasmid from Addgene [Addgene plasmid 16405; submitted by He et al. (1998)]. Adenoviruses were produced according to the protocol previously established in our laboratory (Zhou et al., 2013).
Overnight p2 hAEC cultures were washed twice with Dulbecco's phosphate buffered saline (DPBS) or plain DMEM-LG, and 50 MOI (multiplicity of infection) of virus containing either mPdx1 or eGFP-vector, diluted in plain DMEM-LG, was added. After 6 hours under standard culture conditions, an equal volume of complete medium was added to the cultures and cells were cultured thus overnight. The next morning, virus containing media was removed taking appropriate precautions and cells were washed twice with plain DMEM-LG. Experimental cells were then cultured as follows.
Differentiation with activin A
A modified protocol of the differentiation of human ES cells was used (D'Amour et al., 2006). The culture medium was completely aspirated from the overnight cultures of p2 hAEC004, and the wells were washed thrice with 1 × DPBS. Day 1 activin differentiation medium (DMEM-LG containing 1% NEAA, 1 × ITS Liquid Media Supplement, 1 × Penicillin–Streptomycin, and 100 ng/mL activin A) was added to all cultures except the control plates. Control plates were cultured in differentiation medium without activin A. The plates were incubated under standard culture conditions for 24 hours. The next day, the culture medium was completely aspirated, and the wells were washed thrice with 1 × DPBS. Day 2 activin differentiation medium (Day 1 differentiation medium containing 0.2% FBS) was then added to the experimental plates. The cells were cultured thus for a further 72 hours.
Cells that were transduced with mPdx1/eGFP-harboring adenovirus were similarly treated 48 hours after the complete removal of the virus-containing media.
Differentiation with nicotinamide
The culture medium was completely aspirated from the overnight cultures of p2 hAEC004, and the wells were washed thrice with 1 × DPBS. The experimental plates were treated with nicotinamide differentiation medium (DMEM-LG containing 10% FBS, 1% NEAA, 1 × ITS Liquid Media Supplement, 1 × Penicillin–Streptomycin, and 10 mM nicotinamide) (Miki et al., 2005). The corresponding control plates were treated with medium lacking nicotinamide. The plates were incubated under standard culture conditions, and the growth medium was completely changed every 4 days. Culture was carried out for a total of 14 days.
Cells that were transduced with mPdx1/eGFP-harboring adenovirus were similarly treated with differentiation medium 48 hours after the complete removal of the virus-containing media. The virally transduced cells were, however, cultured only for a further 4 days after their first exposure to nicotinamide.
RNA extraction
At the end of the culture period, RNA was isolated from the experimental and control cells using the 5 PRIME Manual PerfectPure RNA Cell Isolation Kit (5 PRIME, Inc.). Since it was found that RNA yield from cells in one well of the six-well plate was low, RNA from three wells was pooled together.
Total RNA content was estimated using a NanoDrop 2000c (Thermo Scientific).
cDNA synthesis
Five hundred nanogram of total RNA was used per 20 μL reaction, to synthesize cDNA using the ABI-RT Kit (Applied Biosystems). Polymerase chain reaction (PCR) conditions were as provided by the manufacturer's protocol, that is, 25°C for 10 minutes, 37°C for 2 hours, and 85°C for 5 minutes.
Primers for quantitative real-time PCR
Based on available RNA-seq data for ES cells differentiating down the pancreatic lineage (Array express ID: E-MTAB-1086), the markers for pancreatic differentiation were categorized as given below for the present study:
Multipotent cell marker: OCT4, NANOG, RFX3; Early/DE markers: SOX17, FOXA2, HNF1B, HNF4A; Primitive gut tube marker: RFX6; Foregut markers: PDX1; Pancreatic endoderm marker: NKX2.2, NKX6.1, NGN3; Pancreatic progenitor markers: NEUROD1, PAX6, PAX4; Endocrine pancreas markers: MAFA, INS, GCG, PPY; Pancreatic duct marker: CFTR; and Housekeeping gene marker: GAPDH.
The sequence of the primers obtained from Integrated DNA technologies (IDT) and purified by standard desalting is given in Table 1. The specificity of the primers for the particular target gene was evaluated in silico using the Primer BLAST tool (Ye et al., 2012). All others were validated hydrolysis probes (Applied Biosystems) (Table 2).
Table 1.
Primers for Genes Whose Expression Was Evaluated by the SYBR Green Method
| Gene (GenBank ID) | F primer sequence (5′–3′) | R primer sequence (5′–3′) | Expected product size |
|---|---|---|---|
| RFX6 (306518575) | TCTCTTTGACCAGCATGTCG | CTGTGCTGCCTGAAATGGTA | 104 bp spanning region within exon 12 |
| CFTR (306514) | CTATGACCCGGATAACAAGGAGG | CAAAAATGGCTGGGTGTAGGA | 107 bp spanning region within exon 4 of all transcript variants |
| RFX3 (Harvard PrimerBank ID 19743882c2) | CCAGGTGACTACCGTGGTCT | GCTGCTGATGAGTTGTCCTCC | 88 bp spanning region within exon 1 |
The table lists the genes whose expression was evaluated by the SYBR® Green method. The corresponding forward (F) and reverse (R) primer sequences and the region it spans in the target gene are also mentioned.
Table 2.
Primers for Genes Whose Expression Was Evaluated by the TaqMan Method
| Serial No. | Gene symbol | Catalog number |
|---|---|---|
| 1 | OCT4 | Hs03005111_g1 |
| 2 | NANOG | Hs02387400_g1 |
| 3 | SOX17 | Hs00751752_s1 |
| 4 | PAX6 | Hs01088112_m1 |
| 5 | FOXA2 | Hs00232764_m1 |
| 6 | PDX1 | Hs00426216_m1 |
| 7 | NGN3 | Hs00360700_g1 |
| 8 | NEUROD1 | Hs00159598_m1 |
| 9 | NKX2.2 | Hs00159616_m1 |
| 10 | NKX6.1 | Hs00232355_m1 |
| 11 | PAX4 | Hs00173014_m1 |
| 12 | MAFA | Hs01651425_s1 |
| 13 | INS | Hs00355773_m1 |
| 14 | GCG | Hs00174967_m1 |
| 15 | PPY | Hs00237001_m1 |
| 16 | GAPDH | Hs99999905_m1 |
The table lists the genes which were evaluated by the TaqMan® method. The corresponding primers were validated hydrolysis probes from Applied Biosystems.
Quantitative real-time PCR
cDNA equivalent to 12.5 ng of initial RNA was used per 20 μL reaction for quantitative real-time PCR (qPCR) analysis. RFX6, RFX3, and CFTR expression were estimated by the SYBR green method (Applied Biosystems). Two hundred fifty nanomolar of each primer (forward or reverse) was used per reaction. In the case of hydrolysis probes, 1 μL of the appropriate 20 × TaqMan® Hydrolysis probe mix was used per reaction. Ten microliter of the 2 × SYBR® Green PCR Master Mix (Applied Biosystems) or 2 × TaqMan Gene Expression Master Mix (Applied Biosystems) was added to the appropriate reaction mixes and made up to 20 μL with nuclease-free water. Reactions were set up in MicroAmp® Optical 96-Well Reaction Plates (Applied Biosystems), and plates were sealed with MicroAmp Optical Adhesive Film (Applied Biosystems).
qPCR was performed on an Applied Biosystems® 7300 Real-time PCR system. Default PCR conditions were used (50°C for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. Dissociation: 95°C for 15 seconds, 60°C for 20 seconds, 95°C for 15 seconds, and 60°C for 15 seconds). All qPCR were carried out in technical duplicates. Relative gene expression was estimated manually using the ddCt method (Real time qRT-PCR expression profiling, ABI PRISM 7700 Sequence Detection System. User Bulletin No. 2). hGAPDH was used as the reference gene for all experiments. Unmanipulated p2 hAEC gene expression was the quantification calibrator (baseline gene expression). All results are represented according to the MIQE guidelines (Bustin et al., 2009).
For the present study, gene expression in terms of quantification cycle (Cq) was classified as high (Cq <25), medium (Cq between 25 and 30), low (Cq between 30 and 35), very low expression (Cq between 35 and 37), and no expression (Cq ≥37).
The relative quantitation (RQ) values were used to plot a heat map using the conditional formatting option in Microsoft Excel. The dCq for a particular gene was calculated as (Cq of sample)−(Cq of p2 hAECs). The fold change in gene expression was calculated as the relative quantitation (RQ; 2^-ddCq) where ddCq was calculated as (dCq for gene X)−(dCq for GAPDH) for the same sample. The RQ value of those genes whose expression was undetectable was given to be 0.
For individual gene expression under each culture condition, the statistical significance was estimated by a two-tailed t-test (assuming equal variances) in Microsoft Excel. The 2^-ddCq values for each replicate were used for the calculations. Alpha was set at 0.05.
Results
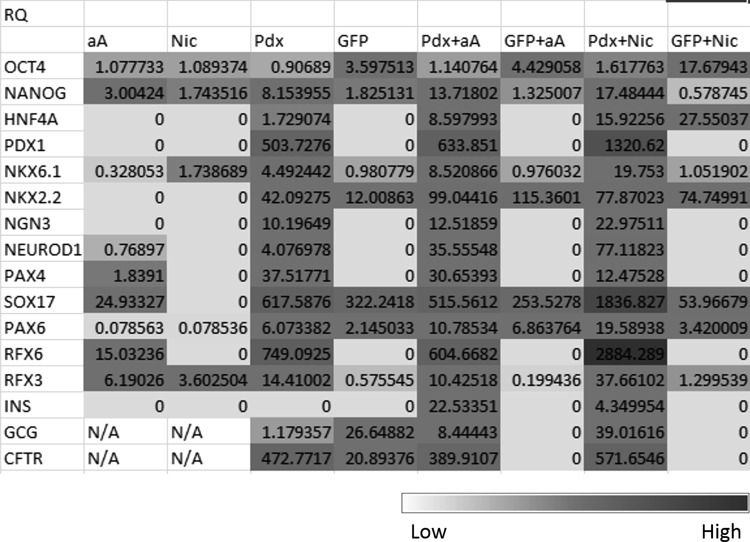
PCR analysis shows that passage 2 (p2) hAECs express stem cell markers, OCT4 (Avg. Cq 28) and NANOG (Avg. Cq 32), alluding to their multipotent progenitor status, as well as RFX3 (Avg. Cq 25), as has been previously described (Balaji et al., 2016). In contrast, hAECs are negative for all stage-specific pancreatic markers, namely PDX1, RFX6, CFTR, PAX4, HNF1B, and NEUROD1 (Avg. Cq >35).
Three sets of culture conditions were set up per exogenous growth factor studied—one set of cultures was cultured in the presence of either activin A or nicotinamide alone, one set of cultures was transduced with the adenovirus expressing murine Pdx1 (mPdx1) and subsequently exposed to activin A/nicotinamide, and a final set of cultures was transduced with adenovirus expressing eGFP and subsequently exposed to activin A/nicotinamide. Pancreatic differentiation was determined by analysis of developmental genes using qPCR.
p2 hAECs were exposed to 100 ng/mL activin A for 24 hours under serum-free conditions, followed by addition of low level of FBS (0.2%) to the medium (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/cell).
Although addition of low dose of FBS or activin A was used to induce DE or mesoderm differentiation of ES cells, respectively (D'Amour et al., 2005; Vallier et al., 2005), our study did not show a significant difference in morphology between cells grown in the presence or absence of activin A (Supplementary Fig. S1A). qPCR revealed that activin A reduces expression of PAX4, SOX17, RFX6, and RFX3 while causing an increased expression of NEUROD1 and PAX6, in comparison to the controls not treated with activin A (Supplementary Table S1). Although there was a significant change compared to p2 hAECs (Fig. 2), there was less than a one cycle difference in expression of most of these genes compared to the aA controls (Supplementary Table S1). Thus we do not consider this a significant change in gene expression.
FIG. 2.
Pancreatic gene expression in hAECs under different conditions. A heat map comparing gene expression under the various culture conditions: aA: Nontransduced cells in the presence of Activin A, Nic: Nontransduced cells in the presence of nicotinamide, Pdx: Pdx1-transduced cells, GFP: eGFP-transduced cells, Pdx+aA: Pdx1-transduced cells in the presence of Activin A, GFP+aA: eGFP-transduced cells in the presence of Activin A, Pdx+Nic: Pdx1-transduced cells in the presence of nicotinamide, GFP+Nic: eGFP-transduced cells in the presence of nicotinamide. Gene expression from low to high is represented as a gradation from white to gray. Gene expression is represented as the RQ values (Please refer methodology for RQ calculations). The RQ value of those genes whose expression was undetectable is represented as 0. RQ values corresponding to 1 indicate baseline gene expression in unmanipulated p2 hAECs. All values greater than 1 indicate expression higher than baseline, whereas values <1 indicate lower than baseline expression. N/A indicates that expression of that gene was not studied for that particular sample. RQ, relative quantitation.
In a second set of experiments, p2 hAECs were grown in the presence of 10 mM nicotinamide in serum-containing media (Supplementary Fig. S1B). Similar to the results obtained with activin A, there was no significant difference in morphology between cells grown in the presence or absence of nicotinamide (Supplementary Fig. S1B). qPCR revealed that nicotinamide alone treatment of p2 hAECs caused a marginal increase in expression of NKX6.1 and RFX3 and reduced expression of stem cell marker OCT4 along with reduced expression of RFX6 from baseline (Fig. 2 and Supplementary Table S2).
We have previously used the genetic modification of AFS cells with forced expression of Pdx1, a key transcription factor in pancreatic cell fate determination, to successfully produce pancreatic progenitors capable of producing insulin (Chun et al., 2015; Zhou et al., 2013). Accordingly, we forced endogenous PDX1 expression by a transduction of the cells with an adenovirus expressing mPdx1. Transduction of cells with Pdx1 was successful with an approximate transduction efficiency of 40% (Balaji et al., 2016). This was sufficient to cause a dramatic change in pancreatic gene expression.
There was a greater than nine cycle difference in endogenous PDX1 expression compared to untransduced (∼500-fold higher expression) and eGFP-transduced cells (Fig. 2). Of the other markers of pancreatic differentiation, the most dramatic expression was seen of the primitive gut tube marker, RFX6. The fold change (2^-ddCq) in expression of this marker was ∼800 times more than the eGFP control. This was followed by a greater than 20-fold increase of the pancreatic progenitor marker PAX4. The existing RFX3 expression also increased by 20 times compared to the eGFP control. Since PDX1 expression was so dramatic, we decided to include CFTR and GCG expression, as markers of the pancreatic duct and endocrine pancreas, to our panel of markers (Fig. 2). A 500-fold increase in the expression of CFTR was observed. The markers of a functional endocrine pancreas continued not to be detected. This is in accordance with results we have previously published (Balaji et al., 2016).
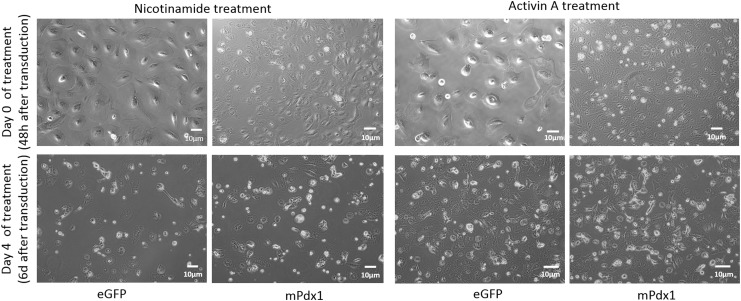
Since Pdx1 transduction resulted in the expression of many stage-specific markers of the developing pancreas, we studied the effects of the combination of Pdx1 transduction with the exogenous growth factors, activin A or nicotinamide. When observed under the microscope 2 days after adenoviral transduction and at the point that activin A was added to the cultures, there seemed to be a large number of floating cells (Day 0 of Fig. 1). Four days after exposure of the transduced cells to activin A, the morphology of the cells did not change significantly (Day 4 of Fig. 1). Since this was also observed in the eGFP controls, this may be indicative of some amount of adenoviral stress or due to the overconfluence of the cultures.
FIG. 1.
Cell phenotype in response to adenoviral transduction and activin A and nicotinamide exposure p2 hAECs that were seeded at a density of 10,000 cells/cm2 were transduced with 50 MOI of adenovirus harboring either a mPdx1- or eGFP-gene vector. The cells were exposed to activin A and nicotinamide 48 hours later, and images were taken before exposure (Day 0) and after 4 days (Day 4). No significant morphological changes were observed between cells in the different conditions. hAECs, human amnion epithelial cells; MOI, multiplicity of infection; Pdx1, pancreatic and duodenal homeobox-1.
Similarly, microscopic observation of cells indicated that at the end of the culture period, the adenovirally transduced cells, which were subsequently exposed to nicotinamide, did not have a significantly different morphology (Day 4 of Fig. 1).
qPCR analysis, however, revealed that the combination of PDX1 and either one of the growth factors caused several genes to be expressed at a higher level than under the separate treatments. Pdx1 transduction followed by exposure to activin A increased the expression of HNF4A, NEUROD1, NKX6.1, NKX2.2, and PAX6 (Fig. 2 and Supplementary Table S1). HNF4A expression was almost twice higher than in eGFP controls where expression of this gene was undetectable.
The increase in HNF4A expression was statistically significant over cells that were just transduced with Pdx1 (p < 0.05). Exposure to activin A subsequent to mPdx1 transduction did not alter RFX6 or PDX1 expression significantly compared to Pdx1 alone cells, although their expression remained undetected in eGFP-transduced control cells. There was more than a twofold increase in Cq and therefore an almost 45-fold increase in expression of NEUROD1, the pancreatic progenitor marker, relative to those cells that were treated with activin A alone (Supplementary Table S1; p < 0.05).
Although INS expression was very low (Cq = 36), exposure to activin A following Pdx1 transduction resulted in detectable gene expression of the same unlike other treatment conditions where the expression of the INS gene was undetectable. Another endocrine cell marker, GCG, also had a 10-fold higher expression compared to the controls. There was less than one cycle increase in expression of NKX6.1, NKX2.2, and PAX6 and less than one cycle decrease in expression of PAX4 and RFX3 compared to the Pdx1 transduced controls (Fig. 2). The loss of RFX3 expression was alone statistically significant (p < 0.05). Combined, the data suggest that treatment of Pdx1-induced cells with high doses of activin A seems to push the hAECs toward the pancreatic progenitor stage.
Exposing the PDX1+ hAECs to nicotinamide increased the expression of most of the markers of the various stages of pancreatic development (Fig. 2). In particular, expression of 8 marker genes, namely RFX3 (p < 0.1), SOX17, RFX6, PDX1, PAX6, NEUROD1, NKX6.1, and GCG, were higher compared with Pdx1-transduced cells treated with activin A. HNF4A, NKX6.1, and NEUROD1 had a greater than 1-cycle difference in expression upon exposure to nicotinamide subsequent to Pdx1 transduction. This was not, however, statistically significant. Only PAX4 expression reduced with the combination of Pdx1 and nicotinamide.
Although INS expression was detected in these cells, the expression was not as high as in cells exposed to activin A. In contrast, GCG was detected significantly upon nicotinamide treatment. In general, mPdx1 transduction followed by treatment with nicotinamide resulted in the higher expression of key pancreatic transcription factors compared with cells exposed to activin A.
Discussion
The present study aimed at exploring an alternative source of cells for diabetes therapy. hAECs were used as the cellular sources for differentiation toward the pancreatic lineage. Since they are easily available and are readily expanded in culture, the continued study of their ability to differentiate into pancreas-committed cells is justified. Endogenous gene expression studies suggested that the hAECs were good candidates for pancreatic differentiation because they were positive for the multipotent cell markers OCT4, NANOG, and RFX3.
Activin A has previously been used to direct ES cells and pluripotent stem cells toward a DE fate (D'Amour et al., 2005, 2006; McLean et al., 2007; Ungrin et al., 2012). The ES cells in these studies were positive for the genetic markers OCT4 and NANOG before exposure and SOX17 and FOXA2 after exposure to activin A.
However, while the hAECs used in the current study were positive for OCT4 and NANOG to begin with, they failed to show any other change in endogenous marker expression upon exposure to activin A, under similar culture conditions. The inhibition of the PI3K signaling pathway is required for activin A-mediated DE differentiation, whereas insulin is an activator of the same signaling pathway (McLean et al., 2007; Yu et al., 2015). Since the culture medium in this study contained both insulin and activin A, it could have led to the nullification of two opposing effects and thus the differentiation process. Since we also failed at previous attempts at the differentiation of AFS cells using β-cell differentiation protocols developed for ES cells (Zhou et al., 2013), it seems that one must exercise caution in trying to use the same differentiation protocol for cells of different types.
In contrast, although previous studies have successfully used nicotinamide alone to differentiate hAECs into functional pancreatic endocrine cells (Hou et al., 2008; Miki et al., 2005), we were unable to reproduce the same results. This could be attributed to an interdonor variation since we found a difference in basal gene expression between hAECs derived from different donors (data not shown). It has also been previously speculated that nicotinamide-induced differentiation does not reproduce a normal pancreatic development pathway in vivo and hence a lack of expression of certain marker genes is to be expected (Hou et al., 2008).
We were able to overcome this lack of differentiation by expressing PDX1 ectopically, as we have previously shown for the successful differentiation of AFS cells and hAECs (Balaji et al., 2016; Chun et al., 2015; Zhou et al., 2013). Although hAECs may have a differentiation bias toward other lineages, since PDX1 is considered a marker for pancreatic lineage commitment (Jennings et al., 2013; Teo et al., 2015), we did not determine this in the current study. Furthermore, while an adenoviral approach may not be well suited for in vivo transplant experiments due to their elevated immune potential (Chuah et al., 2003), this work has shown conclusively that turning on the master regulator of pancreatic development can indeed drive multiprogenitor cells from tissues as diverse as the amnion, down the pancreatic lineage. Future work can be carried out with less immunogenic helper dependent vectors.
The present study showed that the process of differentiation that is turned on by PDX1 expression can be reinforced by exposure to exogenous growth factors such as activin A or nicotinamide. It seems that turning on the master regulator is important to overcome the initial barrier that the chemical methods alone could not cross. However since the transduction efficiency was low when using 50 MOI of adenovirus, it may provide a possible reason as to why the addition of activin A or nicotinamide could not cause an elevated action on these cells since by themselves they were unable to turn on pancreas-associated genes. Since we have previously shown that a higher adenoviral titer is cytotoxic (Balaji et al., 2016), alternate methods of forcing PDX1 expression such as a lentiviral system can be tested.
Of the two growth factors, nicotinamide increased the overall expression of genes turned on by PDX1. While mPdx1 transduction followed by activin A treatment alone was able to turn on the expression of INS, nicotinamide treatment led to the significant expression of GCG.
Another advantage of this method of mPdx1 transduction followed by activin A or nicotinamide treatment is that the gene expression changes seen were achieved in a matter of 6 days compared to currently existing protocols that take a minimum of 2 weeks (D'Amour et al., 2006; McLean et al., 2007; Otonkoski et al., 1993). Perhaps a sequential differentiation protocol involving these two exogenous growth factors along with the use of different cell culture matrices as tested previously (Balaji et al., 2016; Chun et al., 2015) would not only yield a higher number of endocrine pancreas-positive cells, but would also be able to further reduce the differentiation time. It would be interesting to see if these cells when transplanted would be able to maintain normoglycemia in diabetic animal models.
Conclusion
A majority of the currently existing protocols follow an entirely chemical approach to pancreatic differentiation. Furthermore, they only focus on the end point markers of pancreatic function such as insulin or c-peptide production. While this is important from a clinical point of view, it is also important to understand the process of cell differentiation to explore the use of progenitor cells from different sources as an insulin-producing source. Furthermore, new methods can be devised to shorten the in vitro culture period so that the differentiated cells may be available for transplantation sooner. Our attempt to understand these changes at the molecular level may be a step in that direction.
Supplementary Material
Acknowledgments
The authors thank Dr. Sean V. Murphy (WFIRM) for kindly providing the human amnion epithelial cells (Approved by the Institutional Review Board, Wake Forest School of Medicine, Winston-Salem, NC, File No. IRB00002852); Drs. Christopher Newgard and Sarah Ferber, Duke University, for kindly providing the mPdx1-expressing adenovirus; the Vogelstein lab, Johns Hopkins Oncology Center, for their submission of the pAdTrack-CMV plasmid to the Addgene repository; and Dr. Anasuya Ganguly (BITS-Pilani KK Birla Goa campus) for a critical review of the article. Funding in support of this work was kindly provided by the National Institutes of Health (Grant No. R01DK080897), and The Vila Rosenfeld Diabetes Research Fund gift to E.C.O. S.B. was supported by a Fulbright-Nehru Doctoral and Professional Research grant for the duration of this work (Grant No. 1669/DPR/2012-2013).
Authors' Contributions
S.B. and Y.Z. designed and carried out the experiments, data analysis, and drafted the article. E.C.O. was involved in drafting the article and revising it critically. S.S. participated in the design and coordination of the study and helped to draft the article. All authors read and approved the final article.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Balaji S., Zhou Y., Ganguly A., Opara E.C., and Soker S. (2016). The combined effect of PDX1, epidermal growth factor and poly-L-ornithine on human amnion epithelial cells' differentiation. BMC Dev. Biol. 16, 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas M., and Showalter S.A. (2013). Thermodynamic and structural determinants of differential Pdx1 binding to elements from the insulin and IAPP promoters. J. Mol. Biol. 425, 3360–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie G.M., Lopez A.D., Bucay N., Hinton A., Firpo M.T., King C.C., and Hayek A. (2005). Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23, 489–495 [DOI] [PubMed] [Google Scholar]
- Bonfanti P., Nobecourt E., Oshima M., Albagli-Curiel O., Laurysens V., Stangé G., Sojoodi M., Heremans Y., Heimberg H., and Scharfmann R. (2015). Ex vivo expansion and differentiation of human and mouse fetal pancreatic progenitors are modulated by epidermal growth factor. Stem Cells Dev. 24, 1766–1778 [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 [DOI] [PubMed] [Google Scholar]
- Chuah M.K., Collen D., and VandenDriessche T. (2003) Biosafety of adenoviral vectors. Curr. Gene Ther. 3, 527–543 [DOI] [PubMed] [Google Scholar]
- Chun S.Y., Mack D.L., Moorefield E., Oh S.H., Kwon T.G., Pettenati M.J., Yoo J.J., Coppi P.D., Atala A., and Soker S. (2015). Pdx1 and controlled culture conditions induced differentiation of human amniotic fluid-derived stem cells to insulin-producing clusters: Pdx1 and controlled culture-induced differentiation of hAFSCs. J. Tissue Eng. Regen. Med. 9, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., and Baetge E.E. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541 [DOI] [PubMed] [Google Scholar]
- D'Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., and Baetge E.E. (2006). Production of pancreatic hormone—Expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 24, 1392–1401 [DOI] [PubMed] [Google Scholar]
- Guariguata L. (2011). Estimating the worldwide burden of type 1 diabetes. Diabetes Voice 56, 6–8 [Google Scholar]
- He T.-C., Zhou S., Da Costa L.T., Yu J., Kinzler K.W., and Vogelstein B. (1998). A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Huang Q., Liu T., and Guo L. (2008). Human amnion epithelial cells can be induced to differentiate into functional insulin-producing cells. Acta Biochim. Biophys. Sin. 40, 830–839 [PubMed] [Google Scholar]
- Jennings R.E., Berry A.A., Kirkwood-Wilson R., Roberts N.A., Hearn T., Salisbury R.J., Blaylock J., Hanley K.P., and Hanley N.A. (2013). Development of the human pancreas from foregut to endocrine commitment. Diabetes 62, 3514–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S., Muthyala S., Nair P., and Bhonde R. (2010). Human placenta-derived mesenchymal stem cells and islet-like cell clusters generated from these cells as a novel source for stem cell therapy in diabetes. Rev. Diabet. Stud. 7, 168–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiyama H., Hamazaki T.S., Tokuhara M., Masui S., Okabayashi K., Ohnuma K., Yabe S., Yasuda K., Ishiura S., Okochi H., et al. (2010). Pdx1-transfected adipose tissue-derived stem cells differentiate into insulin-producing cells in vivo and reduce hyperglycemia in diabetic mice. Int. J. Dev. Biol. 54, 699–705 [DOI] [PubMed] [Google Scholar]
- Kelly O.G., Chan M.Y., Martinson L.A., Kadoya K., Ostertag T.M., Ross K.G., Richardson M., Carpenter M.K., D'Amour K.A., Kroon E., et al. (2011). Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat. Biotechnol. 29, 750–756 [DOI] [PubMed] [Google Scholar]
- Khoo C., Yang J., Weinrott S.A., Kaestner K.H., Naji A., Schug J., and Stoffers D.A. (2012). Research resource: The Pdx1 cistrome of pancreatic islets. Mol. Endocrinol. 26, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauda-Havakuk M., Litichever N., Chernichovski E., Nakar O., Winkler E., Mazkereth R., Orenstein A., Bar-Meir E., Ravassard P., Meivar-Levy I., et al. (2011). Ectopic PDX-1 expression directly reprograms human keratinocytes along pancreatic insulin-producing cells fate. PLoS One 6, e26298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean A.B., D'Amour K.A., Jones K.L., Krishnamoorthy M., Kulik M.J., Reynolds D.M., Sheppard A.M., Liu H., Xu Y., Baetge E.E., et al. (2007). Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells 25, 29–38 [DOI] [PubMed] [Google Scholar]
- Miki T., Lehmann T., Cai H., Stolz D.B., and Strom S.C. (2005). Stem cell characteristics of amniotic epithelial cells. Stem Cells 23, 1549–1559 [DOI] [PubMed] [Google Scholar]
- Ohlsson H., Karlsson K., and Edlund T. (1993). IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12, 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otonkoski T., Beattie G.M., Mally M.I., Ricordi C., and Hayek A. (1993). Nicotinamide is a potent inducer of endocrine differentiation in cultured human fetal pancreatic cells. J. Clin. Invest. 92, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E.A., Paty B.W., Senior P.A., Bigam D., Alfadhli E., Kneteman N.M., Lakey J.R.T., and Shapiro A.M.J. (2005). Five-year follow-up after clinical islet transplantation. Diabetes 54, 2060–2069 [DOI] [PubMed] [Google Scholar]
- Shapiro A.M.J., Lakey J.R.T., Ryan E.A., Korbutt G.S., Toth E., Warnock G.L., Kneteman N.M., and Rajotte R.V. (2000). Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 343, 230–238 [DOI] [PubMed] [Google Scholar]
- Shapiro A.M.J., Ricordi C., Hering B.J., Auchincloss H., Lindblad R., Robertson R.P., Secchi A., Brendel M.D., Berney T., Brennan D.C., et al. (2006). International trial of the edmonton protocol for islet transplantation. N. Engl. J. Med. 355, 1318–1330 [DOI] [PubMed] [Google Scholar]
- Tang D.-Q., Cao L.-Z., Chou W., Shun L., Farag C., Atkinson M.A., Li S.-W., Chang L.-J., and Yang L.-J. (2006). Role of Pax4 in Pdx1-VP16-mediated liver-to-endocrine pancreas transdifferentiation. Lab. Investig. J. Tech. Methods Pathol. 86, 829–841 [DOI] [PubMed] [Google Scholar]
- Teo A.K.K., Tsuneyoshi N., Hoon S., Tan E.K., Stanton L.W., Wright C.V.E., and Dunn N.R. (2015). PDX1 binds and represses hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Rep. 4, 578–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungrin MD, Clarke G, Yin T, Niebrugge S, Nostro MC, Sarangi F, Wood G, Keller G, and Zandstra PW. (2012). Rational bioprocess design for human pluripotent stem cell expansion and endoderm differentiation based on cellular dynamics. Biotechnol. Bioeng. 109, 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Alexander M., and Pedersen R.A. (2005). Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., and Madden T.L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.S.L., Ramasamy T.S., Murphy N., Holt M.K., Czapiewski R., Wei S.-K., and Cui W. (2015). PI3K/mTORC2 regulates TGF-β/Activin signalling by modulating Smad2/3 activity via linker phosphorylation. Nat. Commun. 6, 7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Mack D.L., Williams J.K., Mirmalek-Sani S.-H., Moorefield E., Chun S.-Y., Wang J., Lorenzetti D., Furth M., Atala A., et al. (2013). Genetic modification of primate amniotic fluid-derived stem cells produces pancreatic progenitor cells in vitro. Cells Tissues Organs 197, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.