Abstract
Background:
Methicillin resistant Staphylococcus aureus (MRSA) has been emerged as a nosocomial and community acquired pathogen worldwide. There are many challenges for laboratory detection of MRSA. The aim of this study was to compare different phenotypic methods with PCR based method as a gold standard for detection of mecA gene.
Methods:
A total of 220 clinical isolates of S. aureus which were isolated from various clinical specimens from September 2013 until the June of 2014 in Milad Hospital of Tehran, Iran was subject of our study. Methicillin resistance was determined by oxacillin and cefoxitin disks, oxacillin screen agar and CHROMagar™ MRSA medium. The results of these methods were compared with mecA gene based PCR method as a gold standard method.
Results:
Among 220 isolates from S. aureus, 105 (47.72%) isolates were positive for mecA gene by PCR method. The results of cefoxitin disk diffusion method with 100% sensitivity and specificity was the same as PCR method .CHROMagar™ MRSA medium had 98.13% sensitivity and 100% specificity. Oxacillin disk diffusion and oxacillin screen agar had 95.42% and 97.22% sensitivity respectively.
Conclusion:
Result of cefoxitin disk diffusion method with 100% sensitivity and specificity was the same as PCR method for detection mecA gene. Cefoxitin disk diffusion method can be used as an alternative method of PCR for detection of MRSA.
Key Words: Staphylococcus aureus, mecA gene, CHOROM Agar MRSA, Oxacillin screen Agar, Methicillin Resistant
Introduction
Staphylococcus aureus is an opportunistic pathogen and as a normal microbiota of the nose, skin, mouth and other parts of human body (1-2). It is an important agent of hospital and community acquired infections worldwide (3-5). Methicillin was first introduced in 1959 for treatment of infections caused by penicillin resistant strains of S. aureus. In 1961, first methicillin- resistant strain of S. aureus (MRSA) was reported from United Kingdom hospitals and recovered from other European countries hospitals afterwards (6-10). Infections caused by MRSA associated with a higher rate of mortality in compaction with Methicillin–susceptible S. aureus isolates (11). Methicillin resistance in S. aureus is mediated by expression of mecA gene, which results in production of modified penicillin–binding proteins (PBP2a).In addition recently other genes such as femA and other auxiliary genes are recently as the one, which can contribute to MRSA resistance (12).
In Iran, the mean prevalence of MRSA was 52.7% ±4.7 (13). For this reason, detection of MRSA in microbiology laboratories is very important in order to choose appreciates antimicrobial agents. Misidentification of MRSA leads to using of unnecessary antibiotics such as vancomycin and linezolid (14).
Appropriate and rapid identification of MRSA in clinical microbiology laboratories is essential issue for treatment and epidemiological purpose. There are many different laboratory methods for detection of MRSA (15, 16). Some of them such as detection of mecA gene is a gold standard method and other phenotypic methods are compared with this procedure.
The main objective of this study was to evaluate different phenotypic methods in relation to detecting mecA gene. This evaluation helps us for choosing a reliable routine method for detection of MRSA in our microbiology laboratories.
Materials and Methods
Strains
During September 2013 and June 2014; 220 clinical isolates of S. aureus were collected these strains were isolated from different clinical specimen such as blood, urine, sputum, tracheal aspirate and others. Gram stain and biochemical test such as growth on Heat-Stable nuclease, Mannitol salt agar, Coagulase, Catalase tests and susceptibility to novobiocin were used for identification of these isolates. All isolates were kept frozen at -70 °C in Trypticase soy broth containing 15% glycerol until for performance of susceptibility testing and MRSA detection.
Detection of the mecA Gene by Polymerase Chain Reaction:
Bacterial DNA was extracted by the rapid cell lysis method (18-19). First microtubes containing the bacteria in PBS were centrifuged at 10000 g for 3 min and then the supernatant was discarded. Sediment of bacteria STES and 20 ml solution 100 ml solution TE (pH = 7.6) was added to the microtube multi-tap to scale well be solved in the solution. Thirty ml lysosome were added to the mixture and were incubated at 37 °C for an hour. Then, 30 ml proteinase (K) for 30 min at 56 °C micro tubes were added. With regard to the total volume, half the volume of phenol and chloroform, add half the volume was centrifuged for 5 min at rpm 13000 rpm. After centrifugation, phase separation and the supernatant was transferred to another vial 0.1 volume of sodium acetate and 2.5 times the volume of cold absolute ethanol was added. Vials gently upside down several times, and then for 30 min in the freezer was - 20° C. Since then were centrifuged for 5 min at rpm 13000 rpm. The supernatant was discarded and 500 ml 70% ethanol was added to the vial. After a few hit again for 5 min in a centrifuge at 13,000 rpm around the supernatant was discarded and vials fixed until completely dry. In the last 100 ml was added to the solution with pH = 8 was for 18-16 h at room temperature.
The bottles after it were preserved in refrigerator. PCR – multiplex reaction for detection of mecA gene primers for replication were provided from Cinagene Company (Cinageneco, Tehran Iran) from this suspension, a 5 µL volume was directly used as the template for the PCR amplification of the mecA gene fragments.
The mecA-F (TCCAGATTACAACTTCACCAGG) and the mecA-R (CCACTTCATATCTTGTAACG) primers were used for the amplification of the 162bp fragment of the Methicillin-resistant gene (mec A) (11). A 50 µl PCR reaction consisted of plus 45 µl of the master mix which contained the PCR buffer (1.5X), dNTP mix (0.25mM of each), the primer (0.3 pmol), Taq DNA polymerase (0.1 U/μlit), and MgCl 2 (1.5mM) with 5 µL of the template DNA. The cycling conditions were as follows: 5 min sat 94 °C, followed by 32 cycles of denaturation at 94 °C for 50 sec, annealing at 58 °C for 50 sec, extension at 72 °C for 50 sec and the final extension step at 72 °C for 10 min.
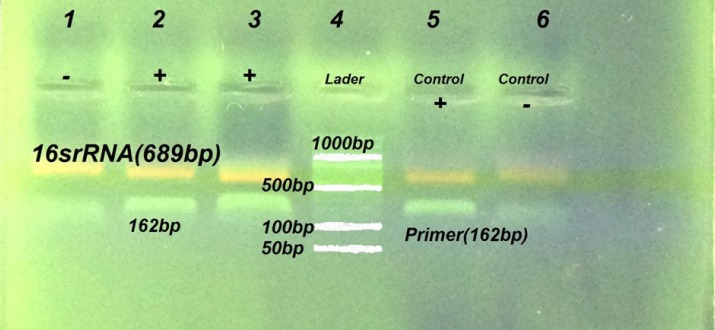
The PCR products were visualized on a 1.5% agarose gel with ethidium bromide dye under a UV transilluminator. Amplicons of 162 bp were consistent with the mecA gene amplification (Fig. 1).
Fig. 1.
Detection of mecA gene by PCR method. Lane6 is negative control strain S. aureus ATCC: 29213, Lane 5 was positive control S. aureus ATCC: 33591, Lanes (1-2-3). Representative MRSA strains isolated from clinical specimens
Detection of MRSA by phenotypic methods
Disk diffusion methods.
Disk diffusion method using oxaciliin and cefoxitin disks were performed for all isolates on Mueller–Hinton agar for detection of MRSA as recommended by CLSI guideline (20). Briefly four to five isolated colonies from overnight growth of S. aureus on Blood agar was suspended in n to 4-5 ml of PBS. The turbidity of suspension was adjusted to 0.5 McFarland standard turbidity and inoculated on two separate Mueller –Hinton agar plates. An oxacillin (1µg) disk aseptically placed on the Mueller–Hinton agar and incubated at 35 for 24 hours. Cefoxitin disk was placed on the other Mueller –Hinton agar and incubated at 35°C for 18 h. The inhibition zone of inhibition for each antibiotic disks were measured and by referring to CLSI guidelines reported as MRSA or methicillin susceptible S. aureus (20). All of antibiotic disks were provided from ROSC Company (ROSCO .Co Denmark)
Oxacillin screen Agar
Muller-Hinton agar plates containing 4% NaCl and 6 μg/ml of oxacillin were prepared. To perform the oxacillin screen test, a swab dipped in 0.5 McFarland’s suspension of the isolate was deposited as a spot on the agar surface and it was incubated at 35 °C for 24 h. Plates were observed carefully in transmitted light for any growth. Any growth after 24 h was considered oxacillin resistant (16, 20).
CHROMagar™ MRSA
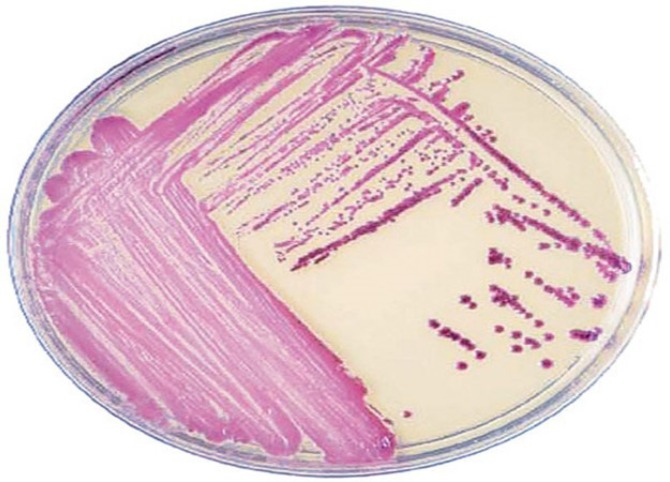
CHROMagarTM MRSA for detection of MRSA was provided from CHROM agar Company, (Paris, France) The medium contained agar (15 g/liter), peptones (40 g/l), NaCl (25 g/l), and a proprietary chromogenic mix (3.5 g/l). The medium was prepared as instruction recommended by manufacturer avoiding heating at over 100 °C. Methicillin or oxacillin (4 μg/ml) was added when the agar was cooled at 48 °C. Each plate contained 20 ml of agar medium dispensed into 90-mm-diameter Petridishes. Fine isolated colony of S. aurues processed by direct streaking on the CHROMagar plate (Fig. 2). Plates were incubated in aerobic conditions at 37 °C for 18-24 h. Growth of intense colonies with Mauve color in 18-24 h considered as MRSA as according manufacture guideline.
Fig. 2.
Colonies of MRSA on CHROMagar™ MRSA
Quality control
The quality control strains including methicillin resistant S aureus ( MRSA ATCC 43300 ) and methicillin sensitive S. aureus (MSSA ATCC 25293) were used as positive and negative controls. These strains were provided from Iranian reference health laboratory.
Statistical analysis
We used descriptive analysis such as percentage for categorical variables. The sensitivity, specificity, positive and negative predictive values and determining of diagnostic value of each method were calculated using Graph Pad in Stat 3.1 software.
Results
From 74572 surveillance specimens submitted to microbiology laboratory of Milad Hospital 220 isolates S. aureus were recovered from clinical specimens. Out of 220 patients, 56.4% were male and 43.6% female. Among 220 S. aureus 105 (47.72%) isolates were positive for the mecA gene by PCR method. Among phenotypic methods, cefoxitin disk diffusion method had the highest sensitivity and specificity. The sensitivity and specificity, of different phenotypic methods in comparison to PCR method as a gold standard procedure for detection of MRSA are shown in Table 1.
Table1.
Comparison of different phenotypic for detection of MRSA
| Method | No of MRSA | Sensitivity | Specificity |
|---|---|---|---|
| Oxacillin Disk (1µg) | 95.45 | 100 | 100 |
| Cefoxitin Disk (30µg) | 100 | 100 | 105 |
| Oxacillin screen agar 6µg/ml | 102 | 97.22 | 100 |
| CHROMagar™ MRSA | 103 | 98.13 | 100 |
| PCR for detection mecA | 105 | 100 | 100 |
Discussion
Recently increase rate of methicillin resistant strains of S. aureus has been posed a great difficulty in choosing suitable antibiotics for the treatment of infection caused by this microorganism (21). In Iran about 52% isolates of S. aureus were resistant to methicillin (13). Timely and correct identification of MRSA is very important issue for prognosis and treatment infections caused by S. aureus (15).There are multiple laboratory methods for detection of MRSA, however many of these methods have not sufficiently sensitivity and specificity. This may be due to the persons carrying the test and material, methods and heterogeneous resistance clinical isolates of S. aureus (15, 23).
Detection of mecA gene by molecular methods such as PCR is a gold standard method for confirmation of MRSA isolates, however, molecular detection of MRSA are not included in routine practice of many clinical microbiology laboratories (9, 24). For this reason, establishment of phenotypic methods with a high sensitivity and specificity for detection of MRSA is very essential. During last decade, there was many attempts by international guidelines such as CLSI to improve and standardization of phenotypic methods for detection of MRSA (25, 26).
In our study, oxacillin disk diffusion method as a routine procedure for detection of MRSA revealed 100 isolatesas MRSA with 95.45% sensitivity and 100%specificity. This finding was in concordance to the results of other studies (25, 27). In spite of high sensitivity and specificity of oxacilin disk diffusion method for detection of MRSA, this method has been encountered with difficulties for detection of hetroresistant strains of S. aureus (26).
Other disk diffusion method in our study was using of cefoxitin disk. In present study, the cefoxitin disk diffusion method detected 105 out of 220 isolates as MRSA. Sensitivity and specificity of this method for detection of MRSA was 100%. Several studies including present study revealed that the results of the cefoxitin disk diffusion testing correlate better with the presence of mecA in comparison with oxacillin disk diffusion method (16 ,26,27). Cefoxitin is a better inducer of mecA gene expression, while oxacillin is a weak inducer of PBP2a production. Recently CLSI recommended usage of cefoxitin instead of oxacillin disk for identification of MRSA (20, 26).
Detection of MRSA by the oxacillin screen agar had 97.22% sensitivity and 100 specificity, which was concordance with other studies (25). In some cases, however heterogeneous MRSA populations of S. aureus are not detected on oxacillin screen agar because of their low expression of resistance. This method also is not recommended for detection coagulase-negative staphylococci by CLSI guideline (20).
Recently chromogenic media are available for detection of MRSA. (28-30). In our study, CHROMagar™ MRSA had a high sensitivity and specificity (Table 1). The results of our study were consistent with other findings for detecting of MRSA from specimens such as nasal swabs. This method is comparable by other methods such as cefoxitin disk diffusion method (31).
Conclusion
The oxacillin disk diffusion method was less sensitive for detection of mrsa, however the results of cefoxitin disk diffusion with 100% sensitivity and specificity was the same as PCR method for detection mecA gene. Cefoxitin disk diffusion method can be used as an alternative method to PCR especially at those microbiology laboratories without molecular facilities.
Acknowledgment
We thank all personnel of Microbiology laboratory at Milad hospital of Tehran for their technical supports.
Conflict of Interest:
The authors declare that there is no Conflict of Interests.
References
- 1.Lozano C, Gómez-Sanz E, Benito D, Aspiroz C, Zarazaga M, Torres C. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int J Med Microbiol. 2011;301(6):500–5. doi: 10.1016/j.ijmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Verhoeven PO, Gagnaire J, Botelho-Nevers E, Grattard F, Carricajo A, Lucht F, Pozzetto B, Berthelot P. Detection and clinical relevance of Staphylococcus aureus nasal carriage: an update. Expert Rev Anti Infect Ther. 2014;12 (1):75–89. doi: 10.1586/14787210.2014.859985. [DOI] [PubMed] [Google Scholar]
- 3.Michiels B, Appelen L, Franck B, den Heijer CD, Bartholomeeusen S, Coenen S. Staphylococcus aureus, Including Meticillin-Resistant Staphylococcus aureus, among General Practitioners and Their Patients: A Cross-Sectional Study. PLoS One. 2015 ;10(10):e0140045. doi: 10.1371/journal.pone.0140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanal R, Sah P, Lamichhane P, Lamsal A, Upadhaya S, Pahwa VK. Nasalcarriage of methicillin resistant Staphylococcus aureus among health care workers at a tertiary care hospital in Western Nepal. Antimicrob Resist Infect Control. 2015 Oct 9;4:39. doi: 10.1186/s13756-015-0082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J, Li CS, Giordani M, Shahlaie K, Klineberg EO, Tripet-Diel JR, Ihara MS, Cohen SH. Swabbing Surgical Sites Does Not Improve the Detection of Staphylococcus aureus Carriage in High-Risk Surgical Patients. Surg Infect (Larchmt) 2015;16(5):523–5. doi: 10.1089/sur.2014.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci U S A. 2002 May 28;99(11):7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand KB, Agrawal P, Kumar S, Kapila K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009 Jan-Mar;27(1):27–9. [PubMed] [Google Scholar]
- 8.Strommenger B, Bartels MD, Kurt K, Layer F, Rohde SM, Boye K, Westh H, Witte W, De Lencastre H, NübelU Evolution of methicillin-resistant Staphylococcus aureustowards increasing resistance. J Antimicrob Chemother. 2014;69(3):616–22. doi: 10.1093/jac/dkt413. [DOI] [PubMed] [Google Scholar]
- 9.Rahbar M, YaghoobiY , Fattahi Comparison of different laboratory methods for detection of methicillin resistant Staphylococcus naureus. Pak J Med Sci. 2006;22(4):442–445. [Google Scholar]
- 10.Bhutia KO, Singh TS, Biswas S, AdhikariL Evaluation of phenotypic with genotypic methods for species identification and detection of methicillin resistant in Staphylococcusaureus. Int J Appl Basic Med Res. 2012;2(2):84–91. doi: 10.4103/2229-516X.106348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You JH, Ip DN, Wong CT, Ling T, Lee N, IpM Meticillin-resistant Staphylococcus aureus bacteraemia in Hong Kong. JHosp Infect. 200;70(4):379–81. doi: 10.1016/j.jhin.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Boşgelmez-Tinaz G, Ulusoy S, Aridoğan B, Coşkun-Ari F. Evaluation of different methods to detect oxacillin resistance in Staphylococcus aureus and their clinical laboratory utility. Eur J ClinMicrobiol Infect Dis. 2006;25(6):410–2. doi: 10.1007/s10096-006-0153-8. [DOI] [PubMed] [Google Scholar]
- 13.Askari E, Soleymani F, Arianpoor A, Tabatabai SM, Amini A, Naderinasab M. analysis. Iran J Basic Med Sci. 2012;15(5):1010–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Rodvold KA, McConeghyKW Methicillin-resistant Staphylococcus aureus therapy: past, present, and future. Clin Infect Dis. 2014 Jan;58(Suppl 1):S20–7. doi: 10.1093/cid/cit614. doi: 10.1093/cid/cit614. Review. [DOI] [PubMed] [Google Scholar]
- 15.Velasco D, del Mar Tomas M, Cartelle M, Beceiro A, Perez A, Molina F, Moure R, Villanueva R, Bou G. Evaluation of different methods for detecting methicillin (oxacillin) resistance in Staphylococcus aureus. JAntimicrob Chemother. 2005;55(3):379–82. doi: 10.1093/jac/dki017. [DOI] [PubMed] [Google Scholar]
- 16.Brown DF1, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, Wren MW Joint Working Party of the British Society for Antimicrobial Chemotherapy Hospital Infection Society Association. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA) J AntimicrobChemother. 2005;56(6):1000–18. doi: 10.1093/jac/dki372. [DOI] [PubMed] [Google Scholar]
- 17.Isenberg HD, editor. Clinical microbiology procedures hand book. 2nd ed. . Washington DC: ASM Press; 2004. [Google Scholar]
- 18.Unal S, Hoskins J, Flokowitsch JE, Wu CY, Preston DA, Skatrud PL. Detection of Methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–91. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Roth E, Claverie-Martin F, Villar J, Mendez-Alvarez S. Multiplex PCR for the simultaneous identification of Staphylococcus aureus and the detection of methicillin and mupirocin resistance. J Clin Microbiol. 2001;39:4037–41. doi: 10.1128/JCM.39.11.4037-4041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute CLSI 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement.
- 21.Velasco V, Sherwood JS, Rojas-García PP, Logue CM. Multiplex real-time PCR for detection of Staphylococcus aureus,mecA and Panton-Valentine Leukocidin (PVL) genes from selective enrichments from animals and retail meat. PLoS One. 2014 ;9(5):e97617. doi: 10.1371/journal.pone.0097617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velasco D1, del Mar Tomas M, Cartelle M, Beceiro A, Perez A, Molina F, Moure R, Villanueva R, BouG Evaluation of different methods for detecting methicillin (oxacillin) resistance in Staphylococcus aureus. J Antimicrob Chemother. 2005;55(3):379–82. doi: 10.1093/jac/dki017. [DOI] [PubMed] [Google Scholar]
- 23.Pramodhini S, Thenmozhivalli PR, Selvi R, Dillirani V, Vasumathi A, Agatha D. Comparison of Various Phenotypic Methods and mecA Based PCR for the Detection of MRSA. J Clin Diag Res. 2011;5:1359–1362. [Google Scholar]
- 24.Kunsang O Bhutia, T Shantikumar Singh, Shilpie Biswas, Luna Adhikari. Evaluation of phenotypic with genotypic methods for species identification and detection of methicillin resistant in Staphylococcus aureus. Int J Appl Basic Med Res. 2012;2(2):84–91. doi: 10.4103/2229-516X.106348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand KB, Agrawal P, Kumar S, Kapila K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009;27(1):27–9. [PubMed] [Google Scholar]
- 26.Abbas Farahani, Parviz Mohajeri, Babak Gholamine, Mansour Rezaei, Hassan Abbasi. 2 Comparison of Different Phenotypic and Genotypic Methods for the Detection of Methicillin-Resistant Staphylococcus Aureus. N Am J Med Sci. 2013;5(11):637–640. doi: 10.4103/1947-2714.122305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahbar M, Safadel NA. Evaluation of Cefoxitin Disk Diffusion Test for Routine Detection of Methicillin-resistant Staphylococcus Aureus. Iranian J Pathol. 206;1:145–148. [Google Scholar]
- 28.Louie L, Soares D, Meaney H, Vearncombe M, Simor AE. Evaluation of a new chromogenic medium, MRSA select, for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006;44:4561–3. doi: 10.1128/JCM.01763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry JD, Davies A, Butterworth LA, Hopley AL, Nicholson A, Gould FK. Development and evaluation of a chromogenicagar medium for methicillin-resistant Staphylococcusaureus. J Clin Microbiol. 2004;42(10):4519–23. doi: 10.1128/JCM.42.10.4519-4523.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Z, Lautenbach E, Fishman N, NachamkinI Evaluation of mannitol salt agar, CHROMagar Staph aureus and CHROMagar MRSA for detection of meticillin-resistant Staphylococcus aureus from nasal swab specimen. J Med Microbiol. 2007;56(Pt 1):43–6. doi: 10.1099/jmm.0.46777-0. [DOI] [PubMed] [Google Scholar]
- 31.Ghasemian A, NajarPeerayeh S, Bakhshi B, Mirzaee M. The Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs) Genes among Clinical Isolates of Staphylococcus aureus from Hospitalized Children. Iran J Pathol. 2015 Fall;10(4):258–64. [PMC free article] [PubMed] [Google Scholar]