Abstract
Background:
MicroRNA-7 (miR-7) has been observed as a potent tumour suppressor in multiple cancer types including breast cancer. The aim of this study was to investigate the response sensitivities of metastatic breast cancer cells to miR-7 and the roles of miR-7 in the interaction of endothelial cells and metastatic cancer cells.
Methods:
Expression profile of miRNAs in a breast cancer specimen cohort and breast cancer cells were determined using real-time quantitative miRNA assays. Effect of the altering expression of miR-7 on migration, invasion, proliferation, interaction and underlying molecular mechanism of breast cancer cells and endothelial cells was investigated after treatment with the synthesised mimic of miR-7. Luciferase activity analysis was performed to validate Wave-3 as a novel target of miR-7.
Results:
miR-7 expression was negatively correlated with the stage, grade and survival of the breast cancer patients. There was also differential expression of miRNAs including miR-7 in the breast cancer cells. The synthesised mimic of miR-7 inhibits the motility and wound healing potential of breast cancer cells. The highly metastatic MDA-MB-231 cells are more sensitive to the miR-7 treatment than the poorly invasive MCF-7 cells. Treatment with miR-7 downregulated the expression of EGFR, IGF1R and Wave3 in MDA-MB-231 cells but not in MCF-7 cells. In addition, we further demonstrated that miR-7 inhibited the proliferation, migration and invasion of endothelial cells. And more importantly, miR-7 suppressed the homing and migration of endothelial cells to more aggressive tumour cell conditions.
Conclusions:
Given the dual inhibitory effect of miR-7 on metastatic breast cancer cells alone and the interaction of endothelial cells with the tumour-conditioned microenvironment, we suggest miR-7 may be a new therapeutic candidate for its capacity not only to prevent breast cancer cell spreading but also to inhibit tumour-associated angiogenesis in the metastatic breast cancer.
Keywords: miR-7, breast cancer, endothelial cells, migration, homing, metastasis
Breast cancer is the most common cancer type among women and the second most common cancer overall, with nearly 1.7 million new cases diagnosed and over 0.5 million death every year worldwide (Ferlay et al, 2015a). This accounts for ∼12% of all new cancer cases and 25% of all cancers in women. The UK is among the top 7 countries with the highest incidence of breast cancer (Ferlay et al, 2015b). Breast cancer starts as a primary tumour, and this progresses to metastasise to distance sites. The most common sites of breast cancer metastasis are bone, liver, brain and distant lymph nodes. Breast cancer is a heterogeneous disease, and can be classified into multiple subtypes according to distinct histopathological features, genetic and genomic variability, and diverse prognostic outcomes (Curtis et al, 2012). Despite intensive investigation, the cellular and molecular mechanisms of breast cancer metastasis is still not clear. Due to the complexity of the cancer onset and progression, it is still a clinical challenge for the early diagnosis and efficient treatment of breast cancer metastasis, which accounts for 90% of all breast cancer deaths. Very often the metastatic breast cancer becomes incurable and the patients can be only treated through a palliative care approach (Sledge et al, 2014). Additional studies are needed to elucidate the molecular mechanisms and identify the histological characteristics which can be useful for the identification of a pre-metastatic state.
MicroRNAs (miRNAs, miRs) are endogenous, non-coding RNAs, which are 18–20 nucleotides and regulate gene expression post-transcriptionally. They bind to the 3′-untranslated region of mRNAs thus blocking mRNA translation. They can also promote mRNA instability and facilitate its degradation (Guo et al, 2010). A single miRNA may target and exert full functional effects on several hundred mRNAs over the entire human genome and show multiple targets in a solid tumour such as breast cancer (Jansson and Lund, 2012). miRNAs are master regulators of cell behaviour under normal and pathological conditions. In metastatic tumours, miRNAs can act as epigenetic regulators of gene expression which control multiple aspects of metastasis (Tavazoie et al, 2008). Regulation by miRNAs can be involved in the multiple stages of dissemination of cancer cells from the primary tumour site including intravasation and extravasation. It also plays a role in tumour cell homing to distant sites, tumour–stroma cell interactions, dormancy and outgrowth (Ell et al, 2013). As there are no effective targeted therapeutic drugs to prevent or halt breast cancer metastasis in the clinic, miRNAs may be a potential avenue for a new therapeutic approach. This is supported by the recent studies with miR-34, a master regulator of tumour suppression (Bader, 2012), which has shown promising results in animal models of cancer and has become the first microRNA to enter multicentre Phase 1 clinical study in solid tumours and haematological malignancies (ClinicalTrials.gov Identifier: NCT01829971).
In this study we hypothesised that certain breast cancer cells may have a distinctive sensitivity to a specific miRNA mimic which regulates critical pathways involved in aggressive tumour cell behaviours, including spreading and invasion. We first screened a panel of miRNAs in a breast cancer cohort, and then determined the differential expression of lead miRNAs in a panel of breast cancer cell lines with varying invasion properties. We observed that the highly metastatic MDA-MB-231 cells show the lowest expression of miR-7 compared to the other breast cancer cells considered. Furthermore, we used chemically synthesised mimics of miRNAs, including miR-7, to evaluate whether they had an inhibitory effect on motility and wound healing potential of breast cancer cells. We then investigated how miR-7 mimic can modulate the behaviours of breast cancer cells, and what were the possible mechanisms by which metastatic cells are more susceptible to the anti-tumoural effect to miR-7. Furthermore, in order to evaluate the potential therapeutic application of miR-7 in the control of tumour metastasis, we evaluated the effect of miR-7 on endothelial cell behaviours including migration, proliferation, invasion and tumour–endothelial interaction.
Our data indicate that miR-7 controls the angiogenic potential of endothelial cells in the presence of aggressive tumour conditions as well. To the best of our knowledge, this is the first report describing the dual inhibitory effect of miR-7 on invasive breast cancer cells and endothelial cells, which are both essential for breast cancer metastasis. We therefore support a new concept for the development of a miR-7 replacement therapy to prevent or delay breast cancer metastasis.
Materials and methods
Human breast cancer samples
Breast cancer patient specimens were acquired in accordance with institutional guidelines after ethical approval by the Bro Taf Health Authority local ethics committee. Written informed consent was obtained from all study subjects. Primary breast cancer tissues (n=127) and adjacent non-cancerous mammary tissue (n=33) were collected immediately after surgical excision and stored at −80 °C until use. All tissues were randomly numbered and the details were only made known after all analyses were completed. The presence of tumour cells in the collected samples was verified by an independent consultant pathologist using haematoxylin and eosin stained frozen sections. A routine follow-up was carried out after surgery and details were stored in a customised database. The median follow-up period was 120 months. The clinical data are provided in Table 1.
Table 1. Primers Used for the Real-Time qRT-PCRa.
| Gene | Sense primer (5′–3′) | Anti-sense primer (5′–3′) |
|---|---|---|
| ANGPT2 | GTCCACATCAAACTCTAAGGA | ACTGAACCTGACCGTACAATGTTAACGTGTAGATGCCA |
| CXCR4 | GCCTCTTTTGCAGATATACAC | ACTGAACCTGACCGTACAGGTGGGCAGGAAGATTTTAT |
| CXCR7 | TTCCAAGACTTTTTCTGCCT | ACTGAACCTGACCGTACATGCGCAAGCTATAAACAAAG |
| EGFR | AGAGTCTCAAAGCCATGTT | ACTGAACCTGACCGTACACCATCCTAAGCATGACTCC |
| ERK | ACACGCAGTTGCAGTACA | ACTGAACCTGACCGTACAGGGGCTGATCTTCTTGAT |
| GAPDH | CTGAGTACGTCGTGGAGTC | ACTGAACCTGACCGTACACAGAGATGATGACCCTTTTG |
| GROα | TCTTCCGCTCCTCTCAC | ACTGAACCTGACCGTACACGGACGCTCCTGCTG |
| IGF1R | GTGTTCTTCTATGTCCAGGC | ACTGAACCTGACCGTACAAGACGTACAGCATAATCACC |
| mTOR | GCTGCAGAAGAAGGTCACT | ACTGAACCTGACCGTACAAAGGAGATGGAACGGAAG |
| STAT3 | CATGGAAGAATCCAACAACG | ACTGAACCTGACCGTACAAATCAGGGAAGCATCACAAT |
| THBS1 | ACCAACCGCATTCCAGAGTC | ACTGAACCTGACCGTACATCAGGTTGGCATCCTCGAT |
| VEGFA | GAGCCGGAGAGGGAG | ACTGAACCTGACCGTACACTGGGACCACTTGGCAT |
| WAVE3 | TGACACCATACAGAGATGAC | ACTGAACCTGACCGTACACTGTTAGGGTTCAGCTTGTG |
The z-sequence (which binds to the 3′-end of the Amplifluor probes) are underlined.
Cells and cell culture
All human breast cell lines used for this study were obtained from the ATCC (Rockville, MD, USA). The human breast cancer cell lines MDA-MB-231, MDA-361, MCF-7, ZR-75.1, BT-474, BT-20, T47D and BT-549 were routinely maintained in Dulbecco’s modified Eagle’s medium/Ham’s F12 (Sigma-Aldrich, Irvine, UK) supplemented with 10% foetal calf serum (FCS), and 1 × penicillin and streptomycin. MCF10A cells were maintained in MEGM (mammary epithelial growth medium) (Lonza, Slough, UK) supplemented with 100 ng ml−1 cholera toxin. The human microvascular endothelial cells (HMVECs) (Life Technologies, Paisley, UK) were maintained using Medium 131 supplemented with the addition of Microvascular Growth Supplement (MVGS) in flasks treated with Attachment Factor Protein (Life Technologies). All cells were incubated at 37 °C, with 5% CO2 and 95% humidity.
RNA isolation
Total cellular RNA was isolated from the cultured breast cancer cells using TRI-Reagent RNA isolation agent (Sigma-Aldrich) according to the manufacturer’s protocol. The quality and concentration of RNA was determined through spectrophotometric measurement (NanoPhotometer, IMPLEN, Munich, Germany).
Real-time PCR-based miRNA detection
Expression of miRNAs in the breast cancer cohort and breast cancer cell lines was quantified using a Sybr-Green-based method described elsewhere (Balcells et al, 2011). Alternatively, the Megaplex TaqMan MicroRNA Arrays v2.0 and Megaplex Primer Pools following the manufacturer’s instructions (Life Technologies) have also been used. Expression of miRNAs in breast cancer cohort was normalised using U48 and calculated using a vector containing Podoplanin gene ranging from 101 to 108 copy numbers. Expression of miRNAs in breast cancer cells was relatively quantified using the 2−▵CT method (Pfaffl, 2001).
Cell treatment with miR mimics or inhibitors
All the synthetic miR inhibitors and mimics were purchased from Sigma–Aldrich (Irvine, UK). After cancer cells or endothelial cells reached the logarithmic growth phase, they were transfected with 25 nM of appropriate miRNA mimics or negative control using Dharmafect transfection reagents (GE Healthcare, Buckinghamshire, UK) following the manufacturer’s instruction. Dharmafect 1 reagent (2 μl ml−1) was used for MCF-7 cells, while Dharmafect 4 reagent (1 μl ml−1) was used for MDA-MB-231 and endothelial cells.
Electric cell-substrate impedance sensing assay
The electric cell-substrate impedance sensing (ECIS) Zθ instrument and 96W1E arrays (Applied Biophysics, Inc., Troy, NY, USA) were used for the measurement of spreading, attachment and migration behaviour of cancer and endothelial cells. Cells were seeded at a density of 4 × 104 cells per well and cultured at 37° for 24 h before the electric impedance of the cell layer was recorded. Each treatment group was set up with 6 repetitions. After differing time periods, the cell monolayer in each well was electrically wounded at 2600 μA for 20 s and post-wound migration was monitored at multiple frequencies in the ECIS system.
In-vitro scratch wound assay
Breast cancer cells were seeded into a 48-well plate at a density of 1 × 105 cells per well and allowed to form a monolayer, which was then scratched with a pipette tip to create a linear wound with a width of around 200 μm wide. Migration of cells to the wounding gap was monitored by serial time-lapse imaging using an EVOS FL imaging system (Life Technologies, Carlsbad, CA, USA) with a × 10 objective. Percentage of wound gap closure was measured using Image J software (National Institutes of Health, Bethesda, MD, USA; http://www.rsb.info.nih.gov/ij) and a customised macro.
Cell proliferation
The proliferation level of the cultured cells with response to various treatments was assessed using the AlamarBlue assay. Cells were seeded into 96-well plates at a density of 3000 cells per well and cultured overnight. After treatment with miR mimics or inhibitors for 48 h, 10 μl of the AlamarBlue reagent (Serotec, Ltd., Oxford, UK) was added to each well containing 100 ul of fresh culture medium. Cells were then incubated for 3 h at 37 °C. The fluorescence was measured with a fluorescence plate reader (Promega, Southampton, UK) with excitation at 544 nm and emission at 590 nm. The fluorescence value was proportional to the numbers of viable cells.
Apoptosis assay
Apoptosis of cells after miR treatment for 24 h was estimated using Caspase-Glo 3/7 Assay Kit (Promega, Madison, WI, USA) following the manufacturer’s instruction.
Luciferase activity analysis
The plasmid reporters which contained the predicted 3′-UTR sites in Wave-3 for miR-7 were analysed using a Dual-Glo luciferase assay system according to the manufacturer’s instructions (Promega). Briefly, plasmids and miR mimics (miR7 or control) were co-transfected into MCF10A cells using DharmaFECT Duo transfection reagent (Dharmacon, GE Healthcare, Hammersmith Imanet, UK). After 48 h, Firefly luciferase activity was measured using the Promega Dual-Glo luciferase kit and normalised to Renilla luciferase activity in GloMax plate reader (Promega).
Flow cytometric analysis
The molecular mechanism by which miRNA mimics regulate cancer cell behaviour were studied using flow cytometric analysis of cellular protein molecules. Cells were collected using HyQtase (Thermo Scientific, St Leon-Rot, Germany). They were then fixed with 1X fixation solution (eBioscience, San Diego, CA, USA) and permeabilised with 90–100% methanol. After blocking with 1%BSA solution, they were then stained with individual primary antibodies (1 : 100) and fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (1 : 600. Sigma) sequentially. The primary antibodies were anti-EGFR (SC-71034, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-IGF1R (SC-712, Santa Cruz Biotechnology), anti-pMDM2Ser166 (3521, Cell Signaling Technology, Inc., Beverly, MA, USA) and anti-Wave3 (2806, Cell Signaling Technology), respectively. Cells were then analysed using a flow cytometer (CANTO II, BD Biosciences, Oxford, UK).
In-vitro angiogenesis assay
A tubule-formation assay was performed to evaluate the effect of miRNA mimics on angiogenesis properties of endothelial cells. Briefly, Matrigel (BD Biosciences), a basement membrane matrix commonly used to study angiogenesis in vitro, was placed in a 24-well plate at 200 μl/well. The plate was then incubated at 37 °C for 1 h to allow Matrigel to solidify. HMVECs were then plated at a density of 1 × 105 cells per well in 450 μl of culture medium. Tubule formation of the endothelial cells Matrigel was monitored using an EVOS FL imaging system (Life technologies, Paisley, UK) with a × 4 objective. Tubule formation was then measured using Image J software (NIH).
Transwell Matrigel invasion and migration assays
An in vitro Matrigel invasion assay was used to assess the invasive capability of breast cancer and endothelial cells respectively. After cancer cells were transfected with miRNA mimic or negative control for 48 h, transwell inserts (8-μm pore sise. Greiner Bio-One Stonehouse, UK) for 24-well plates were pre-coated with 50 μl per insert of 1 mg ml−1 Matrigel (BD Bioscience), for 1 h at 37 °C. Cells were seeded into the upper chamber of each insert in 100 μl basal medium and 650 μl culture medium was added to each lower chamber. For investigation of tumour-endothelial cell interaction, tumour cell conditioned medium (TCM) was prepared from MCF-7 or MDA-MB-231 cells by collecting normal growth medium from 75 cm flasks of confluent cells. Medium was clarified by centrifugation.
To understand the transwell invasion (‘homing’) potential of endothelial cells after pre-treatment with miR-7 mimic with responding to TCM from breast cancer cells, HMVECs were pre-treated with 20 nM of mimics of negative control, miR-7 and miR-140-3p and incubated for 24 h, they were loaded onto the Matrigel-coated inserts as described above. And in the lower chamber of each insert, 50% of TCM was mixed with 50% of endothelial cell growth medium and used as a ‘chemoattractant’. After incubation at 37 °C for 24 h, HMVECs that penetrated the Matrigel-coated membrane and adhered to underside of the inserts were dissociated with cell dissociation solution (MerkMillipore, Watford, UK) containing 4 μg ml−1 Calcein AM (eBiosciences, Hatfield, UK) for 1 h at 37 °C. The solution containing invaded cells was transferred to a 96-well black-well plate at a volume of 100 μl per well. Invaded cells labelled with Calcein AM were then quantified using a fluorescence plate reader (Promega, Southampton, UK) with excitation at 490 nm and emission at 520 nm. Migration assay was performed similarly to the invasion assay described above, but in the absence of Matrigel.
RT–qPCR
Synthesis of cDNA was performed using 0.5 μg of RNA and a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Quantitative analysis of the gene transcripts was carried out using Amplifluor-based technologies, in which a 6-carboxy-fluorescine-tagged Uniprimer (Biosearch Technologies, Novato, CA, USA) was used as a probe. A pair of target specific primers were designed and the anti-sense primer incorporated a Z-sequence (5′-ACTGAACCTGACCGTACA-3′) which was specifically recognised by the probe (Myakishev et al, 2001). The primer sequences for the qRT-PCR are listed in Table 2. Real-time quantitative PCR was carried in an iCycler IQ5 thermocycler (Bio-Rad, Hemel-Hempstead, UK) using a PrecisionFAST qPCR MasterMixes (Primerdesign Ltd, Southampton, UK). Cycling conditions were 95 °C for 10 min, and 50 cycles of 95 °C for 10 s, 55 °C for 35 s and 72 °C for 10 s. An internal standard was used as control and GAPDH was used as the housekeeping gene for normalisation.
Table 2. Expression of miR-7, miR-221-5p and miR-339-5p in Human Breast Cancer.
|
miR-7 |
miR-221-5p |
miR-339-5p |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | N | Mean | s.d. | s.e. | P-value | N | Mean | s.d. | s.e. | P-value | N | Mean | s.d. | s.e. | P-value |
| Normal | 15 | 87.90 | 141.10 | 36.40 | 20 | 73.30 | 114.10 | 25.50 | 22 | 73.80 | 105.20 | 22.40 | |||
| Tumour | 66 | 57.89 | 67.39 | 8.29 | 80 | 56.73 | 62.08 | 6.94 | 80 | 43.45 | 49.18 | 5.50 | |||
| NPI1 | 34 | 69.60 | 82.70 | 14.20 | 38 | 66.00 | 78.40 | 12.70 | 39 | 51.20 | 62.80 | 10.10 | |||
| NPI2 | 18 | 54.10 | 56.60 | 13.30 | 26 | 51.57 | 48.08 | 9.43 | 25 | 38.89 | 35.57 | 7.11 | |||
| NPI3 | 12 | 35.68 | 15.10 | 4.36 | 0.028 vs NPI1 | 14 | 45.50 | 29.93 | 8.00 | 13 | 33.79 | 21.06 | 5.84 | ||
| Grade1 | 11 | 74.10 | 93.40 | 28.10 | 12 | 71.50 | 93.30 | 26.90 | 12 | 56.20 | 75.40 | 21.80 | |||
| Grade2 | 14 | 91.30 | 94.20 | 25.20 | 0.0106 vs Grade3 | 21 | 83.40 | 77.00 | 16.80 | 0.028 vs Grade3 | 22 | 63.00 | 61.00 | 13.00 | 0.026 vs Grade3 |
| Grade3 | 40 | 41.66 | 40.36 | 6.38 | 0.0382 vs Grade1 | 46 | 40.59 | 36.64 | 5.40 | 44 | 30.63 | 27.53 | 4.15 | ||
| TNM1 | 30 | 76.40 | 86.80 | 15.90 | 39 | 72.90 | 77.90 | 12.50 | 39 | 57.36 | 62.33 | 9.98 | |||
| TNM2 | 26 | 47.23 | 47.06 | 9.23 | 28 | 45.46 | 43.85 | 8.29 | 0.086 vs TNM3 | 28 | 33.20 | 32.04 | 6.05 | 0.043 vs TNM1 | |
| TNM3 | 6 | 29.96 | 4.44 | 1.81 | 7 | 30.54 | 3.30 | 1.25 | 0.0017 vs TNM1 | 6 | 22.38 | 2.03 | 0.83 | 0.0012 vs TNM1 | |
| TNM4 | 2 | 29.43 | 1.21 | 0.86 | 2 | 29.42 | 1.42 | 1.01 | 3 | 23.65 | 3.75 | 2.16 | 0.0021 vs TNM1 | ||
| Alive&Well | 48 | 64.10 | 77.30 | 11.20 | 58 | 59.75 | 70.18 | 9.21 | 59 | 45.97 | 55.57 | 7.23 | |||
| With distant metastasis | 4 | 44.03 | 16.68 | 8.34 | 5 | 39.33 | 12.25 | 5.48 | 5 | 29.59 | 10.52 | 4.71 | |||
| With local recurrence | 3 | 46.00 | 20.50 | 11.90 | 3 | 47.70 | 20.80 | 12.00 | 3 | 35.82 | 15.99 | 9.23 | |||
| Died of breast cancer | 8 | 32.63 | 12.75 | 4.51 | 11 | 50.30 | 38.80 | 11.70 | 10 | 37.71 | 27.90 | 8.82 | |||
| With incidence (incl. local recurrence) | 15 | 38.35 | 15.55 | 4.01 | 0.034 vs Alive and Well | 19 | 46.99 | 30.68 | 7.04 | 18 | 35.14 | 21.94 | 5.17 | ||
| With metastasis and death | 12 | 36.43 | 14.52 | 4.19 | 0.024 vs Alive and well | 16 | 46.85 | 32.74 | 8.19 | 15 | 35 | 23.4 | 6.04 | ||
Abbreviations: NPI=Nottingham Prognostic Index (1: <3.4 (good prognosis); 2: 3.4–5.4 (moderate prognosis); 3: >5.4 (poor prognosis)); TNM=tnm classification of malignant tumours.
Statistical analysis
All experimental data were presented as mean±s.d. unless indicated otherwise. The statistical analysis of miR expression in human breast cancer specimens was carried out with Minitab version 14.1 (Minitab Ltd., Coventry, UK) using a custom written macro and two sample comparison. The statistical comparisons of other assays were performed using the SPSS version 20 for Windows (SPSS, Chicago, IL, USA). The significance of differences in the ECIS data was analysed using the repeated-measures (RM) ANOVA. One-way analysis of variance (ANOVA) for other multiple group data. Two group comparison was analysed using Student’s t-test if data were normally distributed (via Shapiro-Wilk W test) or Mann–Whitney U-test if data are not normally distributed. Differences were considered statistically significant when P-values<0.05.
Results
Expression of miR-7 is negative associated with the stage, grade and survival of the breast cancer patients
We evaluated expression profile of a panel of miRs in a breast cancer cohort with 10-year follow-up clinical data. As shown in Table 2, the expression of miR-7 in the breast cancer cohort is negatively associated with the Nottingham Prognostic Index (NPI)-indicated prognosis (P=0.028, NPI3 vs NPI1), grade (P=0.0382, Grade 3 vs Grade 1), and survival (P=0.024, MET+DEAD vs SURV1), respectively. We also observed that miR-221-5p expression appeared to be negatively associated with higher grade (P=0.028, Grade 3 vs Grade 2), and later stage (P=0.017, TNM3 vs TNM1) although the sample size of TNM3 was limited (n=7). Likewise, miR-339-5p expression was also negatively associated with higher grade (P=0.026, Grade 3 vs Grade 2), and later stage (P=0.043, TNM2 vs TNM1), respectively. Therefore, these three miRNAs were considered as lead candidates in the following research.
Differential expression of miRNAs in the breast cancer cells
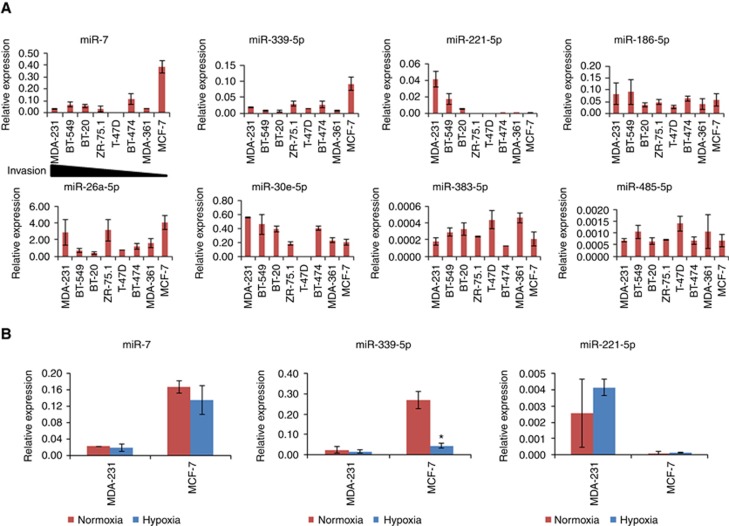
Through multiplex miRNA gene expression assays, we found that there was differential expression of eight miRNAs in a panel of 8 breast cancer cell lines with different invasion potential (Figure 1A, ordered with their invasion potential). MCF-7 (luminal) cells have the least invasion potential but showed at least a threefold increase in miR-7 expression compared to all the other breast cancer cells screened (for example, P<0.0001 MCF-7 vs MDA-MB-231 cells). miR-339-5p was also expressed strongly in MCF-7 cells compared to other breast cancer cells (P<0.0001 MCF-7 vs MDA-MB-231 cells). miR-221-5p was only detected in the highly invasive cells MDA-MB-231 (basal-like), BT-549 and BT-20 cells. miR-186-5p was expressed at similar levels in all the breast cancer cells, miR-26a-5p was expressed relatively strongly in MCF-7, ZR-75.1 and MDA-MB-231 cells compared to the other cells tested. miR-30e-5p expression level was higher in more invasive MDA-MB-231, BT-549 and BT-20 cells than the less invasive cells. T-47D did not show expression of miR-30e-5p and miR-7, but did show high expression of miR-383-5p and miR-485-5p.
Figure 1.
miRNA expression in breast cancer cells and post-wound invasion of breast cancer cells following treatment with miRNA mimics monitored by the ECIS system. (A) Differential expression of miRNAs in breast cancer cells was quantified using the Megaplex TaqMan MicroRNA Arrays v2.0 and Megaplex Primer Pools following the manufacturer’s instructions. Small RNA U48 was used to normalise the expression data. (B) post-wound invasion of breast cancer cells. Cells were transfected in an ECIS plate with miRNA mimics and negative controls (25 nM each), respectively. The electric wound was performed at 25 h and 36 h (2600 μA for 20 s). The signal of impedance was continuously recorded in the ECIS system. *P<0.05. A full colour version of this figure is available at the British Journal of Cancer journal online.
As hypoxia may play a key role in a tumour microenvironment, we investigated whether it could alter the expression of miRNA in invasive and non-invasive breast cancer cells (MDA-MB-231 and MCF-7). We found that hypoxia did not significantly alter miR-7 or miR-221-5p expression in MDA-MB-231 and MCF-7 breast cancer cells. In contrast, although miR-339-5p expression was not changed in MDA-MB-231 cells, the high expression in MCF-7 cells was significantly down-regulated (P<0.05 vs normoxia) in response to hypoxia (Figure 1B).
MDA-MB-231 cells are more sensitive than MCF-7 with response to the treatment of miR-7 which exerts inhibitory effect on motility and wound healing potentials
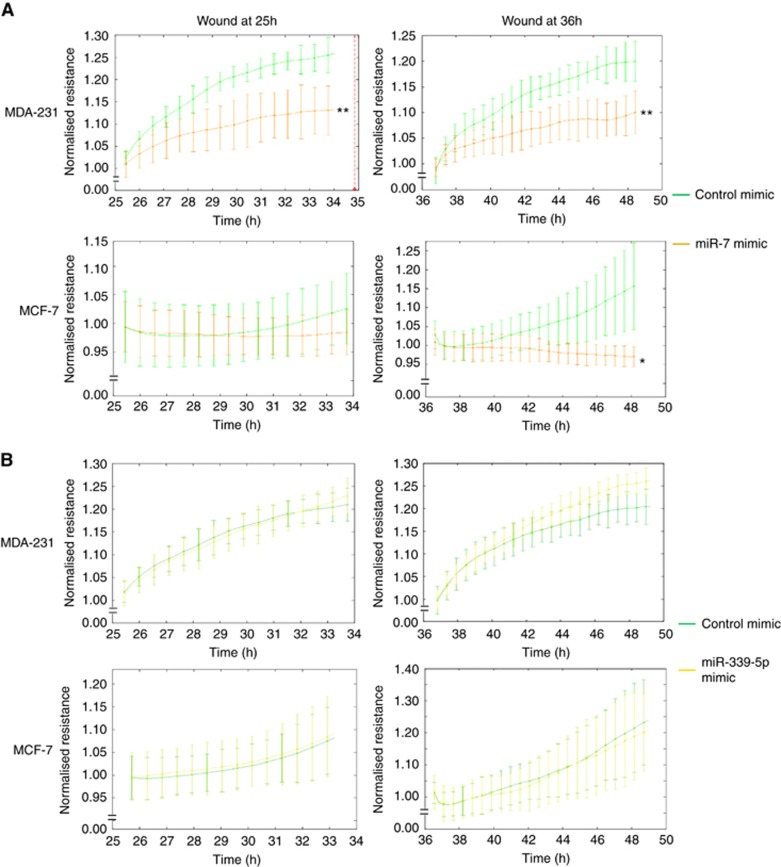
To understand whether miRNAs can have an effect on invasion of breast cancer cells, we used the ECIS system to monitor cells behaviour following transfection of cells with synthetic miRNA mimics. As shown in Figure 2A, we found that miR-7 inhibited the post-wound migration of MDA-MB-231 cells after electric wound at 25 h (P<0.01 vs mimic control) and at 36 h (P<0.01 vs mimic control), respectively. miR-7 mimic did not show any effect on migration of MCF-7 after electric wound at 25 h, but began to show inhibitory effect on migration of MCF-7 cells after electric wound at 36 h (P<0.05 vs mimic control). In contrast, miR-339-5p mimic did not show any effect on migration of MDA-MB-231 cells and MCF-7 cells after wound at the two time points, respectively (Figure 2B).
Figure 2.
Post-wound invasion of breast cancer cells following treatment with miRNA mimics monitored by the ECIS system. The electric wound was performed at 25 h and 36 h (2600 μA for 20 s) in the system. The signal of impedance was continuously recorded in the ECIS system. (A) Treatment with miR-7 and control mimics, respectively. (B) Treatment with miR-339-5p and control mimics, respectively. *P<0.05, **P<0.01. A full colour version of this figure is available at the British Journal of Cancer journal online.
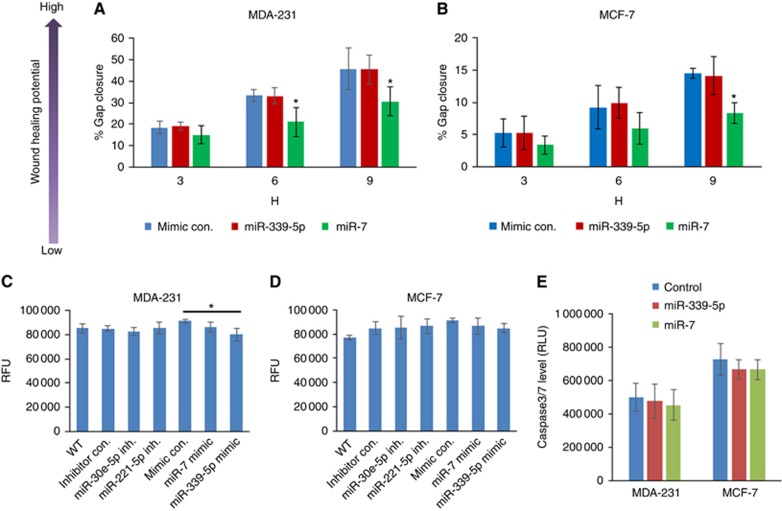
The ability of the miR-7 mimic to inhibit the wound healing potential of cancer cells was confirmed using a manual scratch assay. As shown in Figure 3A and B, after treatment for 48 h using miR mimics, both MDA-MB-231 and MCF-7 cells showed reduced wound healing capacity. However, miR-7 appeared to exert inhibitory effect earlier on MDA-MB-231 cells than on MCF-7 cells. The scratch assay also confirmed that miR-339-5p had no effect on wound healing in both MDA-MB-231 and MCF-7 cells in contrast.
Figure 3.
Scratch wound healing, proliferation and apoptosis of breast cancer MDA-MB-231 and MCF-7 cells responding to the miRNA mimic treatment. (A and B) Scratch wound healing capacity. Cells were treated with individual miRNA mimics for 48 h. The monolayer of the cells were then scratched with a pipette tip for a line gap. The 48-well plate containing scratched cells were loaded on to an EVOS system and monitored by time lapse. The gap closure was then automatically measured using a home-created macros in ImageJ. (C and D) Proliferation determined with AlamarBlue assay. (E) Apoptosis estimated using Caspase-Glo 3/7 Assay. The data shown are means±s.d. (n=4). *P<0.05 vs mimic control at the same time point. A full colour version of this figure is available at the British Journal of Cancer journal online.
miR-7 does not have an effect on proliferation and apoptosis in MDA-MB-231 and MCF-7 cells
We investigated whether the observed inhibitory effect on cell migration could also have been attributable to the miRNA effect on cell proliferation or viability after transfection. 48 h treatment with synthetic miR-7 did not inhibit proliferation in MDA-MB-231 or MCF-7 cells (Figure 3C and D, respectively). However, 48 h treatment with the miR-339-5p mimic showed a small inhibitory effect on proliferation of MDA-MB-231 cells but not MCF-7 cells.
We further estimated the apoptosis level of breast cancer cells with response to the treatment of miRNA mimics for 48 h. Caspase 3/7 Glo assay showed that miR-7 and miR-339-5p had no effect on apoptosis of the breast cancer cells (Figure 3E).
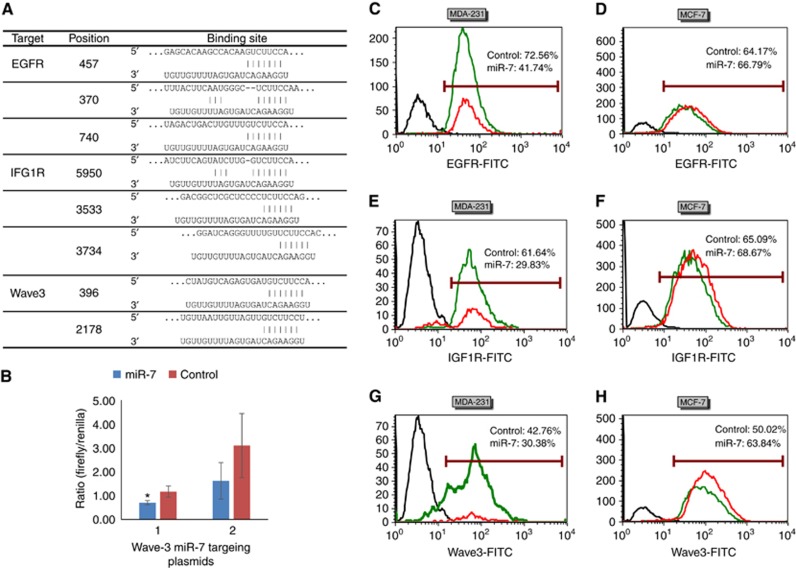
miR-7 regulates expression of IGF-1R, EGFR and WAVE3 in MDA-MB-231 but not MCF-7 cells
Using online searching tools including TargetScanHuman 7.0 (http://www.targetscan.org) and miRBase 21 (http://www.mirbase.org), the predicted binding sites of miR-7 in potential signalling targets were identified (Figure 4A). As the targeting of miR-7 to IGFR and IGF1R has been previously reported (Li and Carthew, 2005; Jiang et al, 2010), we therefore only performed the validation of the predicted target sites in Wave3 for miR-7 using the luciferase assay. As shown in Figure 4B, in the presence of the luciferase plasmid containing the target site at Position 396 in Wave3, The luciferase signal was reduced significantly with the response to miR-7 mimic (P<0.05, vs control). In the presence of the luciferase plasmid containing the target site at Position 2178 in Wave3, although it appeared that the luciferase signal was reduced with the response to miR-7 mimic, there was no significance.
Figure 4.
Protein level of IGF1R, EGFR and wave3 in MDA-MB-231 and MCF-7 cells with response to the treatment of miR-7 Mimic for 48 h. (A) Predicted 3’-UTR sites in the three genes for miR-7 using online tools. (B) Validation of the miR-7 target sites in Wave3 using luciferase assay. (C–H) Expression of the cellular protein molecules determined using flow cytometry after immunostaining. The primary antibodies used were anti-EGFR (SC-71034. Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-IGF1R (SC-712. Santa Cruz Biotechnology), and anti-Wave3 (2806. Cell Signaling Technology), respectively. Isotype antibodies were used as background control. Black histogram: background signal after staining with an isotype primary antibody. Green histogram: Protein expression after treatment with mimic control. Red histogram: protein expression after treatment with miR-7 mimic. Percentages of protein expression was shown after gating against background noise in the overlay plots. (C and D) EGFR expression. (E and F) IGF1R expression. (G and H) Wave3 expression. *P<0.05. A full colour version of this figure is available at the British Journal of Cancer journal online.
FACS analysis showed that the tyrosine kinase receptors EGFR, IGF1-R and WAVE 3 (downstream effector of actin polymerisation) were expressed in both MDA-MB-231 and MCF-7 cells. 48 h treatment of these breast cancer cells with 25 nM miR-7 mimic compared to a mimic control caused the down-regulation of EGFR (30%, Figure 4C), IGF-1R (32%, Figure 4E) and WAVE-3 (12%, Figure 4G) in MDA-MB-231 cells, but had no effect on these proteins in MCF-7 cells (Figure 4D, F and H).
We further evaluated the endogenous gene expression of IGF-1, EGFR and WAVE3 in the two cell lines by real-time qPCR. The gene expression of miR-7 targets, EGFR and WAVE3, were significantly higher in MDA-MB-231 cells compared to MCF-7 cells (P<0.05, respectively). This may partially explain why miR-7 mimic was more potent in MDA-MB-231. However, we cannot exclude other possible mechanisms, as in our gene expression data, the gene expression level of IGF-1R was the same in two cell lines, and also ERK, STAT3, mTOR genes were all overexpressed in MDA-MB-231 cells compared to MCF-7 cells (Supplementary Figure 1).
miR-7 mimic inhibits post-wound migration, proliferation and tubule formation of HMVEC endothelial cells
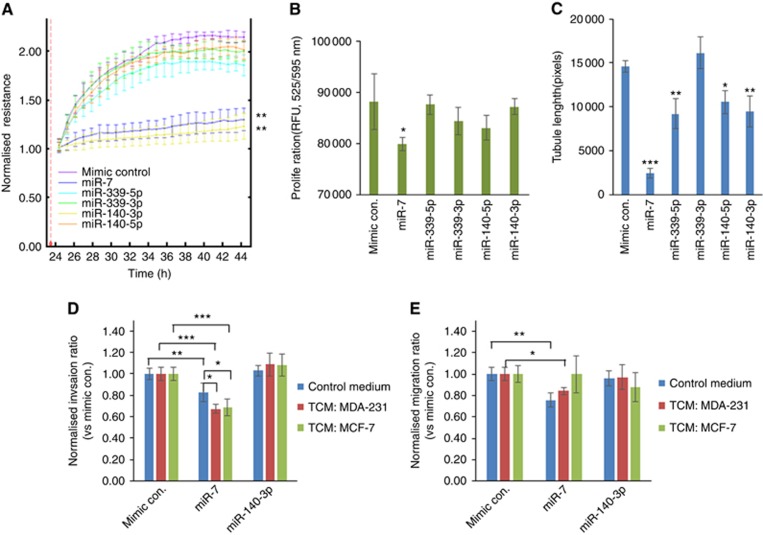
We used the ECIS system to investigate whether miR-7 had an effect on the migration capacity of human endothelial cells which plays the key role in tumour angiogenesis. Confluent monolayers of HMVEC cells were transfected with miRNA mimic or appropriate control for 48 h prior to the creation of an electric wound. As shown in Figure 5A, miR-7 and miR-140-3p mimics significantly inhibited the post-wound migration (RM ANOVA P<0.0001 vs mimic control, respectively). However, the mimics of miR-339-5p, miR-339-3p and miR-140-3p did not show any effect on HMVEC migration.
Figure 5.
miR-7 mimic exerts an inhibitory effect on post-wound migration, proliferation and In-Vitro angiogenic potential of HMVEC endothelial cells. Cells were pre-treated with miR mimics before the cellular assays. (A) Effect of miR mimics on post-wound migration of HMVECs indicated by the ECIS system. Normalised resistance is proportional to cell migration capacity after electric wound (2600 μA for 20 s). Data are means±s.d. with 6 duplication. (B) Effect of miR mimics on proliferation of HMVECs. AlamarBlue assay was used to estimate proliferation. Data are means±s.d. with 6 duplication. (C) Effect of miR mimics on in-vitro angiogenesis of HMVECs. Tubule formation of the HMVECs on Matrigel was monitored using an EVOS FL imaging system with a × 4 objective. Data are means±s.d. with triple tests with 4 captured images per test. (D and E) Pre-treatment with miR-7 reduces transwell (8-μm pores) invasion and migration capacities of HMVECs with response to the Tumour conditioned medium (TCM) of MDA-MB-231. *P<0.05, **P<0.01, ***P<0.001 vs the mimic control or as indicated with a line. A full colour version of this figure is available at the British Journal of Cancer journal online.
We further evaluated the effect of miR mimics on the proliferation of HMVECs using the AlamarBlue cell proliferation assay. As shown in Figure 5B, 48 h treatment with miR-7 inhibited the proliferation of HMVECs by approximately 12% (P<0.05 vs mimic control). Other miR mimics examined including miR-339-5p, miR-339-3p, miR-140-5p, miR-140-3p did not show any effect on proliferation.
As tubule formation on Matrigel can indicate the angiogenic capacity of endothelial cells in vitro, we performed this assay on HMVECs following 48 h treatment with miR mimics. HMVEC control cells formed large tubules on Matrigel after 18 h. As shown in Figure 5C, miR-7 significantly inhibited the tubule formation of HMVECs on Matrigel by approximately 83% (P<0.0001 vs mimic control). Tubule formation of HMVECs was also inhibited by other miR mimics including miR-339-5p (∼36%, P<0.001), miR-140-5p (∼21%, P<0.05) and miR-140-3p (∼32%, P<0.01) at different levels. miR-339-3p did not show any effect on HMVEC tubule formation.
Endothelial cell migratory and invasive behaviours in response to TCM from breast cancer cells can be inhibited by the miR7 mimic
We further investigated the indirect tumour-endothelium interaction using the transwell insert system. As shown in Figure 5D, HMVECs did not show difference of transwell invasion through Matrigel with response to basal control medium, TCM from MDA-MB-231 and TCM from MCF-7, respectively. However, HMVECs treated with miR-7 mimic showed a decreased level of transwell invasion with response to basal control medium (P<0.01), TCM from MDA-MB-231 (P<0.0001) and TCM from MCF-7 (P<0.0001), respectively. Interestingly, miR-7 showed an additional inhibitory effect of HMVEC invasion in the presence of TCM from MDA-MB-231 (P<0.05 vs control medium) and MCF-7 cells (P<0.05 vs control medium), respectively. In contrast, we investigated the miR-140-3p mimic for its effect on invasion of HMVECs, and found no significance, which was independent of the medium used as chemo attractant.
In a parallel experimental manner, we investigated the transwell migration of HMVECs after miRNA treatment. As shown in Figure 5E, after the treatment with miRNA mimic, we did not find any change in the migration of HMVEC s with response to TCM from MDA-MB-231 and MCF-7 cells. On the contrary, miR-7 decreased the migration capacity of HMVECs with response to basal medium (P<0.01 vs mimic control) and TCM from MDA-MB-231 cells (P<0.05 vs mimic control), respectively. In the presence of TCM from MCF-7 cells, no inhibitory effect of miR-7 on HMVEC migration was observed. Again, miR-140-3p mimic did not show significant effect on transwell migration of HMVECs independently of the medium used as chemo attractant.
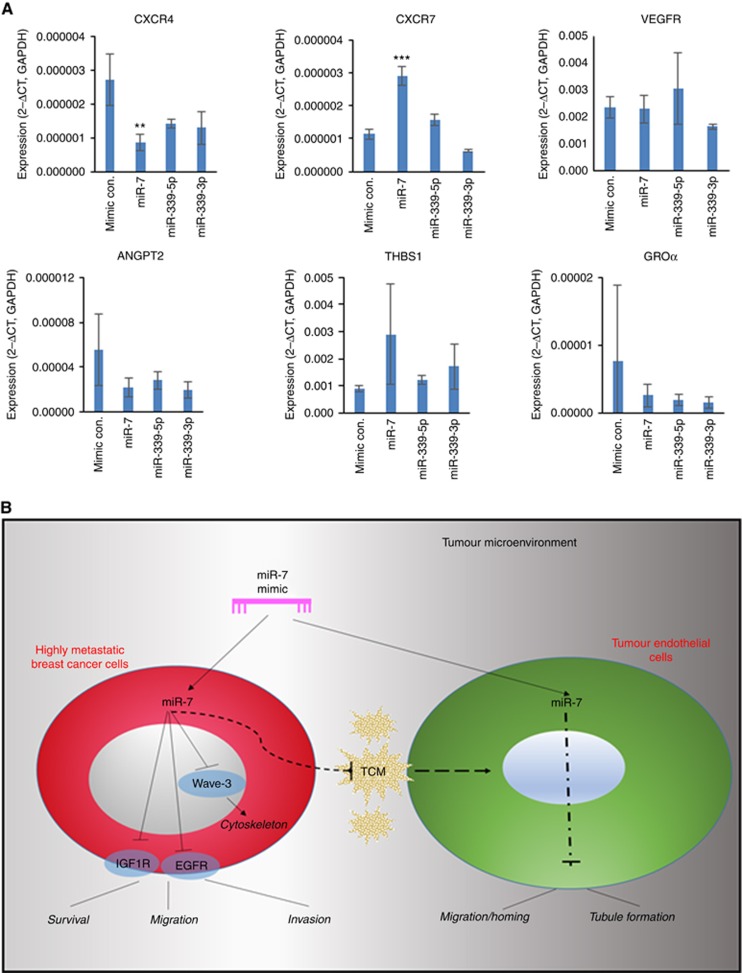
miR-7 modulates transcription of CXCR4 and CXCR7 in HUVEC cells
As the angiogenic potential of endothelial cells is regulated by a variety of angiogenic-associated molecules, we used qPCR to analyse the gene expression profile of those molecules in HMVEC cells with response to the treatments of miR mimics. As shown in Figure 6A, basal HMVEC cells expressed genes including the chemokine receptors CXCR4 and CXCR7, VEGFR, angiopoietin-2 (ANGPT2), thrombospondin-1 (THBS1) and the chemokine GRO alpha (CXCL1). Interestingly, the qPCR data indicated that miR-7 downregulated expression of the CXCR4 gene (P<0.01 vs mimic control) but upregulated expression of CXCR7 (P<0.01 vs mimic control). Treatment with miR-339-3p or miR-339-5p did not significantly affect expression of any of the genes investigated.
Figure 6.
Expression of angiogenic-related genes in HMVECs following microRNA mimic treatment and the proposed mechanism underlying the dual inhibitory effect of miR-7 under tumour microenvironmental conditions. (A) Gene expression in HMVECs. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalise gene expression results. Data are means±s.d. with triple tests. Significance of the data among multiple groups was tested using ANOVA. **P<0.01, ***P<0.001 vs the mimic control. (B) Proposed molecular mechanism. A full colour version of this figure is available at the British Journal of Cancer journal online.
Discussion
Breast cancer is a heterogeneous disease and the development of personalised treatments in the future will depend on the identification of prognostic and predictive gene expression signatures which may complement traditional clinic-pathological parameters (Reis-Filho and Pusztai, 2011). Non-coding miRNAs are a large group of molecules with over 1400 human miRNAs currently identified. Our study focussed on the differential expression of miRNAs in breast cancer types and aimed to identify a link between specific miRNAs and the invasiveness or metastatic potential of the tumour cells. Gene expression assays showed that miR-7 and miR-339-5p had high expression in MCF-7 cells, but very low expression in MDA-MB-231 cells. Our cohort data also showed that miR-7 is negatively associated with prognosis, grade, survival and metastasis of breast cancer patients, whereas miR-339-5p is negatively associated with grade and stage of the patients. This pattern of miR-7 expression has similarly been reported in a breast cancer cell line miRNA expression profiling study (Riaz et al, 2013). This large study showed that the miRNA transcriptome is complex, but can be broadly sub-divided into miRNAs expressed in luminal-type hormone receptor positive cells and ER negative, often triple negative, epithelial-mesenchymal transition type (EMT) cells in typically more aggressive tumours. As miRNA expression predominantly mirrors ER expression, it has been suggested that a significant number of miRNAs are under control of ER regulation and in MCF-7 cells, miR-7 has been identified as an important miRNA significantly induced by oestradiol, the most potent oestrogenic hormone (Masuda et al, 2012). Since miR-7 and miR-339-5p seem to be expressed at higher levels in the non-invasive MCF-7 cells, this suggests that high levels of these miRNAs may play a role in inhibiting invasion. In breast cancer cells, miR-7 has been shown to inhibit the expression of Pak-1, an invasion promoting kinase and transfection with miR-7 was found to decrease invasion in MDA-MB-231 cells (Reddy et al, 2008). However, the role of miR-7 is likely to be more complex, as other reports describing detailed analysis in primary ER positive breast cancers have shown that high expression of four miRNAs, including miR-7, were in contrast, linked to early relapse and reduced time to distant metastasis (Foekens et al, 2008). Hypoxia is the consequence of an aggressive tumour microenvironment and a functional link has previously been demonstrated between hypoxia and micro RNA expression, with a defined group of miRNAs showing increased expression in response to low oxygen (Kulkarni et al, 2012). Despite this, we did not observe any effect of hypoxia on the expression of miR-7 or miR-339-5p in MDA-MB-231 cells.
Our finding of increased miR-339-5p in MCF-7 cells has similarly been reported in a study where it was shown to be expressed 5-10 fold higher in these poorly invasive cells compared to highly invasive MDA-MB-231 and MDA-MB-468 cells, where increased miR-339-5p inhibited cell migration (Wu et al, 2010). miR-339-5p has been shown to be down-regulated in several cancers, including breast cancer, lung cancer and hepatocellular carcinoma and its expression level may be used as a biomarker to predict lymph node metastasis and prognosis in cancers (Rani et al, 2013; Wang et al, 2014, 2015). Our results have also shown that miR-339-5p expression could be reduced in MCF-7 cells grown under hypoxic stress as may be expected in an aggressive tumour environment.
Expression of miR-221-5p appeared to be expressed more strongly in ER negative cells but is also correlated strongly with invasive potential, with the most invasive ER negative cell lines, MDA-MB-231 and BT-549 expressing the highest levels in our study. This result fits in reassuringly with previous findings that miR-221 is negatively regulated by ERα, hence the low expression we found in ER positive cells. Increased expression of miR-221 has been shown to induce genes involved in invasion and survival, generating a more metastatic tumour-type. It has even been suggested that the direct interaction of miR-221 with ER can cause a phenotypic shift of ER-positive cells to ER-negative more aggressive cells (Di Leva et al, 2010; Riaz et al, 2013). Analysis of breast cancer tissue has also confirmed that high miRNA-221 expression correlates with advanced clinical stage and poor prognosis, suggesting its possible role as a biomarker for predicting patient survival (Eissa et al, 2015). Our results suggest that expression of miR-7, miR-339-5p and miR-221-5p may represent a powerful diagnostic tool for predicting metastasis in breast cancer patients.
Like miR-221, miR-30e-5p was also found in our study to be upregulated in MDA-MB-231 cells compared to MCF-7, however these results do not closely mirror reported findings in tissue samples. In contrast, in breast cancer tissue, downregulation of miR-30e has been defined as ‘risk-associated’ and shown to correlate with worse patient outcome (Gasparini et al, 2014). Increased miR-30e-5p expression has therefore been associated with good prognosis and considered as a protective prognostic factor in lymph node-negative breast cancer patients with ER+/ERBB2− tumours (D’Aiuto et al, 2015) and its expression level is believed to be useful as a powerful diagnostic tool (Gasparini et al, 2014).
Our study enabled us to compare the functional effect of increased miR-7 or miR-339-5p expression in an invasive triple negative (ER−, PR− and HER−) MDA-MB-231 and a non-invasive double positive (ER+, PR+ and HER−) MCF-7 cell line. Cell migration was measured in a label-free, real-time fashion using the ECIS system and with a wound-healing (scratch) assay to evaluate the overall physical gap-closure properties of cancer cells. Increasing the miR-7 level by transfection with synthetic miR-7 mimic caused a significant decrease in MDA-MB-231 migration measured by both ECIS and scratch assays, whereas an increase in miR-339-5p had no effect. Post-wound migration in MCF-7 cells, as measured by ECIS, was not changed by addition of extra miR-7 mimic at an earlier period. The inhibition detected in MCF-7 cells in the scratch assay can be observed later than that in MDA-MB-231 cells. This confirms that miR-7 expression can play a role in inhibiting breast cancer invasive capability and acting as a tumour suppressor as has been previously described (Reddy et al, 2008), but also suggests that miR-339-5p does not have the same effect in our MDA-MD-231 cells. We propose that invasive breast cancers may be more sensitive than less aggressive cells to treatment by miR-7 mimic as a micro RNA-based anti-tumour therapy.
Bioinformatic approaches were used to find signalling proteins with potential binding targets for miR-7, enabling EGFR, IGF-1R and WAVE3 to be identified. Both human EGFR and IGF-1R mRNA 3′-untranslated regions contain three miR-7 target sites. We observed that transfection with mimic miR-7 suppressed expression of EGFR, IGF-1R and WAVE3 in MDA-MB-231 cells but did not change expression of these proteins in MCF-7 cells. The tyrosine kinase receptors, EGFR and IGF-1R are oncogenes frequently observed to be mutated or amplified in a wide range of solid tumours and have been considered as an important therapeutic target for clinical studies partly because of their involvement in human cancer progression and metastasis (van der Noll et al, 2015). In cancers including breast, lung, colorectal and glioblastoma it has been reported that miR-7 inhibits EGFR and its downstream signalling pathways [31–33]. These include raf1-erk1/2, crucial for proliferation, and PI3K/AKT and PAK1 pathways that both play a crucial role in cancer cell migration and invasion (Liu et al, 2014; Zhou et al, 2014; Suto et al, 2015). Interestingly, our data suggest that miR-7 is more likely to regulate mechanisms controlling migration and invasion instead of proliferation and apoptosis in aggressive breast cancer cells. In ovarian and gastric cancer studies, overexpression of miR-7 has been shown to inhibit metastasis and to reverse epithelial-mesenchymal transition (EMT) caused by EGFR and IGF1-R downregulation (Zhou et al, 2014). In addition to directly inhibiting EGFR expression, reports have also demonstrated that miR-7 transfection can independently inhibit downstream signalling pathways including PAK1 expression by targeting the 3′-untranslated region of Pak1 mRNA (Reddy et al, 2008) and AKT through targeting upstream regulators (Kefas et al, 2008). WAVE3 is part of a family of scaffold proteins that can link upstream receptor tyrosine kinase signals to the activation of the ARP2/3 complex, leading to a burst of actin polymerisation. This is crucial for the reorganisation of the cytoskeleton in cell movement and hence plays a role in cancer cell invasion (Takenawa and Suetsugu, 2007). It interacts with the PI3kinase pathway and acts as a pro-metastatic molecule in breast cancer (Sossey-Alaoui et al, 2005; Kulkarni et al, 2012; Taylor et al, 2013) since knock-down of WAVE3 results in decreased cell invasion and metastasis (Sossey-Alaoui et al, 2007). Our results have shown for the first time that WAVE3 is another downstream signalling protein involved in cell migration that appears to be directly targeted by miR-7.
Expression of WAVE3, IGF-1R and EGFR were all significantly inhibited by the addition of mimic miR-7 to MDA-MB-231, while it had no effect on the levels of these proteins expressed in MCF-7 cells. One explanation for the difference in sensitivity to miR-7 may be that MCF-7 cells express high levels of miR-7 possibly via ER signalling (Masuda et al, 2012) and have developed compensatory mechanisms that enable the cells to maintain WAVE3, IGF-1R and EGFR protein levels. The ability of miR-7 to reduce the migratory capability in MDA-MB-231 cells but not MCF-7 cells may also be partly explained by the fact that miR-7 only reduces the level of these proteins, which have a role in cell migration, in MDA-MB-231 cells.
Tumour-associated angiogenesis is a critical step during the metastatic process, we therefore investigated whether microRNAs could also be used as anti-angiogenic strategies to control tumour spread. We found that adding mimic miR-7 significantly inhibited the ability of the HMVECs to migrate, proliferate and reduced tubule formation by ∼80%, suggesting this microRNA may be used to inhibit angiogenesis. In vivo models have previously identified miR-7 as a potent negative regulator of angiogenesis producing effects similar to the anti-angiogenic drug sunitinib (Babae et al, 2014). Our results also showed that although having no effect on proliferation, miR-140-30p was another microRNA with anti-angiogenesis potential as it also demonstrated significant inhibition of HMVEC migration and tubule forming ability. Pre-treatment of HMVECs with miR-7 reduced the ability of the cells to migrate and invade in response to signals from breast cancer cells. Importantly, these results imply that in addition to inhibiting angiogenic potential, miR-7 may be an inhibitor of the homing and migration of endothelial cells in response to factors released by aggressive breast tumours. Results showed that expression of a selection of pro or anti-angiogenic factors (ANGPT-2, GROa (CXCL1), TSP-1) normally secreted by endothelial cells did not appear to be directly regulated by treatment with miR-7. However, treatment of endothelial cells with miR-7 significantly regulates expression of the chemokine receptors CXCR4 and CXCR7. Both of these transmembrane G-protein coupled proteins are receptors for the chemokine ligand CXCL12 (SDF-1α) which triggers signalling via PI3kinase/Akt and erk1/2 to induce chemotaxis (Würth et al, 2014). CXCR4/CXCL12 axis is recognised to play an important role in breast cancer metastasis and angiogenesis, as CXCL12 is produced by stromal cells and is expressed at high levels in the main sites of breast cancer metastasis (lymph nodes, bone marrow, liver and lung; Müller et al, 2001). Chemokine receptors are widely expressed on hematopoietic and endothelial cells, but CXCR4 is also known to be overexpressed on breast cancer cells, a factor that has been correlated with metastasis and poor patient survival (Müller et al, 2001; Zhang et al, 2014). CXCR4 has a predicted binding site for miR-7 in the 3’untranslated region, therefore the downregulation of this protein in endothelial cells by the addition of miR-7 may be considered to be one of the mechanisms that decreased the HMVEC ability to migrate in response to chemoattractants produced by breast cancer cells. The upregulation of CXCR7 by miR-7 did not prevent the reduced chemotaxis in HUVEC cells caused by miR-7, indeed it may be postulated that since CXCR7 has been reported to have a function in scavenging CXCL12 and activating b-arrestin to internalise CXCR4, upregulation by miR-7 may have augmented the reduction in CXCR4 signalling (Würth et al, 2014). A number of microRNAs have previously been shown to target CXCR4 and to supress migration in cancer cells including nasopharangeal carcinoma (Lu et al, 2014), colon cancer (Li et al, 2013) and breast cancer (Liang et al, 2007). miR-126 has also been shown to up and down-regulate CXCR4 and CXCL12 in HUVECs (Mondadori dos Santos et al, 2015). To the best of our knowledge this is the first report to show that miR-7 can downregulate CXCR4 and inhibit endothelial cell chemotactic response to breast cancer cells.
In conclusion, we identify that miR-7 is expressed at a low level in highly invasive cells, and when added as a form of mimic, it appears to play a more potent role in inhibition of invasive breast cancer cell migration. More importantly, miR-7 mimic indeed has a dual effect as it also significantly inhibits the proliferative, chemotactic and angiogenic-like homing characteristics of endothelial cells especially in response to chemoattractant factors produced by aggressive breast cancer cells. As illustrated in Figure 6B, our findings in this study suggests that miR-7 may be developed as an anti-cancer therapeutic potentially capable of supressing breast cancer metastasis and tumour-associated angiogenesis simultaneously.
Conclusions
This is the first report demonstrating that the highly metastatic MDA-MB-231 cells are more sensitive to the inhibitory effect of miR-7 mimic treatment than the poorly invasive MCF-7 cells. This distinguishable response may be due to the downregulation of EGFR, IGF1R and Wave3 in breast cancer cells. miR-7 also inhibits the proliferation, migration and invasion of endothelial cells. More importantly, miR-7 suppresses the homing and migration of endothelial cells to more aggressive tumour cell conditions, suggesting its dual-inhibitory role in tumour microenvironment and potential therapeutic value.
Acknowledgments
We thank the University Hospital of Wales for the collection of the breast cancer patient specimens and the Central Biotechnology Services for providing the flow cytometry facility. This work was supported by Cancer Research Wales (201109) and Welsh Life Science National Research Network (02072015CMMP).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Babae N, Bourajjaj M, Liu Y, Van Beijnum JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH, Van Haastert RJ (2014) Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget 5(16): 6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG (2012) miR-34–a microRNA replacement therapy is headed to the clinic. Front Genet 3: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells I, Cirera S, Busk PK (2011) Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol 11(1): 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y (2012) The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486(7403): 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aiuto F, Callari M, Dugo M, Merlino G, Musella V, Miodini P, Paolini B, Cappelletti V, Daidone M (2015) miR-30e* is an independent subtype-specific prognostic marker in breast cancer. Br J Cancer 113(2): 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H (2010) MicroRNA cluster 221-222 and estrogen receptor α interactions in breast cancer. J Natl Cancer Inst 102(10): 706–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa S, Matboli M, Sharawy A, El-Sharkawi F (2015) Prognostic and biological significance of microRNA-221 in breast cancer. Gene 574(1): 163–167. [DOI] [PubMed] [Google Scholar]
- Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y (2013) Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 24(4): 542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015. a) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5): E359–E386. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015. b) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136(5): E359–E386. [DOI] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW (2008) Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA 105(35): 13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, Huebner K (2014) microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 5(5): 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466(7308): 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol Oncol 6(6): 590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, Kolokythas A, Wang A, Dai Y, Zhou X (2010) MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J 432(1): 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68(10): 3566–3572. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Augoff K, Rivera L, McCue B, Khoury T, Groman A, Zhang L, Tian L, Sossey-Alaoui K (2012) Increased expression levels of WAVE3 are associated with the progression and metastasis of triple negative breast cancer. PLoS One 7(8): e42895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Carthew RW (2005) A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123(7): 1267–1277. [DOI] [PubMed] [Google Scholar]
- Li Z, Li N, Wu M, Li X, Luo Z, Wang X (2013) Expression of miR-126 suppresses migration and invasion of colon cancer cells by targeting CXCR4. Mol Cell Biochem 381(1-2): 233–242. [DOI] [PubMed] [Google Scholar]
- Liang Z, Wu H, Reddy S, Zhu A, Wang S, Blevins D, Yoon Y, Zhang Y, Shim H (2007) Blockade of invasion and metastasis of breast cancer cells via targeting CXCR4 with an artificial microRNA. Biochem Biophys Res Commun 363(3): 542–546. [DOI] [PubMed] [Google Scholar]
- Liu Z, Jiang Z, Huang J, Huang S, Li Y, Yu S, Yu S, Liu X (2014) miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int J Oncol 44(5): 1571–1580. [DOI] [PubMed] [Google Scholar]
- Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang L, Xu X, Peng X, Li G, Tian W (2014) miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis 35(3): 554–563. [DOI] [PubMed] [Google Scholar]
- Masuda M, Miki Y, Hata S, Takagi K, Sakurai M, Ono K, Suzuki K, Yang Y, Abe E, Hirakawa H (2012) An induction of microRNA, miR-7 through estrogen treatment in breast carcinoma. J Transl Med 10(Suppl 1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondadori dos Santos A, Metzinger L, Haddad O, M’Baya-Moutoula E, Taibi F, Charnaux N, Massy ZA, Hlawaty H, Metzinger-Le Meuth V (2015) miR-126 Is Involved in Vascular Remodeling under Laminar Shear Stress. BioMed Res Int 2015: 497280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410(6824): 50–56. [DOI] [PubMed] [Google Scholar]
- Myakishev MV, Khripin Y, Hu S, Hamer DH (2001) High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res 11(1): 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29(9): e45–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S, Gately K, Crown J, O’Byrne K, O’Driscoll L (2013) Global analysis of serum microRNAs as potential biomarkers for lung adenocarcinoma. Cancer Biol Ther 14(12): 1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SDN, Ohshiro K, Rayala SK, Kumar R (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res 68(20): 8195–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Pusztai L (2011) Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet 378(9805): 1812–1823. [DOI] [PubMed] [Google Scholar]
- Riaz M, van Jaarsveld M, Hollestelle A, Prager-van der Smissen W, Heine A, Boersma A, Liu J, Helmijr J, Ozturk B, Smid M (2013) miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res 15(2): R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledge GW, Mamounas EP, Hortobagyi GN, Burstein HJ, Goodwin PJ, Wolff AC (2014) Past, present, and future challenges in breast cancer treatment. J Clin Oncol 32(19): 1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Ranalli TA, Li X, Bakin AV, Cowell JK (2005) WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp Cell Res 308(1): 135–145. [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, Cowell JK (2007) Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am J Pathol 170(6): 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, Altan B, Tsutsumi S, Asao T, Kuwano H (2015) MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 36(3): 338–345. [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S (2007) The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8(1): 37–48. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451(7175): 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, Davuluri G, Parvani JG, Schiemann BJ, Wendt MK, Plow EF, Schiemann WP, Sossey-Alaoui K (2013) Upregulated WAVE3 expression is essential for TGF-beta-mediated EMT and metastasis of triple-negative breast cancer cells. Breast Cancer Res Treat 142(2): 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Noll R, Smit W, Wymenga A, Boss D, Grob M, Huitema A, Rosing H, Tibben M, Keessen M, Rehorst H, Beijnen JH, Schellens JH (2015) Phase I and pharmacological trial of lapatinib in combination with gemcitabine in patients with advanced breast cancer. Invest New Drugs 33(6): 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li J, Sun M, Sun L, Zhang X (2014) MiRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life 66(5): 371–377. [DOI] [PubMed] [Google Scholar]
- Wang Y-L, Chen C-M, Wang X-M, Wang L (2015) Effects of miR-339-5p on invasion and prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 40(1): 51–56. [DOI] [PubMed] [Google Scholar]
- Wu Z-S, Wu Q, Wang C-Q, Wang X-N, Wang Y, Zhao J-J, Mao S-S, Zhang G-H, Zhang N, Xu X-C (2010) MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer 10(1): 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würth R, Bajetto A, Harrison JK, Barbieri F, Florio T (2014) CXCL12 modulation of CXCR4 and CXCR7 activity in human glioblastoma stem-like cells and regulation of the tumor microenvironment. Front Cell Neurosci 8: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ni C, Chen W, Wu P, Wang Z, Yin J, Huang J, Qiu F (2014) Expression of CXCR4 and breast cancer prognosis: a systematic review and meta-analysis. BMC Cancer 14(1): 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L (2014) MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One 9(5): e96718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.