Abstract
Aims: Nonalcoholic fatty liver (NAFL) is a common liver disease associated with metabolic syndrome, obesity, and diabetes that is rising in prevalence worldwide. Various molecular perturbations of key regulators and enzymes in hepatic lipid metabolism cause NAFL. However, redox regulation through glutathione (GSH) adducts in NAFL remains largely elusive. Glutaredoxin-1 (Glrx) is a small thioltransferase that removes protein GSH adducts without having direct antioxidant properties. The liver contains abundant Glrx but its metabolic function is unknown.
Results: Here we report that normal diet-fed Glrx-deficient mice (Glrx−/−) spontaneously develop obesity, hyperlipidemia, and hepatic steatosis by 8 months of age. Adenoviral Glrx repletion in the liver of Glrx−/− mice corrected lipid metabolism. Glrx−/− mice exhibited decreased sirtuin-1 (SirT1) activity that leads to hyperacetylation and activation of SREBP-1 and upregulation of key hepatic enzymes involved in lipid synthesis. We found that GSH adducts inhibited SirT1 activity in Glrx−/− mice. Hepatic expression of nonoxidizable cysteine mutant SirT1 corrected hepatic lipids in Glrx−/− mice. Wild-type mice fed high-fat diet develop metabolic syndrome, diabetes, and NAFL within several months. Glrx deficiency accelerated high-fat-induced NAFL and progression to steatohepatitis, manifested by hepatic damage and inflammation.
Innovation: These data suggest an essential role of hepatic Glrx in regulating SirT1, which controls protein glutathione adducts in the pathogenesis of hepatic steatosis.
Conclusion: We provide a novel redox-dependent mechanism for regulation of hepatic lipid metabolism, and propose that upregulation of hepatic Glrx may be a beneficial strategy for NAFL. Antioxid. Redox Signal. 27, 313–327.
Keywords: : glutathione, glutaredoxin, lipids, sirtuin
Introduction
Nonalcoholic fatty liver (NAFL) is the most common form of chronic liver disease affecting an increasing population worldwide. It represents a spectrum of liver pathology ranging from steatosis to inflammatory nonalcoholic steatohepatitis (NASH) with or without fibrosis (13). Clinically, NAFL is strongly associated with metabolic syndrome, obesity, type-2 diabetes, and dyslipidemia (2, 52, 53, 69).
Innovation.
Nonalcoholic fatty liver (NAFL) is a common liver disease associated with oxidative stress. However, the effects of oxidative post-translational modifications including protein glutathione (GSH) adducts on hepatic lipid metabolism are unknown. Ablation of glutaredoxin-1 (Glrx) increased protein GSH adducts, hepatic lipid synthesis, and steatosis in mouse liver. Sirtuin-1, an important metabolic regulator orchestrating hepatic lipid metabolism, was inactivated by GSH adducts and promoted fatty acid synthase expression. Overexpression of Glrx or a nonoxidizable Cys-mutant SirT1 in vivo normalized hepatic lipid synthesis. These data suggest that Glrx deficiency and oxidative inactivation of SirT1 play an important role in the pathogenesis of NAFL.
Hepatic lipid accumulation (hepatic steatosis) is the initial step in the pathogenesis of NAFL, arising from an imbalance of anabolic and catabolic processes in lipid metabolism (21), including lipid uptake, de novo lipogenesis, excretion, and oxidation. These processes are under tight transcriptional control through well-characterized networks of transcription factors such as the sterol regulatory element-binding proteins (SREBPs) regulating de novo fatty acid and cholesterol biosynthesis, and peroxisome proliferator-activated receptor gamma coactivator (PGC) 1α controlling β-oxidation (25, 60, 63, 76).
An emerging paradigm in disease processes is signaling pathways modulated by reactive oxygen and nitrogen species. In the presence of oxidants, reactive cysteines of proteins form reversible modifications that regulate enzyme activity, localization, protein interactions, and stability (17, 32, 33). Owing to abundant intracellular glutathione (GSH), protein GSH adducts are a key modification (referred to as protein S-glutathionylation [Prot-SG]) that is reversed by the enzyme glutaredoxin-1 (Glrx). Although Glrx has reactive thiols, deficient mice exhibited no aggravated oxidative damage upon angiotensin II infusion, ischemia-reperfusion, or hyperoxia (7, 34). Recent studies have demonstrated that GSH adducts, controlled by Glrx, participate in various processes, including cellular growth, apoptosis, cytoskeletal regulation, angiogenesis, and inflammation (1, 3, 4, 51, 68, 74). Glrx is an abundant liver protein that affects numerous proteins, but in the context of hepatic metabolism, only a few are identified and functionally studied.
Sirtuin-1 (SirT1), an NAD+-dependent class III histone deacetylase, modulates key transcription factors orchestrating hepatic lipid metabolism (30, 57, 60). Activation of SirT1 improved NAFL and conversely hepatic SirT1 deficiency led to steatosis (56). Inhibition of SirT1 activity by reversible GSH adducts has recently been described by our group and was confirmed by other investigators (11, 67, 71, 77). Because NAFL (58) is associated with oxidative stress, increased protein GSH adducts may play an important role in the pathogenesis of steatotic livers.
We report here that Glrx knockout mice (Glrx−/−) fed normal diet (ND) develop spontaneous fatty liver and hyperlipidemia, suggesting mechanistic importance of Glrx in the development of NAFL. Furthermore, we demonstrate that inactivation of SirT1 by GSH adducts may be a major contributor to steatosis induced by Glrx deficiency.
Results
Glrx−/− mice fed normal diet develop metabolic disorders
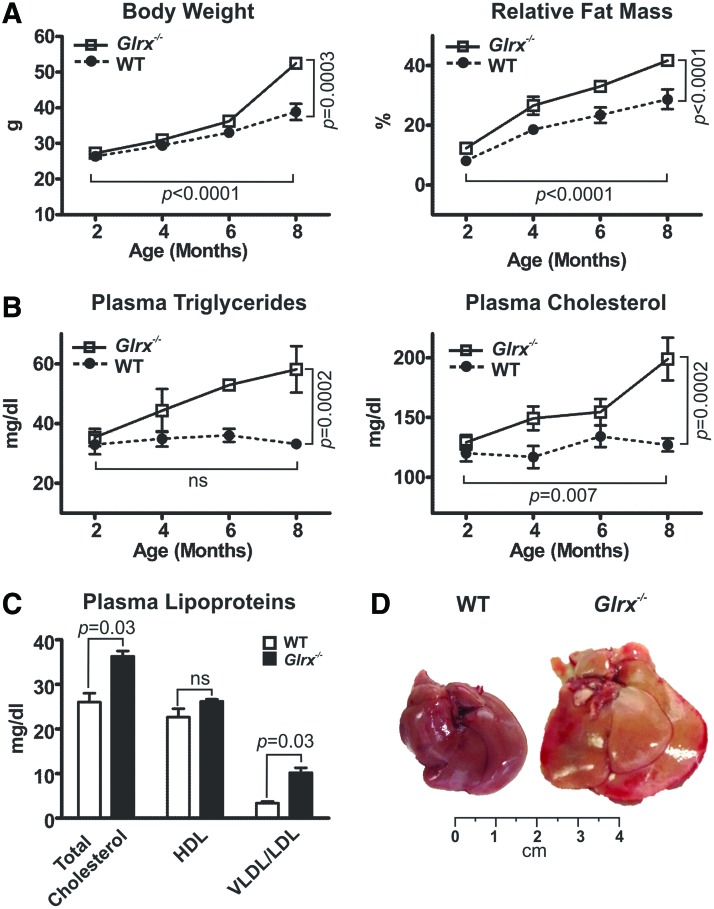
Glrx−/− mice fed ND became obese by 8 months of age compared with age-matched wild type (WT) mice. Body weight (BW) and relative fat mass ratio increased significantly by about 20% in Glrx−/− mice (Fig. 1A and Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/ars). Furthermore, Glrx−/− mice developed hyperlipidemia measured by total plasma triglycerides and cholesterol levels (Fig. 1B). Food intake was comparable between WT and Glrx−/− mice and thus is unlikely to cause increase in fat mass (Supplementary Table S1). Importantly, the lipoprotein profile of Glrx−/− mice at 8 months showed elevated levels of low- and very low-density lipoprotein (LDL/VLDL) cholesterol and unchanged high-density lipoprotein (HDL) (Fig. 1C), suggesting an altered hepatic lipid metabolism. Consistent with an increase in plasma lipoproteins, Glrx−/− mice exhibited significantly enlarged fatty liver at 8 months of age (Fig. 1D and Supplementary Table S1).
FIG. 1.
Glrx−/− mice fed normal diet develop metabolic disorders. (A) Changes in body weight (left) and percentage of fat mass (right) in WT and Glrx−/− mice fed ND. The body fat mass was measured with noninvasive quantitative magnetic resonance (means ± SEM, N = 8–10). (B) Plasma triglycerides (left) and cholesterol (right) concentrations of WT and Glrx−/− mice fed ND (means ± SEM, N = 8–10). Two-way ANOVA for age and genotype was used to determine statistical significance. (C) Cholesterol content of plasma lipoproteins HDL and VLDL/LDL at 8 months of age (means ± SEM, N = 8–10). The nonparametric Mann–Whitney U test was used to determine statistical significance. (D) Representative pictures of livers from ND-fed WT and Glrx−/− mice at 8 months of age. ANOVA, analysis of variance; HDL, high-density lipoprotein; LDL, low-density lipoprotein; ND, normal diet; WT, wild type. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Plasma glucose and insulin levels at 8 months of age were similar between WT and Glrx−/− mice under fasting or fed conditions (Supplementary Table S1). Glrx−/− mice, however, did exhibit mild but significant glucose intolerance and insulin resistance measured by glucose and insulin tolerance tests (Supplementary Fig. S1), consistent with a prediabetic phenotype.
Glrx−/− mice fed ND develop NAFL disease
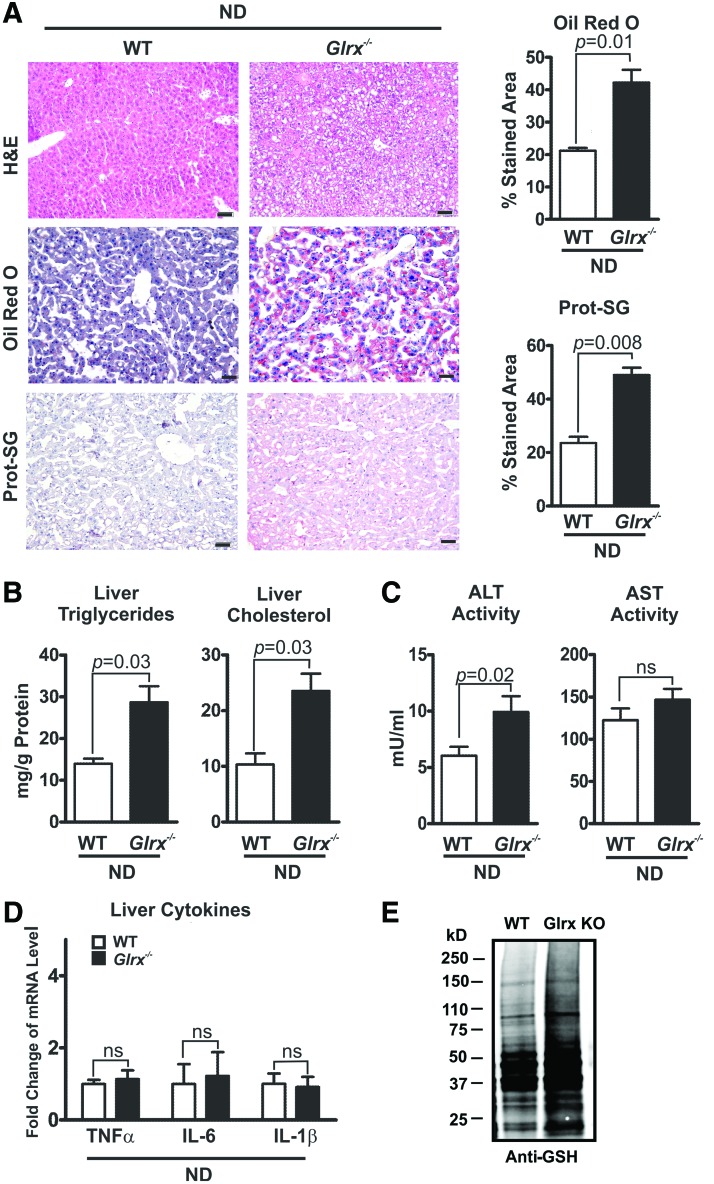
The hepatic lipid content of Glrx−/− mice at 8 months of age was significantly increased compared with WT littermate controls, as measured by Oil Red O and hematoxylin and eosin (H&E) staining of liver sections (Fig. 2A upper and middle rows) and quantification of extracted liver triglycerides and cholesterol (Fig. 2B). These data indicate a typical pathology of liver steatosis. Besides a mildly increased plasma alanine aminotransferase (ALT) activity, Glrx−/− mice showed no other signs of liver damage or inflammation, including changes in plasma aspartate aminotransferase (AST) activity and inflammatory cytokines (Fig. 2C, D and Supplementary Table S1). Liver proteins of Glrx−/− mice had significantly more GSH adducts (Fig. 2A lower rows, E and Supplementary Fig. S14) and reversible oxidation (Supplementary Fig. S2). Hepatic oxidized glutathione (GSSG) was below the detection limit by HPLC, and GSH levels in livers of Glrx−/− mice were comparable with those of WT (Supplementary Fig. S3). Taken together, these data suggest that increased reversible oxidative modifications of liver proteins because of the lack of Glrx may promote hepatic lipid accumulation and contribute to the pathogenesis of NAFL.
FIG. 2.
Glrx−/− mice fed normal diet develop nonalcoholic fatty liver. (A) Representative histological sections obtained from livers of WT (left column) and Glrx−/− mice at 8 months of age (right column) were stained with H&E (upper row), Oil Red O for lipids (middle row), and an antibody against protein GSH adducts (Prot-SG) (lower row). The Oil Red O-stained liver lipids and Prot-SG were quantified with the color deconvolution plugin in ImageJ (N = 5/group). Scale bars denote 100 μm. (B) Levels of liver triglycerides (left) and cholesterol (right) of WT and Glrx−/− mice (means ± SEM, N = 8–10). (C) Plasma levels of AST and ALT in WT and Glrx−/− mice (means ± SEM, N = 8–10). (D) Levels of the proinflammatory cytokines TNFα, IL-6, and IL-1β measured by RT-qPCR in livers of WT and Glrx−/− mice. (E) Representative Western blot of protein GSH adducts of WT and Glrx−/− mouse livers. GAPDH served as the lysate input control for immunoprecipitation (means ± SEM, N = 8–10) (Supplementary Fig. S2). The nonparametric Mann–Whitney U test was used to determine statistical significance. The original Western blot is provided in Supplementary Figure S14. ALT, alanine aminotransferase; AST, aspartate aminotransferase; GSH, glutathione; H&E, hematoxylin and eosin; IL, interleukin; RT-qPCR, quantitative reverse transcriptase–polymerase chain reaction; TNF, tumor necrosis factor. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Hepatic Glrx regulates lipid metabolism and controls plasma lipid levels
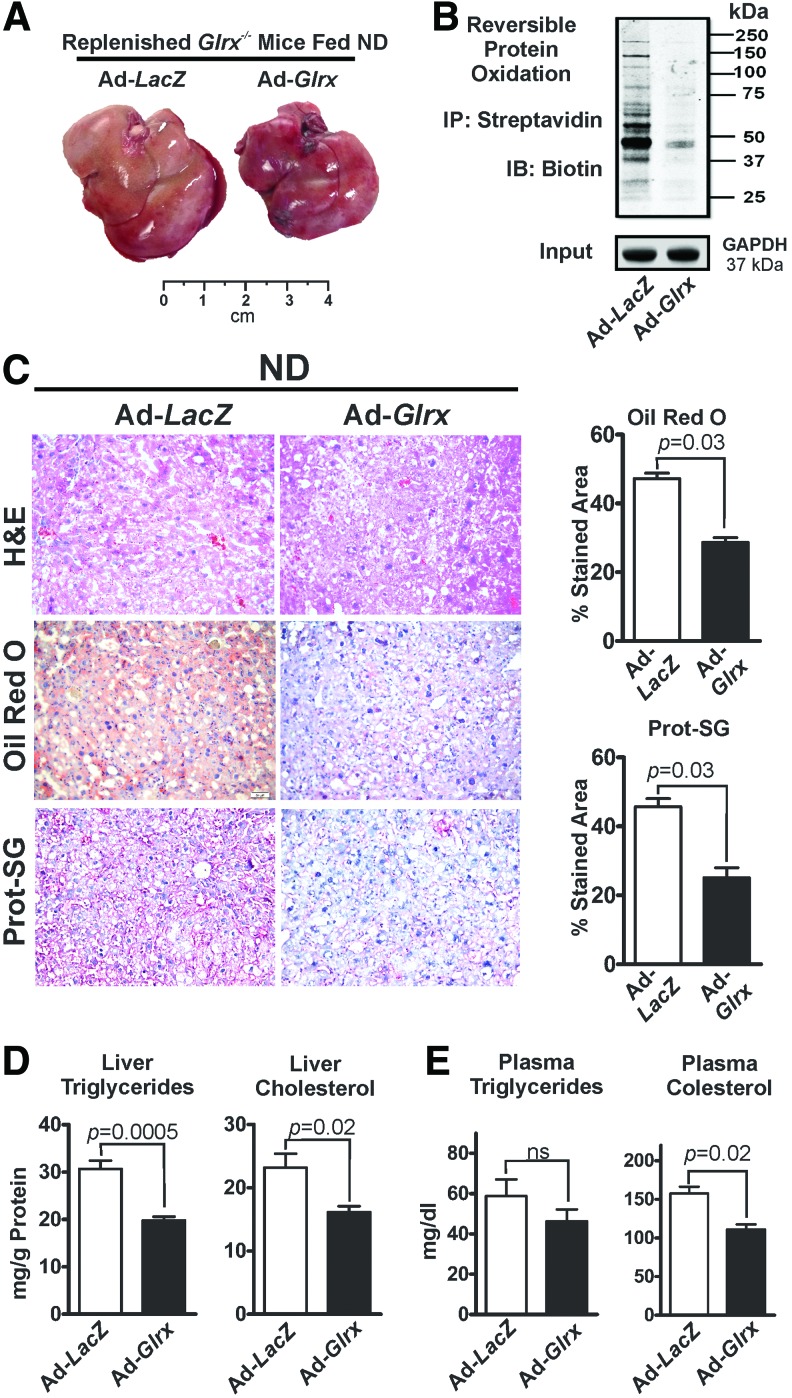
To evaluate the ability of hepatic Glrx to maintain lipid homeostasis, adenovirus-mediated gene repletion (8, 43) of Glrx or LacZ (control) was employed in Glrx−/− mice with hepatic steatosis at 8 months of age. Ten days postadenovirus injection, Western blot analysis confirmed repletion of the liver with Glrx (Supplementary Fig. S4). Glrx expression in other tissues was unaffected by the adenovirus (Supplementary Fig. S4).
Glrx-replenished Glrx−/− mice had significantly decreased levels of GSH adducts (Fig. 3C lower rows) and reversibly oxidized proteins (Fig. 3B and Supplementary Fig. S14), consistent with reacquired Glrx function. Strikingly, Glrx repletion for 10 days markedly diminished liver mass (Fig. 3A and Supplementary Fig. S5) and alleviated steatosis (Fig. 3C upper and middle panels, D) compared with LacZ-transduced mice. BW, however, remained unchanged (Supplementary Fig. S5). In addition, decreased plasma cholesterol level reflected the improved liver function in Glrx-replenished Glrx−/− mice (Fig. 3E). In summary, these data strongly support a critical role for hepatic Glrx in controlling liver and plasma lipids.
FIG. 3.
Glrx repletion in Glrx−/− mice normalizes hepatic lipid metabolism. (A) Representative pictures of livers. (B) Representative biotin-switch assay of reversible cysteine oxidation in liver proteins of Glrx−/− and Glrx-replenished Glrx−/− mice fed ND. GAPDH served as the lysate input control. (C) Liver sections stained with H&E (upper row), Oil Red O for lipids (middle row), and an antibody against protein GSH adducts (Prot-SG; lower row) of ND-fed Glrx−/− and Glrx-replenished Glrx−/− mice, 10 days postadenovirus injection. Scale bars denote 100 μm. The Oil Red O-stained liver lipids and protein GSH adducts were quantified by the color deconvolution plugin in ImageJ (N = 4–5/group). (D) Levels of liver triglycerides (left) and cholesterol (right) and (E) plasma triglycerides (left) and cholesterol (right) levels of Glrx−/− and Glrx-replenished Glrx−/− mice fed ND, 10 days postadenovirus injection (means ± SEM, N = 4–5). The nonparametric Mann–Whitney U test was used to determine statistical significance. The original Western blot is provided in Supplementary Figure S14. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Glrx deficiency increases lipogenesis and cholesterol synthesis
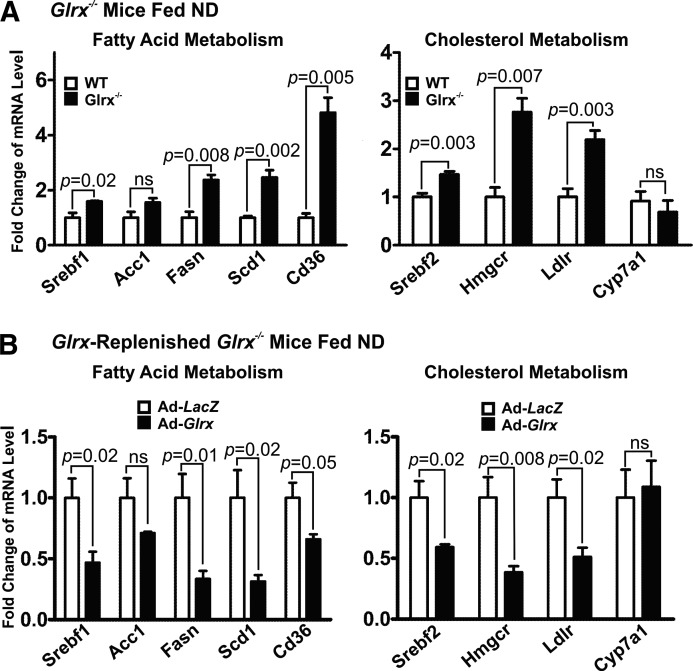
Hepatic lipid metabolism is regulated by a delicate balance of anabolic and catabolic processes (9, 14, 21, 39, 40, 52, 70). Hence, we analyzed the expression of hepatic genes involved in lipid synthesis, uptake, degradation, and transport in WT and Glrx−/− mice fed ND at 8 months of age. Glrx−/− mice expressed significantly higher levels of fatty acid metabolism genes, including sterol regulatory element-binding transcription factor 1 (Srebf1), fatty acid synthase (Fasn), stearoyl-CoA desaturase (Scd1), and fatty acid translocase/CD36 (Cd36) (Fig. 4A). Although expression of acetyl-CoA carboxylase (ACC)—the rate-limiting enzyme in fatty acid synthesis producing the precursor malonyl CoA—was unchanged, Glrx−/− mice had lower levels of active dephosphorylated ACC. Active phosphorylated AMPK, which is upstream of ACC and controls its phosphorylation, was downregulated in Glrx−/− mice consistent with increased fatty acid synthesis (Supplementary Fig. S6).
FIG. 4.
Liver lipid metabolism genes are upregulated in Glrx−/− mice and normalized by Glrx repletion. Gene expression was measured 10 days postadenovirus injection by RT-qPCR analysis in livers of (A) WT and Glrx−/− mice, and (B) adenoviral Glrx-replenished Glrx−/− mice fed ND. Adenoviruses coding for LacZ as control or human Glrx for repletion were used. Genes involved in liver fatty acid metabolism include sterol regulatory element-binding transcription factor 1 (Srebf1), acetyl-CoA carboxylase (Acc1), fatty acid synthase (Fasn), acyl-CoA desaturase (Scd1), and the fatty acid transporter Cd36. Genes participating in liver cholesterol metabolism include the sterol regulatory element-binding transcription factor 2 (Srebf2), HMG-CoA reductase (Hmgcr), the LDL receptor (Ldlr), and cytochrome P-450 7A1 (Cyp7a1). Relative mRNA expression was standardized by β-actin and normalized to the control group (means ± SEM, N = 4–8). The nonparametric Mann–Whitney U test was used to determine statistical significance.
Glrx−/− mice also exhibited higher expression levels of genes related to cholesterol metabolism, including HMG-CoA reductase (Hmgcr), LDL-receptor (Ldlr), and sterol regulatory element-binding transcription factor 2 (Srebf2) (Fig. 4A). The Cytochrome P-450 7A1 (Cyp7a1), which lowers cholesterol by catalysis of the rate-limiting step in bile acid biosynthesis, remained unchanged in Glrx−/− mice (Fig. 4A).
However, Glrx−/− mice exhibited no expression changes of genes involved in hepatic fatty acid oxidation, including the mitochondrial fatty acid transporter carnitine plamitoyl-transferase (Cpt) 1a, peroxisome proliferator-activated receptor alpha (Ppara), acyl-coenzyme A dehydrogenase for mitochondrial β-oxidation (Acadm), and cytochrome P450 (Cyp) 4a10 for peroxisomal β-oxidation (Supplementary Fig. S7). Thus, hepatic lipid accumulation in Glrx−/− mice is unlikely increased to result from decreased lipid oxidation, but rather from de novo biosynthesis.
Expression of Glrx2—the mitochondrial Glrx isoform—and members of the thioredoxin system including thioredoxin (Trx) 1, 2 and thioredoxin-interacting protein (Txnip) (20) also remained unaltered in Glrx−/− mice (Supplementary Fig. S8) and likely did not contribute to the metabolic phenotype.
Consistent with our hypothesis that Glrx directly influences lipid metabolism, Glrx repletion consistently decreased the expression of all genes that were induced in the Glrx−/− mice livers (Fig. 4B), indicating a pivotal regulatory role of liver Glrx in lipid homeostasis.
Glrx deficiency induces hepatic steatosis through inhibition of SirT1 by reversible cysteine oxidation
SirT1, an NAD+-dependent class III histone deacetylase, has emerged as a central regulator of hepatic lipid metabolism (46, 60, 63, 67, 75), and we have recently described it can be modified by GSH adducts (67).
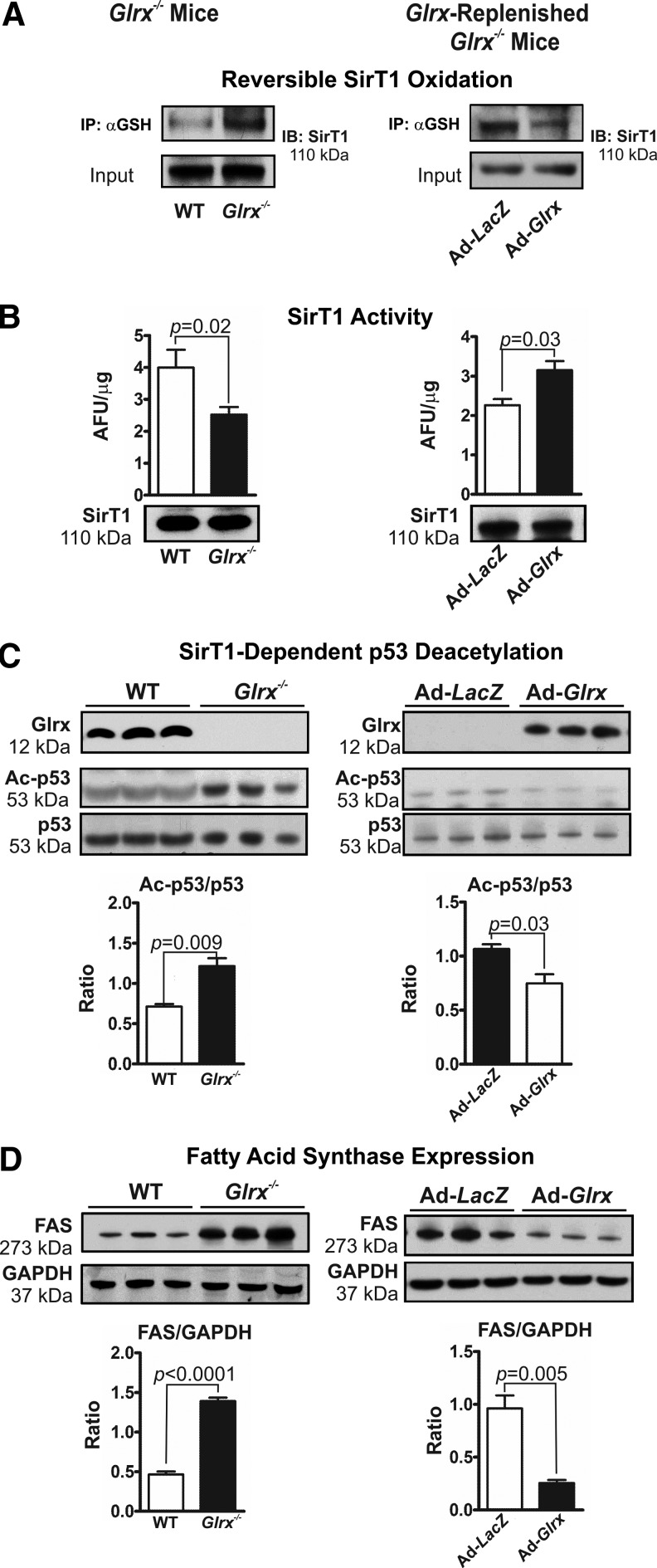
Glrx deficiency in mice markedly increased GSH adducts on hepatic SirT1 (Fig. 5A left panel and Supplementary Figs. 9A and S15), impairing the enzymes deacetylase activity (Fig. 5B left panel and Supplementary Fig. S16). Acetylation of p53 at lysine-379, a deacetylase substrate of SirT1 (30, 42, 48, 60, 72), was increased in livers of Glrx−/− mice, indicating the inhibition of SirT1 activity (Fig. 5C left panel and Supplementary Fig. S16). Consistent with this finding, Glrx deficiency in mice also increased acetylation of lysine-289 and 309 of SREBP1C, another SirT1 substrate that regulates transcription of fatty acid metabolism genes (60) (Supplementary Fig. S10A). Expression level of fatty acid synthase (FAS), which is a key enzyme in de novo lipogenesis and that is downregulated by active SirT1 via SREBP1C (60), was induced in livers of Glrx−/− mice (Fig. 5D left panel and Supplementary Fig. S17). Conversely, Glrx-replenished Glrx−/− mice showed decreased SirT1 GSH adducts (Fig. 5A right panel and Supplementary Figs. S9B and S15), increased SirT1 deacetylase activity (Fig. 5B right panel), decreased acetylated p53 (Fig. 5C right panel and Supplementary Fig. S16) and SREBP1C (Supplementary Fig. S10B), and diminished expression levels of FAS (Fig. 5D right panel and Supplementary Fig. S17). Of importance, neither Glrx gene deletion nor repletion altered hepatic SirT1 protein expression (Fig. 5B and Supplementary Fig. S15). Collectively, these data suggest that SirT1 is an important redox target of Glrx, and may mediate the effect of Glrx on lipid metabolism.
FIG. 5.
SirT1 is inhibited by reversible cysteine oxidation in Glrx−/− mice. Livers of WT and Glrx−/− mice (left column) and Glrx-replenished Glrx−/− mice (8 months of age, right column) were used for the experiments as follows. (A) Reversible cysteine oxidation of endogenous SirT1 detected by the biotin-switch assay in liver proteins. (B) SirT1 activity measured with the Fluor-de-Lys assay in hepatic nuclear extracts. Equal SirT1 protein levels were present in nuclear extracts as measured by immunoblotting. (C) Western blot analysis of Glrx, total p53, and acetylated p53 (Ac-p53) as a marker of biological SirT1 deacetylase activity. The semiquantitative ratios of acetylated to total p53 were determined by densitometry with ImageJ. (D) Western blot analysis of fatty acid synthase expression (FAS). The semiquantitative ratios of FAS to GAPDH expression were determined by densitometry with ImageJ (means ± SEM, N = 3–5). The nonparametric Mann–Whitney U test was used to determine statistical significance. Original Western blots are provided in Supplementary Figure S15 through S17.
Glrx deficiency accumulates lipids through increased reversible oxidation of SirT1 cysteines
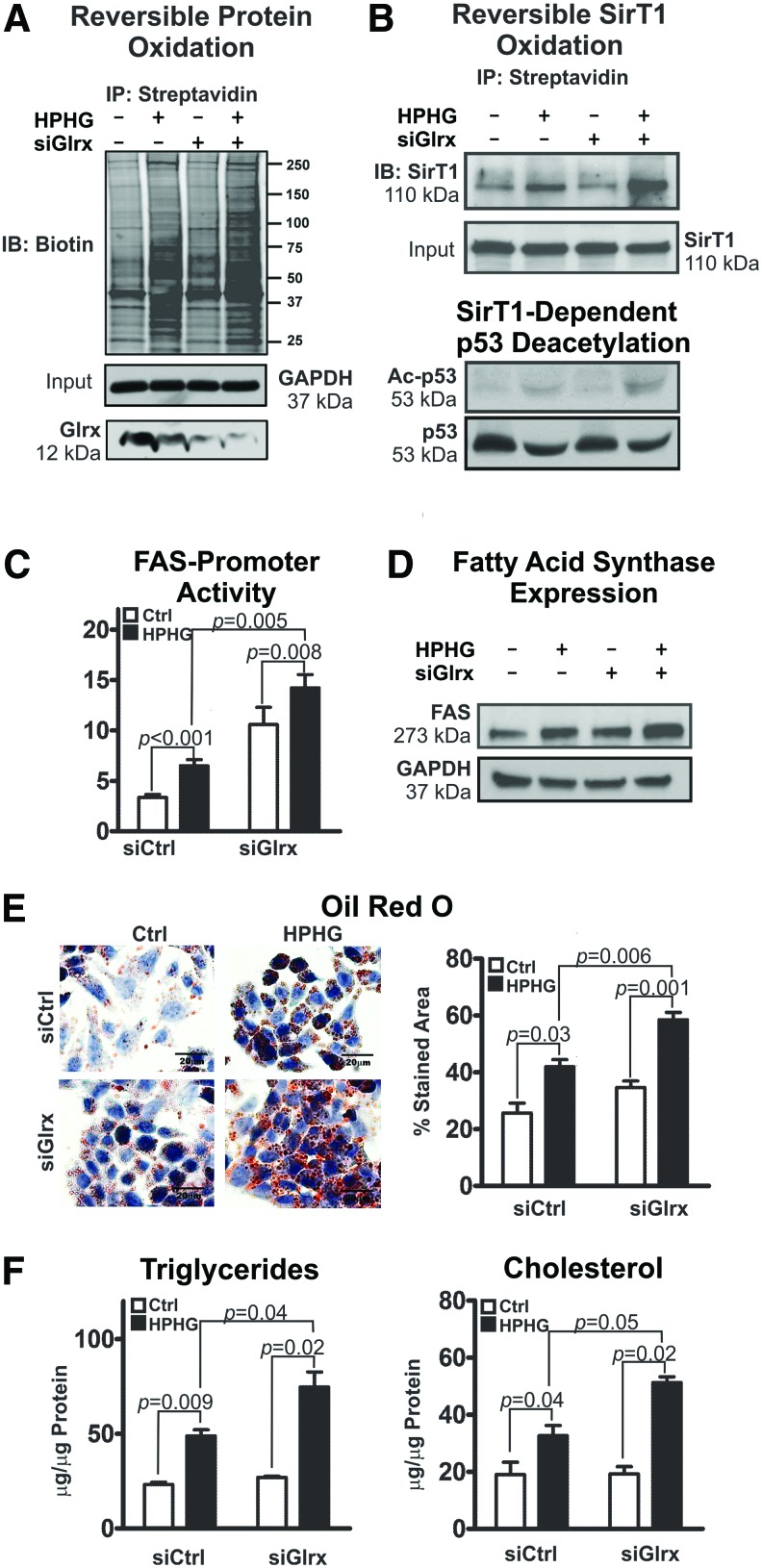
Employing HepG2 cells, a well-established human hepatocellular in vitro model, we further investigated molecular mechanisms by which Glrx regulates hepatic lipid metabolism. High-palmitate and high-glucose (HPHG) treatment increases intracellular oxidants (67) and subsequent protein cysteine oxidation (Fig. 6A and Supplementary Figs. S18 and S19). siRNA-mediated Glrx ablation exacerbated this effect. Consistent with the high-fat diet (HFD)-fed mouse model, Glrx depletion and HPHG treatment further increased reversible cysteine oxidation of SirT1 and acetylated-p53 (Fig. 6B and Supplementary Figs. S18 and S19), indicative of SirT1 inhibition. Transcriptional regulation of FAS, which is a major regulator of lipogenesis and is suppressed by active SirT1 (60), was measured with the FAS-promoter luciferase reporter assay. FAS-promoter activity increased in response to HPHG treatment and Glrx ablation (Fig. 6C left panel), leading to higher FAS protein expression (Fig. 6C right panel and Supplementary Figs. S18 and S19) and accumulation of lipids—triglycerides and cholesterol—in HepG2 cells (Fig. 6E).
FIG. 6.
Glrx regulates lipid homeostasis in HepG2 cells. HepG2 cells were cotransfected with either scrambled (siCtrl) or Glrx siRNA (siGlrx) for 48 h followed by 16 h of incubation in either standard culture medium or medium supplemented with high-palmitate high-glucose (HPHG). (A) Representative biotin-switch assay of reversible cysteine oxidation in cellular proteins. GAPDH served as the lysate input control. (B) Reversible cysteine oxidation of endogenous SirT1 detected by the biotin-switch assay and normalized for total SirT1 expression. Western blot analysis of total p53 and acetylated p53 (Ac-p53) was used as a marker for biological SirT1 deacetylase activity. Numbers under the Western blot indicate the fold change of the corresponding protein bands compared with control. (C) Regulation of FAS expression was measured by transfecting HepG2 cell with a fatty acid synthase (FAS) promoter-luciferase reporter (left). Western blot analysis of FAS expression in HepG2 cells (right). (D) Representative images of Oil Red O-stained HepG2 cells to visualize intracellular lipid accumulation as droplets (left). Scale bars denote 20 μm. Intracellular lipids stained with Oil Red O were quantified with ImageJ using the color deconvolution plugin (n = 5/group) (right). (E) Levels of intracellular triglycerides and cholesterol in HepG2 cells. (F) HepG2 cells were transiently transfected for 12 h with siGlrx and 48 h with WT FLAG-SirT1 (SirT1), mutant FLAG-SirT1 (Mut SirT1), or empty pcDNA3.1 vector followed by 16 h HPHG treatment (means ± SEM, N = 3–5). The nonparametric Mann–Whitney U test was used to determine statistical significance. Original Western blots are provided in Supplementary Figures S18 and S19. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In previous work, we have created and characterized a nonoxidizable mutant SirT1 (Mut SirT1) (C61S+C318S+C613S), in which we replaced three essential cysteine residues by serine. Under oxidative and metabolic stress, mutant SirT1 maintains full activity and exhibits no reversible oxidative modifications (67). Overexpression of the mutant SirT1, as compared with WT, markedly attenuated lipid accumulation—triglycerides and cholesterol—in HepG2 cells under HPHG treatment and Glrx ablation (Fig. 6F). Adenoviral gene transfer of WT (Ad-SirT1), mutant SirT1 (Ad-Mut SirT1), or control (Ad-LacZ) into Glrx−/− mice at around 10 months of age (Supplementary Fig. S11D) was performed to compare the effects on lipid accumulation in vivo. Liver lipids (Supplementary Fig. S11A, B) and plasma cholesterol (Supplementary Fig. S11C) were significantly decreased in mutant SirT1-injected mice. Lipids in WT SirT1-injected mice also improved, but to a lesser degree, consistent with partial oxidative inhibition of WT SirT1. These data together indicate that Glrx through SirT1 also regulate liver lipid metabolism in vivo.
Glrx deficiency accelerates diet-induced NAFL
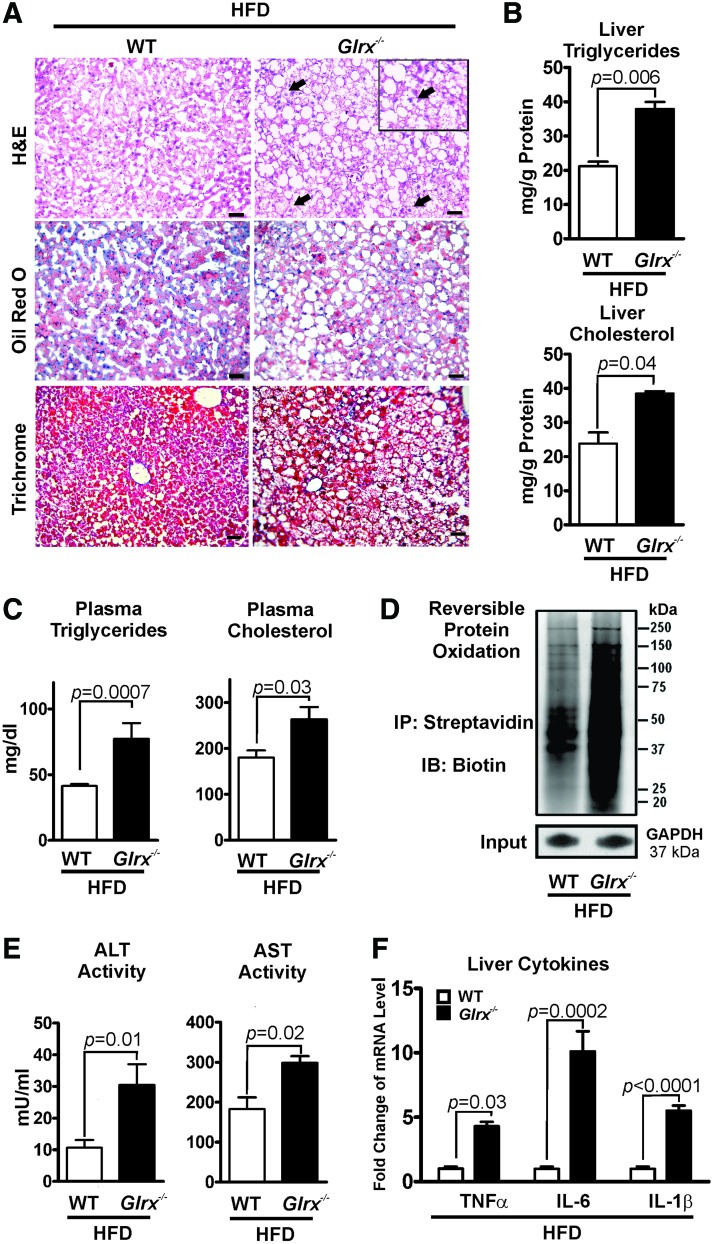
Our previous study (67) showed that HFD induced metabolic syndrome in mice and increased reversible cysteine oxidation of proteins in the liver. To determine whether elevated oxidative cysteine modifications caused by HFD can accelerate the pathogenesis of NAFL in Glrx−/− mice, 2 months old WT and Glrx−/− mice were fed HFD for 12 weeks (Supplementary Table S2). As expected, hepatic and plasma lipids (Fig. 7A–C) and reversible cysteine oxidation of hepatic proteins (Fig. 7D and Supplementary Figs. S18 and S19) were significantly increased in HFD-fed Glrx−/− mice.
FIG. 7.
Glrx−/− mice fed a HFD show nonalcoholic steatohepatitis, which results from accelerated steatosis. (A) Representative sections from livers of WT (left column) and Glrx−/− mice (right column) fed a HFD for 3 months. Liver sections were stained with H&E (upper row), Oil Red O for lipids (middle row), and Masson's trichrome (lower row) for collagen. Ballooned hepatocytes are recognized as swollen hepatocytes with rarefied cytoplasm (black arrows). Scale bars denote 100 μm. (B) Levels of hepatic triglycerides and cholesterol and (C) plasma triglycerides and cholesterol in WT and Glrx−/− mice fed HFD. (D) Representative biotin-switch assay of reversible cysteine oxidation in liver proteins of WT and Glrx−/− mice fed HFD. GAPDH served as the lysate input control for immunoprecipitation. (E) Plasma levels of AST and ALT and (F) gene expression levels of the proinflammatory cytokines TNFα, IL-6, and IL-1β in livers of WT and Glrx−/− mice fed HFD. Data are presented as means ± SEM of N = 8–10. The nonparametric Mann–Whitney U test was used to determine statistical significance. The original Western blot (WB for GAPDH lower right corner) is provided in Supplementary Figures S18 and S19. HFD, high-fat diet. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In contrast to the Glrx−/− mice fed ND (Fig. 2), the plasma levels of ALT and AST (Fig. 7E and Supplementary Table S2) as well as inflammatory cytokines were markedly induced (Fig. 7F). In addition, hepatocellular ballooning was observed (Fig. 7A right upper row), demonstrating an accelerated NAFL to NASH progression in HFD-fed Glrx−/− mice. To further investigate the effect of Glrx deficiency on fibrosis, liver sections were stained with Masson's trichrome, a marker of collagen deposition (Fig. 7A left lower row). Very mild hepatic fibrosis was detected with no significant difference between WT and Glrx−/− mice.
To investigate whether GSH adducts and Glrx level are related to NAFL, GSH adducts and Glrx expression levels were measured in liver biopsy sections. Patients diagnosed with hepatic steatosis (Supplementary Fig. S12) showed diminished Glrx protein expression and increased protein GSH adducts, although we need more samples to conclude significance in human liver.
Collectively, these data suggest that decreased Glrx level and accumulation of GSH adducts of hepatic proteins may contribute to the pathogenesis of NAFL, providing a rationale to increase Glrx expression in treating NAFL.
Discussion
Our results define a novel role of Glrx and protein GSH adducts in regulating hepatic lipid homeostasis.
Molecular mechanisms of NAFL
Various molecular mechanisms causing NAFL have been previously described (5). We investigated the pathogenesis-causing steatosis in Glrx−/− mice. Hepatic lipid content, including cholesterol and fatty acids, is controlled by a delicate balance between lipid uptake, synthesis, degradation, and excretion (18, 38, 41). In many cases, hepatic lipogenesis and cholesterol synthesis greatly contribute to liver steatosis (61). However, diminished mitochondrial lipid uptake by inhibition of the long chain fatty acid transporter Cpt1a (6) or attenuated hydrolysis of tryglycerides by adipose triglyceride lipase also causes hepatic steatosis (28, 29).
Perturbations in hepatic lipid metabolism, as demonstrated for Glrx−/− mice, can severely affect plasma lipids and is a risk factor for atherosclerosis and cardiovascular disease (2, 53, 69).
Glrx−/− mice increased and liver-specific Glrx gene repletion-corrected mRNA expression levels of all three rate-limiting enzymes: fatty acid synthase, acyl-CoA desaturase (monounsaturated fatty acids), and hydroxy-methylglutaryl-CoA reductase (cholesterol) (Fig. 4A, B). The transcription factors, SREBP 1c for fatty acids and SREBP 2 for cholesterol, respectively, regulate the expression of these enzymes. Both transcription factors are associated with NAFL (12, 18, 23, 50) and were upregulated in Glrx−/− mice. Importantly, SirT1 also regulates SREBPs (56, 63, 75). Activation or overexpression of SirT1 can alleviate diet-induced NAFL (35, 46, 56) through downregulation of SREBP1 (56). Conversely, hepatocyte-specific deletion of SirT1 upregulated SREBPs, hydroxy-methylglutaryl-CoA reductase, fatty acid synthase, acyl-CoA desaturase, and induced weight gain and hepatic steatosis in mice (46, 73).
We have previously shown that metabolic or nitrosative stress increases SirT1 reversible oxidative modification, which inhibits its enzyme activity and promotes hepatocyte apoptosis (67). Consistent with these findings, here we demonstrated that Glrx deficiency decreases SirT1 activity by GSH adducts and consequently increases acetylation of Srebp1 and expression of lipid synthesis genes, including Fasn and Scd1 in the liver (Fig. 4A). Importantly, Glrx gene repletion restored liver SirT1 activity, decreased Srebp1 acetylation, suppressed the downstream genes, and ameliorated the fatty liver phenotype (Figs. 3 and 4B). Meanwhile, metabolic pathways for degrading hepatic lipids including fatty acid β-oxidation (Supplementary Fig. S7) were unaltered at mRNA levels in Glrx−/− mice.
Because plasma cholesterol was elevated in Glrx−/− mice, we measured hepatic-free cholesterol (12, 49, 62) and expression of enzymes involved in the classic (cytoplasmic) or “alternative” (acidic) mitochondrial pathway of bile acid formation (27, 55). Expression of microsomal cholesterol 7α-hydroxylase (CYP7A1) (27), the key enzyme of classic bile acid formation, was unchanged (Fig. 4). The alternative bile acid synthesis pathway in hepatocytes utilizes the mitochondrial transporter “steroidogenic acute regulatory protein” to transport cholesterol into mitochondria, which is then metabolized by cytochrome p450 sterol 27-hydroxylase (CYP27). Expression of both enzymes was unaltered. Thus, lipid uptake as shown by increased CD36 and LDL receptor expression and de novo cholesterol synthesis are likely to play a major role in Glrx−/− mice.
The glutaredoxin system may affect the thioredoxin system as demonstrated in Escherichia coli. Ablation of Txnip or Trx reductase-1 (TrxR1) in mice caused altered lipid metabolism (20, 36). However, gene expression of the thioredoxin system was unaltered in Glrx−/− mice. Furthermore, the mitochondrial isoform glutaredoxin-2, which is exclusively mitochondrial matrix localized (54) and controls iron–sulfur clusters (24) as well as mitochondrial protein GSH adducts (19), showed no changes in gene expression.
Upregulation of SirT1 deacetylase activity could mediate the beneficial metabolic effects of caloric restriction through rising cosubstrate NAD+ levels (47). Conversely, decreased SirT1 activity may not necessarily reflect changes of cellular NAD+ concentration (22, 26). We also found that hepatic NAD+ concentrations of Glrx−/− mice were similar to those of WT mice (Supplementary Fig. S13).
The two hit hypothesis
NAFL encompasses a spectrum of liver pathology that includes hepatic steatosis, inflammatory NASH, and fibrosis. The progression of NAFL is delineated by the “two-hit hypothesis” (5, 15, 16). The “first hit” causes hepatic lipid accumulation and leaves stressed hepatocytes susceptible to injury. The “second hit” advances hepatic steatosis to NASH by induction of inflammation and oxidative stress, causing hepatocyte damage and death (10, 56, 59, 66). NASH can occur with or without fibrosis and then progress to end-stage liver disease, cirrhosis, and hepatocellular carcinoma (13). Damaged hepatocytes release specific enzymes such as ALT and AST into the blood. Therefore, NASH diagnostics require measurement of plasma biomarkers, at times in combination with histological assessment of liver tissue.
In this study, we found that Glrx deficiency increases hepatic lipid content without causing damage to hepatocytes that otherwise would have resulted in inflammation and release of liver enzymes into the blood. Thus, Glrx deficiency results in a “first hit” causing hepatic steatosis and plasma dyslipidemia by SirT1-dependent upregulation of de novo fatty acid and cholesterol synthesis. We tested whether feeding a HFD for 3 months to Glrx−/− mice would aggravate NAFL as a “second hit.” HFD-fed Glrx−/− mice exhibited signs of tissue damage, resulting in elevated plasma levels of ALT and AST, as well as hepatitis associated with increased inflammatory cytokines and hepatocellular ballooning (Fig. 7). Increased reversible oxidative modifications, mainly GSH adducts, coincided with HFD feeding and Glrx deficiency further augmented them. Thus, these experiments suggest that HFD in Glrx−/− mice aggravates NAFL to NASH by causing a greater increase in GSH adducts induced by metabolic stress compared with HFD-fed WT mice (Fig. 7), which may further impair SirT1 function.
Furthermore, increased mitochondrial cholesterol can promote inflammation by mitochondrial GSH depletion and sensitize tumor necrosis factor and FAS signaling (44). Glrx−/− mice fed HFD, in particular, may have increased mitochondrial cholesterol levels and thus show increased inflammation and progression to NASH.
Role of Glrx in liver
Our studies used global Glrx−/− mice, therefore, Glrx ablation in other tissues was a concern. Using an adenovirus transgene coding for Glrx, we selectively replenished the liver of Glrx−/− mice to test whether hepatic Glrx deficiency directly caused steatosis. Surprisingly, replenished Glrx expression nearly normalized liver weight, hepatic lipids, and plasma lipid content after only 10 days. Thus, hepatic Glrx deficiency, and no other systemic metabolic abnormality, causes steatosis. Furthermore, liver biopsies from patients diagnosed with NAFL showed first evidence of decreased Glrx expression and increased protein GSH adducts. This finding is supported by a previous study of Piemonte et al. that observed increased protein GSH adducts in children with NAFL using the same monoclonal antibody (58). Glrx in other tissues such as adipose tissue, however, may also have effects on metabolism and requires further investigation.
Conversely, overexpression of Glrx protected the heart from diabetic complications (44) and preserved function of high-glucose exposed cells (45). Glrx also improves insulin secretion of beta cells (65). Thus, overexpression or activation of liver Glrx could be a strategy to normalize hepatic and plasma lipid metabolism.
In conclusion, our findings indicate that reversible thiol modification in the liver is a major mechanism of lipid metabolism and the development of NAFL. Hepatic Glrx controls lipid homeostasis by regulating protein GSH adducts and specifically those on SirT1, which regulate its activity and downstream lipid regulators. Our results assign a novel role for Glrx and Glrx-mediated regulation of reversible thiol modifications in lipid homeostasis and protection from hepatic steatosis. This provides a new clue into the molecular mechanisms underlying NAFL and opens the possibility for new modes of therapy.
Materials and Methods
Reagents, materials, and antibodies
N-(biotinoyl)-N′-iodoacetyl ethylenediamine (BIAM, B-1591), Zeba™ spin desalting columns (40K MWCO, 87767), Lipofectamine™, and cell culture media were obtained from Life Technologies (Grand Island, NY). Anti-SirT1 mouse monoclonal antibody (ab110304) was from Abcam (Cambridge, MA) 1:5000 dilution for Western blot. Antiacetylated p53 (K382) rabbit polyclonal (#2525), antiacetylated p53 (K379) rabbit polyclonal (#2570), and antibiotin HRP-linked goat antibody (#7075) were from Cell Signaling (Danvers, MA), 1:1000 dilution for Western blot. Antitotal p53 (sc-126) mouse monoclonal, anti-SREBP-1C (k-10 [rabbit polyclonal], 2A4 [mouse monoclonal], and H160 [rabbit polyclonal]) antibodies were from Santa Cruz (Dallas, TX), 1:1000 dilution for Western blot. Anti-Glrx rabbit polyclonal antibody was custom ordered by Bethyl Laboratories (Montgomery, TX), 1:1000 dilution for Western blot. Anti-GSH mouse monoclonal antibody (101-A-100) was from Virogen, 1:200 dilution for immunostaining. For details on antibodies and working dilutions please refer to Supplementary Table S3. The luciferase assay kit was obtained from Promega (Madison, WI). Fluor-de-Lys™ SirT1 activity assay was from Enzo Life Sciences (Farmingdale, NY). Polyvinylidene fluoride membrane, polyacrylamide electrophoresis gels, and other reagents for immunoblotting were obtained from Bio-Rad (Hercules, CA). Western blots were corrected for brightness and contrast. The “Precision Plus Protein Standards—All Blue” were used as molecular mass maker for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Cat #161-0373; Bio-Rad, Hercules, CA). Western blots were either developed using ECL or the Odyseey infrared scanner (LI-Cor, NE) equipped with two IR channels—700 and 800 nm—as previously published (67). The 700 nm channels visualized the molecular mass marker that was superimposed over the 800 nm channel. Both channels are provided as supplemental information.
Experimental animals
Glrx−/− mice were originally generated by Dr. Y.S. Ho (Wayne State University, Detroit, MI) (18), and backcrossed to C57BL/6NJ background in Dr. Janssen-Heininger's laboratory (University of Vermont). Male mice were used for all experiments. The mouse colony has been maintained in the animal facility at Boston University Medical Campus. For metabolic characterization, Glrx−/− mice and WT littermates were fed ND (4.5% fat, 0.02% cholesterol by weight).
To investigate the effects of metabolic stress, a cohort of 2 months old Glrx−/− mice and WT littermates were fed a HFD (21% fat representing 42% calories, 34% sucrose, and 0.2% cholesterol, TD.88137; Harlan, South Easton, MA) for 3 months. Mice were housed in rooms with 12 h light–dark cycle and in groups of 3–4 whenever possible. The protocol was approved by the Institutional Animal Care and Use Committee at Boston University School of Medicine.
Metabolic phenotyping
The mouse body composition including fat mass, lean tissue mass, free water, and total body water was assessed with noninvasive quantitative magnetic resonance in an EchoMRI700 instrument. Values are expressed as a percentage of BW. All studies were performed at the Boston University Metabolic Phenotyping Core.
Homogenization and protein extraction from liver tissue—homogenization and extraction of individual liver pieces were carried out in NP-40 lysis buffer containing 50 mM Tris, 150 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail (Roche Applied Science) at pH 7.4.
Cell culture and treatments
HepG2 cells (ATCC, Manassas, VA) were maintained in DMEM containing 10% FBS and penicillin/streptomycin (Gibco, Grand Island, NY). Transfected cells were treated with control medium containing 5 mM glucose and 0.67% bovine serum albumin (BSA, fatty acid free; Sigma-Aldrich St. Louis, MO) or medium in HPHG (25 mM glucose, 0.4 mM palmitic acid, and 0.67% BSA) for 16 h.
Glrx knockdown in HepG2 cells was achieved using on-target plus siRNA (Dharmacon, Lafayette, CO).
ShRNA lentivirial vector against human SirT1 (RHS4533-EG23411, Dharmacon, Lafayette, CO) was packed into lentiviral particles following manufacturers protocol. In brief, 293T cells were transfected with pLKO-shSirT1 or scrambled control pLKO-pGL2 together with the packaging plasmids encoding Δ8.9 and VSV-G. Supernatants containing lentiviral shRNA against SirT1 were collected 48 h post-transfection. HepG2 cells were incubated with collected medium containing lentiviral particles coding for shSirT1. A stable SirT1 knockdown HepG2 cell line was generated by selection with puromycin (2 μg/ml).
Fasn-promoter luciferase reporter
The luciferase reporter vector containing the promoter region of the human Fasn gene was obtained from Addgene (#8890) (Cambridge, MA). Luciferase activity was measured 24–48 h post-transfection in HepG2 cells according to the manufacturer's protocol using a TECAN Infinite M1000 Pro Microplate Reader (TECAN, San Jose, CA).
SirT1 activity measurement
SirT1 activity was tested by Fluor-de-Lys assay. Then 90 μl of 30 μg of nuclear extraction from mouse liver was incubated with 100 μM acetylated p53 peptide (Arg-His-Lys-Lys[Ac]-AMC) for 30 min at 37°C with 100 μM NAD+ in activity assay buffer (50 mM Tris-HCl, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, pH 8.0). Then 100 μl of 1 mg/ml concentrated trypsin solution was added to release the AMC fluorophore, which allows quantification of the amount of substrate deacetylated by SirT1. The fluorescence intensity was recorded over 60 min using a Fluoroscan Ascent microplate reader (Thermo Fisher, Cambridge, MA) with excitation set to 375 nm and emission to 460 nm.
Biotin-switch assay for labeling of reversibly oxidized cysteines
Labeling with N-(biotinoyl)-N′-iodoacetyl ethylenediamine was used in a biotin-switch assay to detect reversibly oxidized cysteines. Cells were lysed in lysis buffer containing 100 mM maleimide. Excess maleimide was removed by passing the lysates over Zeba spin columns. Lysates were incubated with 5 mM DTT for 1 h and reduced cysteines were labeled with 1 mM N-(biotinoyl)-N′-iodoacetyl ethylenediamine for 1 h. Streptavidin beads were added into the lysates and beads were boiled in 30 μl of 2 × reducing Laemmli buffer and loaded on an SDS Tris-glycine gel. SirT1 was detected by immunoblotting with a total SirT1 antibody (Santa Cruz). Reversible cysteine modifications were detected by antibiotin antibody (Cell signaling).
Liver histology and analysis
For H&E staining, liver tissue was fixed in 4% phosphate-buffered formalin, embedded in paraffin, and cut into 5 μm sections. For Oil Red O staining, livers were embedded in optimal cutting temperature compound, cut into 5 μm cryosections, and stained with Oil Red O. Slides were mounted with aqueous mountant. For Masson's trichrome staining, 5 μm liver sections were stained to assess the hepatic collagen deposition (fibrosis). For immunostaining of Glrx, liver tissue was fixed in 4% phosphate-buffered formalin, embedded in paraffin, and cut into 5 μm sections. GSH adducts staining method of liver sections was also performed as previously described (31, 67).
Tissue and plasma biochemical measurements
Three hundred microliters of liver homogenate was extracted with 5 ml of chloroform–methanol (2:1) and 0.5 ml of 0.1% sulfuric acid (55). An aliquot of the organic phase was collected, dried under nitrogen, and resuspended in 2% Triton X-100. Hepatic triglycerides and cholesterol and plasma triglycerides were measured using the infinity triglycerides and total cholesterol reagent kit (TR13421, TR-22421) (Thermo Fisher). Hepatic lipid contents were normalized for differences in protein concentration. Plasma HDL, LDL/VLDL cholesterol was measured using HDL and LDL/VLDL cholesterol assay kit (ab65390) (Abcam). Plasma alanine (ALT) and AST were detected using ALT and AST activity assay kits (K752, K753) (BioVision, San Francisco, CA). NAD NAD+/NADH ratio in tissues was measured using an assay kit (ab65348) (Abcam) according to the manufacturer's instructions. Tissue GSH and GSSG levels were measured using a modified HPLC-based method as established by Reed et al. (37, 64).
Quantitative reverse transcriptase–polymerase chain reaction
Total RNA was isolated from tissues or cells using TRIzol™ reagent and cDNA generated utilizing High Capacity RNA-to-cDNA kit. Quantitative PCR was conducted using inventory gene-specific TaqMan™ primers (Life Technologies): Fasn (Mm00662319_m1), Acc1 (Mm01304257_m1), Scd1 (Mm00772290_m1), Srebf1 (Mm00550338_m1), Cd36 (Mm01135198_m1), Hmgcr (Mm01282499_m1), Srebf2 (Mm01306292_m1), Ldlr (Mm01177349_m1), Cyp7a1 (Mm00484150_m1), Cpt1a (Mm01231183_m1), Acadm (Mm01323360_g1), Ppara (Mm00440939_m1), Cyp4a10 (Mm01188913_g1), Tnfa (Mm00443258_m1), Il1b (Mm00434228_m1), Il6 (Mm00446190_m1), Glrx (Mm00728386_m1), Trx-1 (Mm00726847_s1), Trx-2 (Mm00444931_m1), Txnip (Mm01265659_g1), Glrx-2 (Mm00469836_m1), and Actb (Mm00607939_s1) (Supplementary Table S4). Expression was obtained and analyzed using comparative Ct (ΔΔCT) with StepOne™ quantitative real-time PCR software (Applied Biosystems, Grand Island, NY), normalized to β-actin.
Liver biopsies
We conducted a pilot investigation aimed at determining reversible oxidative protein modifications in liver biopsies of patients with NAFL disease. The study population included two groups: normal liver histology and nonalcoholic hepatic steatosis, all obtained through the Boston University Biospecimen Archive Research Core (BARC). Each of the two groups consisted of three individual patient samples. A single pathologist with specialized training in liver histology reviewed all samples and confirmed the diagnoses in previously specified groups. The Boston University School of Medicine Institutional Review Board (IRB) reviewed the study protocol as “IRB exempt.” All patient studies were conducted in compliance with the principles of the “Declaration of Helsinki.”
Statistical analysis
Statistical analysis was performed using Prism 6.0 (GraphPad Software). Means were compared between two groups by the Mann–Whitney U test. Mann–Whitney U test with Dunn's post-test, paired-test was used in small number animal experiments. Multiple comparisons were conducted with ANOVA. A p value of <0.05 was considered statistically significant.
Supplementary Material
Abbreviations Used
- ACC
acetyl-CoA carboxylase
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BSA
bovine serum albumin
- Cd36
fatty acid translocase/CD36
- Cyp
cytochrome P-450
- Cys
cysteine
- FAS
fatty acid synthase
- Glrx
glutaredoxin-1
- GSH
glutathione
- GSSG
oxidized glutathione
- HDL
high-density lipoprotein
- HPHG
high-palmitate high-glucose supplemented cell culture medium
- HFD
high-fat diet
- Hmgcr
3-hydroxy-3-methyl-glutaryl-CoA reductase
- H&E
hematoxylin and eosin
- LacZ
beta-galactosidase
- LDL
low-density lipoprotein
- IL
interleukin
- NAD
nicotinamide adenine dinucleotide
- NAFL
nonalcoholic fatty liver
- NASH
nonalcoholic steatohepatitis
- ND
normal diet
- p53
tumor suppressor p53
- RT-qPCR
quantitative reverse transcriptase polymerase chain reaction
- Scd1
stearoyl-CoA desaturase
- SirT1
Sirtuin-1
- TNF
tumor necrosis factor
Acknowledgments
This work was supported by NIH grants P01 HL068758, R37 HL104017, R01 DK076942, and R01 DK103750, R01 HL133013, R01 HL115955, NIH CTSI award 1UL1TR001430, NHLBI, National Institutes of Health, Department of Health and Human Services, under contract Nos. HHSN268201000031C and N01-HV-00239, American Heart Association “Grant in Aid” 16GRNT27660006, European Cooperation in Science and Technology (COST Action BM1203/EU-ROS), and the Metabolic Clinical Research Collaborative. The article contents are solely the responsibility of the authors and do not necessarily represent the official views of the awarding offices. D.S. was supported by an American Heart Association Scientist Postdoctoral Fellowship Award 15POST21790006. J.H. was supported by an NRSA grant T32 HL70024 through the Whitaker Cardiovascular Institute postdoctoral training grant program, 1UL1TR001430 (BU CTSI), and an American Heart Association Scientist Development Grant 14SDG20140036. This work was supported by a Strategic Alliance with Institut de Recherche Servier. M.M.B. was supported by the Evans Junior Faculty Research Award by the Department of Medicine of Boston University. We thank Drs. M. Zang, M. Kirber, T. Balon, and L. Deng and the Boston University School of Medicine “Analytical Instrumentation,” “Immunohistochemistry,” “Cellular Imaging,” and “Metabolic Phenotyping” Cores for their technical support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aesif SW, Kuipers I, van der Velden J, Tully JE, Guala AS, Anathy V, Sheely JI, Reynaert NL, Wouters EFM, van der Vliet A, and Janssen-Heininger YMW. Activation of the glutaredoxin-1 gene by nuclear factor κB enhances signaling. Free Radic Biol Med 51: 1249–1257, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed MH, Barakat S, and Almobarak AO. Nonalcoholic fatty liver disease and cardiovascular disease: has the time come for cardiologists to be hepatologists? J Obes 2012: 483135, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anathy V, Aesif SW, Guala AS, Havermans M, Reynaert NL, Ho Y-S, Budd RC, and Janssen-Heininger YMW. Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J Cell Biol 184: 241–252, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anathy V, Aesif SW, Hoffman SM, Bement JL, Guala AS, Lahue KG, Leclair LW, Suratt BT, Cool CD, Wargo MJ, and Janssen-Heininger YMW. Glutaredoxin-1 attenuates S-glutathionylation of the death receptor fas and decreases resolution of Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 189: 463–474, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anstee QM. and Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol 87: 1–16, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auinger A, Rubin D, Sabandal M, Helwig U, Rüther A, Schreiber S, Foelsch UR, Döring F, and Schrezenmeir J. A common haplotype of carnitine palmitoyltransferase 1b is associated with the metabolic syndrome. Br J Nutr 109: 810–815, 2013 [DOI] [PubMed] [Google Scholar]
- 7.Bachschmid MM, Xu S, Maitland-Toolan KA, Ho Y-S, Cohen RA, and Matsui R. Attenuated cardiovascular hypertrophy and oxidant generation in response to angiotensin II infusion in glutaredoxin-1 knockout mice. Free Radic Biol Med 49: 1221–1229, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bécard D, Hainault I, Azzout-Marniche D, Bertry-Coussot L, Ferré P, and Foufelle F. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes 50: 2425–2430, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Bertolotti M, Spady DK, and Dietschy JM. Regulation of hepatic cholesterol metabolism in the rat in vivo: effect of a synthetic fat-free diet on sterol synthesis and low-density lipoprotein transport. Biochim Biophys Acta 1255: 293–300, 1995 [DOI] [PubMed] [Google Scholar]
- 10.Bhardwaj P, Madan K, Thareja S, Joshi YK, and Saraya A. Comparative redox status in alcoholic liver disease and nonalcoholic fatty liver disease. Hepatol Int 2: 202–208, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bräutigam L, Jensen LDE, Poschmann G, Nyström S, Bannenberg S, Dreij K, Lepka K, Prozorovski T, Montano SJ, Aktas O, Uhlén P, Stühler K, Cao Y, Holmgren A, and Berndt C. Glutaredoxin regulates vascular development by reversible glutathionylation of sirtuin 1. Proc Natl Acad Sci U S A 110: 20057–20062, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caballero F, Fernández A, De Lacy AM, Fernández-Checa JC, Caballería J, and García-Ruiz C. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. J Hepatol 50: 789–796, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, and Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 55: 2005–2023, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Chiang JYL. Bile acid metabolism and signaling. Compr Physiol 3: 1191–1212, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen JC, Horton JD, and Hobbs HH. Human fatty liver disease: old questions and new insights. Science 332: 1519–1523, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Day CP. and James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology 114: 842–845, 1998 [DOI] [PubMed] [Google Scholar]
- 17.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta 1830: 3217–3266, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Desvergne B, Michalik L, and Wahli W. Transcriptional regulation of metabolism. Physiol Rev 86: 465–514, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Diotte NM, Xiong Y, Gao J, Chua BHL, and Ho Y-S. Attenuation of doxorubicin-induced cardiac injury by mitochondrial glutaredoxin 2. Biochim Biophys Act 1793: 427–438, 2009 [DOI] [PubMed] [Google Scholar]
- 20.Donnelly KL, Margosian MR, Sheth SS, Lusis AJ, and Parks EJ. Increased lipogenesis and fatty acid reesterification contribute to hepatic triacylglycerol stores in hyperlipidemic Txnip-/- mice. J Nutr 134: 1475–1480, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Ebbert JO. and Jensen MD. Fat depots, free fatty acids, and dyslipidemia. Nutrients 5: 498–508, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Escande C, Chini CCS, Nin V, Dykhouse KM, Novak CM, Levine J, van Deursen J, Gores GJ, Chen J, Lou Z, and Chini EN. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest 120: 545–558, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frederico MJ, Vitto MF, Cesconetto PA, Engelmann J, De Souza DR, Luz G, Pinho RA, Ropelle ER, Cintra DE, and De Souza CT. Short-term inhibition of SREBP-1c expression reverses diet-induced non-alcoholic fatty liver disease in mice. Scand J Gastroenterol 46: 1381–1388, 2011 [DOI] [PubMed] [Google Scholar]
- 24.Gao X-H, Qanungo S, Pai H V, Starke DW, Steller KM, Fujioka H, Lesnefsky EJ, Kerner J, Rosca MG, Hoppel CL, and Mieyal JJ. Aging-dependent changes in rat heart mitochondrial glutaredoxins—implications for redox regulation. Redox Biol 1: 586–598, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim S-H, Mostoslavsky R, Alt FW, Wu Z, and Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J 26: 1913–1923, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerhart-Hines Z, Dominy JE, Jr., Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, and Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 44: 851–863, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodwin B. and Kliewer SA. Nuclear receptors. I. Nuclear receptors and bile acid homeostasis. Am J Physiol Gastrointest Liver Physiol 282: G926–G931, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, Kratky D, Wagner EF, Klingenspor M, Hoefler G, and Zechner R. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312: 734–737, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Haemmerle G, Moustafa T, Woelkart G, Büttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, Schreiber R, Eichmann T, Kolb D, Kotzbeck P, Schweiger M, Kumari M, Eder S, Schoiswohl G, Wongsiriroj N, Pollak NM, Radner FPW, Preiss-Landl K, Kolbe T, Rülicke T, Pieske B, Trauner M, Lass A, Zimmermann R, Hoefler G, Cinti S, Kershaw EE, Schrauwen P, Madeo F, Mayer B, and Zechner R. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat Med 17: 1076–1085, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haigis MC. and Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5: 253–295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J, Weisbrod RM, Shao D, Watanabe Y, Yin X, Bachschmid MM, Seta F, Janssen-Heininger YMW, Matsui R, Zang M, Hamburg NM, and Cohen RA. The redox mechanism for vascular barrier dysfunction associated with metabolic disorders: glutathionylation of Rac1 in endothelial cells. Redox Biol 9: 306–319, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock JT, Desikan R, and Neill SJ. Role of reactive oxygen species in cell signalling pathways. Biochem Soc Trans 29: 345–350, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Hensley K, Robinson KA, Gabbita SP, Salsman S, and Floyd RA. Reactive oxygen species, cell signaling, and cell injury. Free Radic Biol Med 28: 1456–1462, 2000 [DOI] [PubMed] [Google Scholar]
- 34.Ho Y-S, Xiong Y, Ho DS, Gao J, Chua BHL, Pai H, and Mieyal JJ. Targeted disruption of the glutaredoxin 1 gene does not sensitize adult mice to tissue injury induced by ischemia/reperfusion and hyperoxia. Free Radic Biol Med 43: 1299–1312, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, Cohen RA, and Zang M. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem 283: 20015–20026, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iverson SV, Eriksson S, Xu J, Prigge JR, Talago EA, Meade TA, Meade ES, Capecchi MR, Arnér ESJ, and Schmidt EE. A Txnrd1-dependent metabolic switch alters hepatic lipogenesis, glycogen storage, and detoxification. Free Radic Biol Med 63: 369–380, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DP, Carlson JL, Samiec PS, Sternberg P, Mody VC, Reed RL, and Brown LA. Glutathione measurement in human plasma. Evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin Chim Acta 275: 175–184, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Kawano Y. and Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol 48: 434–441, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller C. Indication of low-density lipoprotein apheresis in severe hypercholesterolemia and its atherosclerotic vascular complications: dextran sulfate cellulose low-density lipoprotein apheresis. Ther Apher Dial 7: 345–349, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Kleemann R, Verschuren L, van Erk MJ, Nikolsky Y, Cnubben NHP, Verheij ER, Smilde AK, Hendriks HFJ, Zadelaar S, Smith GJ, Kaznacheev V, Nikolskaya T, Melnikov A, Hurt-Camejo E, van der Greef J, van Ommen B, and Kooistra T. Atherosclerosis and liver inflammation induced by increased dietary cholesterol intake: a combined transcriptomics and metabolomics analysis. Genome Biol 8: R200, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koo S-H. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clin Mol Hepatol 19: 210–215, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langley E, Pearson M, Faretta M, Bauer U-MM, Frye RA, Minucci S, Pelicci PG, and Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J 21: 2383–2396, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leja J, Nilsson B, Yu D, Gustafson E, Akerström G, Oberg K, Giandomenico V, and Essand M. Double-detargeted oncolytic adenovirus shows replication arrest in liver cells and retains neuroendocrine cell killing ability. PLoS One 5: e8916, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lekli I, Mukherjee S, Ray D, Gurusamy N, Kim YH, Tosaki A, Engelman RM, Ho Y-S, and Das DK. Functional recovery of diabetic mouse hearts by glutaredoxin-1 gene therapy: role of Akt-FoxO-signaling network. Gene Ther 17: 478–485, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, Sun Y, Qi X, Shi Y, Gao H, Wu Q, Liu X, Yu H, and Zhang C. Protective effect and mechanism of glutaredoxin 1 on coronary arteries endothelial cells damage induced by high glucose. Biomed Mater Eng 24: 3897–3903, 2014 [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Wong K, Giles A, Jiang J, Lee JW, Adams AC, Kharitonenkov A, Yang Q, Gao B, Guarente L, and Zang M. Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology 146: 539–549.e7, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin S-J, Ford E, Haigis M, Liszt G, and Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18: 12–16, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, and Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107: 137–148, 2001 [DOI] [PubMed] [Google Scholar]
- 49.Maxfield FR. and Tabas I. Role of cholesterol and lipid organization in disease. Nature 438: 612–621, 2005 [DOI] [PubMed] [Google Scholar]
- 50.Min H-K, Kapoor A, Fuchs M, Mirshahi F, Zhou H, Maher J, Kellum J, Warnick R, Contos MJ, and Sanyal AJ. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab 15: 665–674, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murdoch CE, Shuler M, Haeussler DJF, Kikuchi R, Bearelly P, Han J, Watanabe Y, Fuster JJ, Walsh K, Ho Y-S, Bachschmid MM, Cohen RA, and Matsui R. Glutaredoxin-1 up-regulation induces soluble vascular endothelial growth factor receptor 1, attenuating post-ischemia limb revascularization. J Biol Chem 289: 8633–8644, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musso G, Gambino R, and Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog Lipid Res 52: 175–191, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Oni ET, Agatston AS, Blaha MJ, Fialkow J, Cury R, Sposito A, Erbel R, Blankstein R, Feldman T, Al-Mallah MH, Santos RD, Budoff MJ, and Nasir K. A systematic review: burden and severity of subclinical cardiovascular disease among those with nonalcoholic fatty liver; should we care? Atherosclerosis 230: 258–267, 2013 [DOI] [PubMed] [Google Scholar]
- 54.Pai H V, Starke DW, Lesnefsky EJ, Hoppel CL, and Mieyal JJ. What is the functional significance of the unique location of glutaredoxin 1 (GRx1) in the intermembrane space of mitochondria? Antioxid Redox Signal 9: 2027–2033, 2007 [DOI] [PubMed] [Google Scholar]
- 55.Pandak WM, Ren S, Marques D, Hall E, Redford K, Mallonee D, Bohdan P, Heuman D, Gil G, and Hylemon P. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J Biol Chem 277: 48158–48164, 2002 [DOI] [PubMed] [Google Scholar]
- 56.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, and Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A 105: 9793–9798, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, and Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429: 771–776, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piemonte F, Petrini S, Gaeta LM, Tozzi G, Bertini E, Devito R, Boldrini R, Marcellini M, Ciacco E, and Nobili V. Protein glutathionylation increases in the liver of patients with non-alcoholic fatty liver disease. J Gastroenterol Hepatol 23: e457–e464, 2008 [DOI] [PubMed] [Google Scholar]
- 59.Podrini C, Borghesan M, Greco A, Pazienza V, Mazzoccoli G, and Vinciguerra M. Redox homeostasis and epigenetics in non-alcoholic fatty liver disease (NAFLD). Curr Pharm Des 19: 2737–2746, 2013 [DOI] [PubMed] [Google Scholar]
- 60.Ponugoti B, Kim D-H, Xiao Z, Smith Z, Miao J, Zang M, Wu S-Y, Chiang C-M, Veenstra TD, and Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem 285: 33959–33970, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Postic C. and Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest 118: 829–838, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min H-K, Contos MJ, Sterling RK, Fuchs M, Zhou H, Watkins SM, and Sanyal AJ. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 50: 1827–1838, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, and Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 9: 327–338, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reed DJ, Babson JR, Beatty PW, Brodie AE, Ellis WW, and Potter DW. High-performance liquid chromatography analysis of nanomole levels of glutathione, glutathione disulfide, and related thiols and disulfides. Anal Biochem 106: 55–62, 1980 [DOI] [PubMed] [Google Scholar]
- 65.Reinbothe TM, Ivarsson R, Li D-Q, Niazi O, Jing X, Zhang E, Stenson L, Bryborn U, and Renström E. Glutaredoxin-1 mediates NADPH-dependent stimulation of calcium-dependent insulin secretion. Mol Endocrinol 23: 893–900, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seki S, Kitada T, Yamada T, Sakaguchi H, Nakatani K, and Wakasa K. In situ detection of lipid peroxidation and oxidative DNA damage in non-alcoholic fatty liver diseases. J Hepatol 37: 56–62, 2002 [DOI] [PubMed] [Google Scholar]
- 67.Shao D, Fry JL, Han J, Hou X, Pimentel DR, Matsui R, Cohen RA, and Bachschmid MM. A redox-resistant sirtuin-1 mutant protects against hepatic metabolic and oxidant stress. J Biol Chem 289: 7293–7306, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shelton MD, Kern TS, and Mieyal JJ. Glutaredoxin regulates nuclear factor kappa-B and intercellular adhesion molecule in Müller cells: model of diabetic retinopathy. J Biol Chem 282: 12467–12474, 2007 [DOI] [PubMed] [Google Scholar]
- 69.Targher G, Day CP, and Bonora E. Risk of cardiovascular disease in patients with nonalcoholic fatty liver disease. N Engl J Med 363: 1341–1350, 2010 [DOI] [PubMed] [Google Scholar]
- 70.Turley SD, Spady DK, and Dietschy JM. Role of liver in the synthesis of cholesterol and the clearance of low density lipoproteins in the cynomolgus monkey. J Lipid Res 36: 67–79, 1995 [PubMed] [Google Scholar]
- 71.Vall-Llaura N, Reverter-Branchat G, Vived C, Weertman N, Rodríguez-Colman MJ, and Cabiscol E. Reversible glutathionylation of Sir2 by monothiol glutaredoxins Grx3/4 regulates stress resistance. Free Radic Biol Med 96: 45–56, 2016 [DOI] [PubMed] [Google Scholar]
- 72.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, and Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159, 2001 [DOI] [PubMed] [Google Scholar]
- 73.Walker AK, Yang F, Jiang K, Ji J-Y, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, and Näär AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev 24: 1403–1417, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watanabe Y, Murdoch CE, Sano S, Ido Y, Bachschmid MM, Cohen RA, and Matsui R. Glutathione adducts induced by ischemia and deletion of glutaredoxin-1 stabilize HIF-1α and improve limb revascularization. Proc Natl Acad Sci U S A 113: 6011–6016, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, and Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/- mice: a role of lipid mobilization and inflammation. Endocrinology 151: 2504–2514, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu X, So J-S, Park J-G, and Lee A-H. Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP. Semin Liver Dis 33: 301–311, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zee RS, Yoo CB, Pimentel DR, Perlman DH, Burgoyne JR, Hou X, McComb ME, Costello CE, Cohen RA, and Bachschmid MM. Redox regulation of sirtuin-1 by S-glutathiolation. Antioxid Redox Signal 13: 1023–1032, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.