Abstract
Seed banks are believed to contribute to compositional changes within and across microbial assemblages, but the application of this concept to natural communities remains challenging. Here we describe the core seed bank of a bacterial metacommunity from a boreal watershed, using the spatial distribution of bacterial operational taxonomic units (OTUs) across 223 heterogeneous terrestrial, aquatic and phyllosphere bacterial assemblages. Taxa were considered potential seeds if they transitioned from rare to abundant somewhere within the metacommunity and if they were ubiquitous and able to persist under unfavorable conditions, the latter assessed by checking their presence in three deeply sequenced samples (one soil, one river and one lake, 2.2–3 million reads per sample). We show that only a small fraction (13%) of all detected OTUs constitute a metacommunity seed bank that is shared between all terrestrial and aquatic communities, but not by phyllosphere assemblages, which seem to recruit from a different taxa pool. Our results suggest directional recruitment driven by the flow of water in the landscape, since most aquatic sequences were associated to OTUs found in a single deeply-sequenced soil sample, but only 45% of terrestrial sequences belonged to OTUs found in the two deeply-sequenced aquatic communities. Finally, we hypothesize that extreme rarity, and its interplay with water residence time and growth rates, may further constrain the size of the potential seed bank.
Introduction
Whether bacterial communities react to environmental or spatio-temporal gradients through shifts in relative abundances of taxa that are always present, or through species replacement driven by immigration or extinction, is a fundamental question that has direct implications on our understanding of microbial community ecology. With the advent of high-throughput sequencing technologies, which allow the detection of increasingly rare taxa that escaped more traditional approaches (Pedrós-Alió, 2012), there is growing evidence that temporal or spatial changes in bacterial taxonomic composition are largely due to shifts in relative abundances of extant taxa, rather than to species turnover (Caporaso et al., 2012a; Staley et al., 2013; Shade et al., 2014; Ruiz-González et al., 2015; Valter de Oliveira and Margis, 2015). This has led to the notion of the existence of a microbial seed bank, a standing reservoir of bacteria that persist at low abundances but which may recruit to higher densities upon changes in environmental conditions (Lennon and Jones, 2011).
Recent work has supported the existence of such microbial seed banks: For example, multiple studies have showed that the recruitment of rare bacteria can partly explain seasonal or spatial taxonomic changes within or across communities (Campbell et al., 2011; Sjöstedt et al., 2012; Caporaso et al., 2012a; Comte et al., 2014; Shade et al., 2014; Aanderud et al., 2015; Neuenschwander et al., 2015; Ruiz-González et al., 2015; Langenheder et al., 2016; Niño-García et al., 2016a). Other studies have considered the widespread dormancy found among bacteria or archaea (Jones and Lennon, 2010; Campbell et al., 2011; Hugoni et al., 2013; Aanderud et al., 2016) or the presence of core taxa that persist spatially and seasonally within or across habitat types (Caporaso et al., 2012a; Gibbons et al., 2013; Valter de Oliveira and Margis, 2015) as evidence of a seed bank. In spite of this evidence, the identification and delineation of the microbial seed bank remains difficult in practice. Microbial dormancy, for example, is not easy to measure (del Giorgio and Gasol, 2008), and the fact that a bacterial cell may appear dormant in a given site does not imply that it can reactivate within the range of environmental conditions that exist in that particular ecosystem. Moreover, a major limitation of current studies is that our perspective on microbial composition depends entirely on the sequencing depth, which largely complicates determining the ubiquity of a given taxon. For example, recent studies using deep sequencing have shown that most marine operational taxonomic units (OTUs) can be found in a single sample (Caporaso et al., 2012a; Gibbons et al., 2013), but this would not be apparent with shallower sequencing efforts. Moreover, there is evidence that the presence of many bacterial taxa within the rarity tail results from passive transport (Galand et al., 2009; Saunders et al., 2016; Niño-García et al., 2016b), and whereas some of these ‘accidental’ taxa may thrive in the new environment, most of them will not (Ruiz-González et al., 2015). Rarity and dormancy are therefore not sufficient conditions to include a taxon within a community’s seed bank, and identifying which taxa within the vast tail of rare bacteria act as seeds, and limiting the temporal and spatial boundaries of a given bacterial seed bank, remain our greatest challenge.
An alternative approach to reconstructing the potential seed bank of bacterial communities is to empirically determine which taxa actually recruit within the temporal and spatial confines of the community, and to discriminate these from those that remain rare and unreactive over space and time (Galand et al., 2009; Campbell et al., 2011; Hugoni et al., 2013; Aanderud et al., 2015; Lindh et al., 2015).Recruitment in this context implies transitioning from rare to abundant, and this exercise requires circumscribing the spatial boundaries of the communities wherein this will happen. This is particularly challenging in landscapes like the boreal biome, characterized by complex networks of interconnected freshwater bacterial communities, which in turn interact closely with the surrounding soils (Ruiz-González et al., 2015). Understanding the structure of the seed bank associated to such bacterial metacommunities thus requires a whole network perspective: Since taxa that are rare in one portion of the network may become dominant elsewhere within the metacommunity, identifying the bacterial seed bank requires extending beyond individual assemblages or ecosystem types. The existence of such a core seed bank of rare taxa that recruit, its size, composition and spatial boundaries have never been addressed for complex metacommunities, and yet this is an essential aspect of the assembly of natural microbial assemblages.
Here we reconstruct the core seed bank of a bacterial metacommunity inhabiting a complex network of interconnected soils, rivers and lakes located in a boreal region of Québec, Canada. We based the analysis on sequencing the 16 S rRNA gene from 223 bacterial communities previously shown to make part of a network metacommunity (Ruiz-González et al., 2015). We isolated OTUs that transitioned from rare to abundant within this metacommunity, in order to differentiate rare taxa that recruit, from those that never recruit within the entire network. We further assessed which of these potential seed taxa were ubiquitous within the network, as opposed to those that are occasional and should not be considered part of the metacommunity seed bank. Because sequencing depth may strongly influence our perception of rare taxa distribution, we addressed the latter question by checking the presence of all potential seed taxa (identified from the spatial study) in three deeply-sequenced communities (2–3 million sequences each) within the network. Twenty bacterial communities from the phyllosphere of trees from the same sites were also included as a way to explore the potential spatial boundaries and connectivity of the seed bank.
Materials and methods
Study sites and sampling design
The study region (Côte-Nord, 44–56ºN,64–80ºW, Québec, Canada), and the sampling design have been previously described in Ruiz-González et al. (2015). Briefly, during July 2013, we collected 223 samples for characterization of bacterial communities using Illumina sequencing (Illumina Inc., San Diego, CA, USA) of the 16 S rRNA gene, including soils (n=36), soilwaters (n=36), small streams (Strahler order ⩽2, n=54), rivers (order >2, n=31) and lakes (n=47, Supplementary Figure S1). Three of those samples were further subjected to a deeper sequencing (ca. 2–3 million sequences/sample): (1) one of the highest altitude (410 m) soils, (2) the deepest lake in Québec (Lake Walker, 42 km2, 280 m depth, 2100 km2 catchment area), and (3) the mouth of the largest river in the network (River Moisie, order 8, 420 km length, 19 000 km2 catchment area, Supplementary Figure S1). In addition, we collected 20 samples from the phyllosphere of 4 different tree species (Abies sp., Alnus sp.,Larix sp. and Picea sp.) from 14 of the sites where soil samples were taken. Soil, soilwater and water sample collection has been detailed in Ruiz-González et al. (2015). Samples from the leaf surfaces of the trees were collected as detailed in Kembel et al. (2014). Briefly, between 1 and 3 individual tree species were sampled at a given site. Each sample consisted of leaves from the subcanopy (1–2 m above ground). 50–100 g fresh leave mass was cut from 3 to 5 individual trees into sterile roll bags with surface-sterilized shears. Bacterial cells were collected from leaf surfaces by agitation in a buffer solution followed by DNA extraction (Kembel et al., 2014).
Bacterial community composition
Either 0.25 g of soil, 300–500 ml of water filtered onto 0.22 μm filters, or the resuspended phyllosphere cells were used for genomic analyses. Genomic DNA was extracted using MoBio PowerWater (lake and river samples) and MoBio PowerSoil (phyllosphere, soil and soilwater samples) DNA extraction kits, after verifying the processing the same sample with both kits yielded similar results. The V3–V4 region of the 16 S rRNA gene was amplified with 515 F and 806 R primers, and sequenced on an Illumina MiSeq2000 following a paired-end approach (Caporaso et al., 2012b) at the Génome Québec Innovation Centre (http://gqinnovationcenter.com/index.aspx, Montréal, QC, Canada). Paired-end reads were assembled with FLASH (Magoc and Salzberg, 2011) and sequences between 250 and 290 bp were used for downstream analyses in QIIME to remove primers, low-quality, archaeal and chloroplast reads (Caporaso et al., 2010). Quality sequences were binned into OTUs using Swarm (Mahé et al., 2014), a recently developed method that avoids a fixed clustering threshold. Calculation of the sequence dissimilarity contained within our Swarm-derived OTUs showed that >98% of OTUs harbored amplicons with >99% similarity, and thus that the OTU ‘width’ varied little among OTUs.
Representative sequences were aligned against the SILVAv108 reference alignment (Pruesse et al., 2007). Chimeric sequences were removed using UCHIME (Edgar et al., 2011). To minimize sequencing errors, we discarded all OTUs present in <10 samples and/or showing <10 sequences in the whole data set. The OTU table was randomly subsampled to ensure an equal number of sequences per sample (n=35 113). Raw sequence data have been deposited in the European Nucleotide Archive (acc. num. PRJEB17975).
Statistical analyses
Differences in taxonomic structure among ecosystems were tested with ANOSIM (Clarke, 1993). Correlations between compositional dissimilarity and environmental distance were calculated using Mantel tests (R Vegan package, Oksanen et al., 2015). Spatial OTU turnover was estimated using the beta.pair function from the R betapart package (Baselga and Orme, 2012), as the turnover-fraction of Jaccard pair-wise dissimilarity. All analyses were done with R3.0.0 software (R Core Team, 2013).
Results
The ranges of physicochemical conditions within and across the different ecosystem types are presented in Supplementary Table S1. Overall, the sequencing of all 223 samples recovered 17 373 868 quality sequences (avg. 77 909 sequences per sample, range 35 113–272 722) that clustered into 172 123 OTUs. After rarefaction to 35 113 reads per sample, 7 830 199 sequences (155 426 OTUs) were retained. The deep sequencing of the three samples resulted in 7 644 961 reads after quality checking and filtering, ranging from 2 155 465 sequences in the soil sample to 3 008 455 in the river sample. The OTU accumulation curves for the 3 deeply sequenced samples (hereafter ‘deep’ samples) approached an asymptote yet did not plateau (Supplementary Figure S2), indicating that this sequencing effort captured a large fraction, but not all, of the bacterial richness.
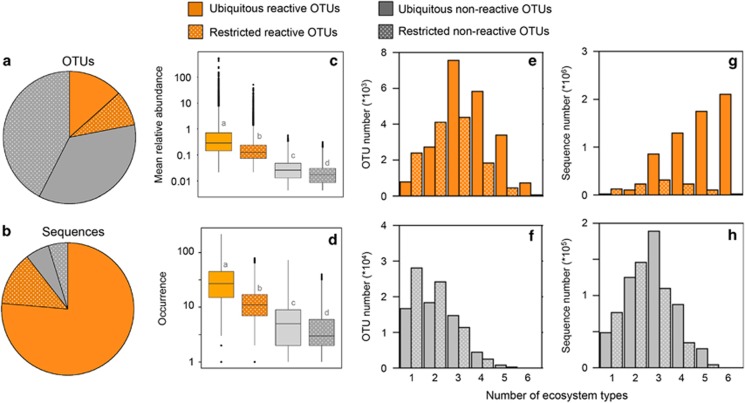
In order to define taxa potentially belonging to the core metacommunity seed bank, we first identified OTUs that showed transitions from rare to abundant within the network, extracting those OTUs that surpassed a minimum local relative abundance threshold of 0.01% in at least one community (hereafter ‘reactive’ pool of taxa, Supplementary Table S2). OTUs that never showed abundance increases across the wide range of habitat types sampled were considered as belonging to the ‘non-reactive’ pool of taxa. Only 22% of all OTUs were classed in this ‘reactive’ pool, yet they accounted for 89% of total sequences (Figures 1a and b,Supplementary Table S2). As expected, the ‘reactive’ pool comprised taxa that were on average more ubiquitous and more abundant than the ‘non-reactive’ pool (Figures 1c and d).
Figure 1.
Characteristics of the four pools of bacteria. Comparison between the regional ‘reactive’ (orange) and the ‘non-reactive’ (grey) pools of taxa, and the fractions within them that were represented (solid) or not (dotted) in the 3 deeply sequenced samples combined. Proportion of OTUs (a) and sequences associated to those OTUs (b) belonging to the 4 pools of bacteria. Mean relative abundance (number of sequences) and occurrence (number of individual sites) of the OTUs belonging to each pool of taxa. Distribution of OTUs (e, f) or sequences (g, h) from each pool depending on whether they where detected in 1,2,3,4,5, or all of the 6 types of ecosystems studied here (soils, soilwaters, streams, rivers, lakes, tree leaves). Note the different scales and magnitudes of the Y axes. Letters on panels c and d refer to results of an ANOVA with a Tukey’s post hoc test. Different letters indicate significant differences (P<0.05) between pools of taxa.
We then assessed the ubiquity of these OTUs by checking their presence in the three deep samples. Whereas 61% of the reactive taxa could be found in some of the 3 deep samples, only 45% of ‘non-reactive’ OTUs were detected by the deep sequencing (termed ‘ubiquitous reactive’ and ‘ubiquitous non-reactive’ OTUs, respectively, Figure 1a). This difference was even larger in terms of sequences, and while the ‘ubiquitous reactive’ taxa accounted for 86% of all ‘reactive’ sequences, the ‘ubiquitous non-reactive’ taxa represented only 56% of the ‘non-reactive’ sequences (Figure 1b, Supplementary Table S2). OTUs from the ‘ubiquitous reactive’ pool were on average more abundant and had higher occurrence than the ‘restricted reactive’ OTUs (that is, ‘reactive’ OTUs not detected in the deep samples, Figures 1c and d). The ‘ubiquitous reactive’ OTUs were distributed across a wider range of ecosystems (Figure 1e), and most of their sequences were associated to OTUs present across four, five or six different ecosystem types (Figure 1g), whereas OTUs from the other bacterial pools were restricted to fewer habitat types (Figures 1f and h).
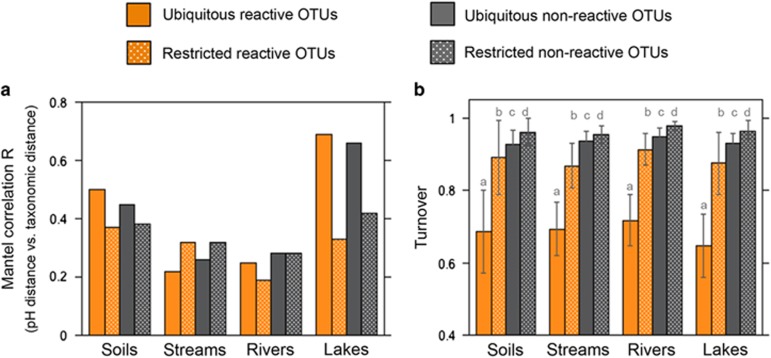
Regardless of these differences between pools, their spatial distribution was similar (Supplementary Figure S3), and differences in taxonomic composition between the six ecosystem types were almost the same for the four pools of taxa considered (ANOSIMbyECOSYSTEM R=0.79–0.81, P<0.0001, Supplementary Figure S3). We further calculated, for each type of ecosystem (except tree leaves), the Mantel correlation between differences in pH and in taxonomic composition, since we have previously shown that pH is the strongest driver of taxonomic structure in this landscape (see Supplementary Figure S2 in Ruiz-González et al., 2015). In lakes and soils, where communities are presumably more subjected to environmental sorting, the responses to pH were stronger for the pools of taxa that were detected in the deep samples (‘ubiquitous’) compared to those OTUs not recovered in the deep samples (‘restricted’, Figure 2a). Although this was true for both the ‘reactive’ and the ‘non-reactive’ ‘ubiquitous’ pools of taxa, the mechanisms underlying their responses to shifts in pH clearly differed: the ‘ubiquitous reactive’ pool showed the lowest taxa turnover across sites (Figure 2b), suggesting that their responses to pH largely reflected shifts in relative abundances of taxa within the group, whereas the pH-driven changes in the other three pools involved an almost complete replacement of OTUs (Figure 2b).
Figure 2.
Environmental responses of the four identified pools of bacteria. Variation in the R coefficients of the Mantel correlations between the taxonomic dissimilarity matrices calculated for the four different subsets of bacteria (see legend), and the differences in pH within each type of ecosystem, excluding the tree leaves (a). Fraction of pairwise dissimilarity between communities due to OTU turnover for each type of ecosystem (b). Soils and soilwaters were considered together because communities aligned along the same pH gradient (see Supplementary Figure S2 from Ruiz-González et al., 2015). Letters on panel (b) refer to an ANOVA with a Tukey’s post hoc test, where different letters indicate significant differences (P<0.05) between pools of taxa for each type of ecosystem.
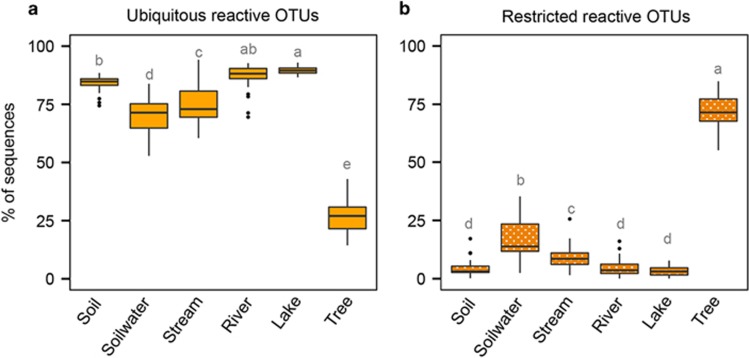
Overall, OTUs from the ‘ubiquitous reactive’ pool accounted for the majority of sequences (average per ecosystem 52–92%) of each of the sampled terrestrial and aquatic bacterial communities (Figure 3a), whereas the ‘restricted reactive’ OTUs accounted for less than 25% (avg. 0.1–35%, Figure 3b). The ‘non-reactive’ pools accounted for a very small percentage of all sequences per community. This would support the notion that the ‘ubiquitous reactive’ pool represents the core seed bank from which the active components of these communities recruit, whereas the ‘restricted reactive’ appear to represent taxa that do recruit but whose distribution within the metacommunity may be more accidental. Interestingly, the ‘ubiquitous reactive’ OTUs accounted for only 27% of the sequences recovered in the phyllosphere, the large majority (72%) represented by ‘restricted reactive’ OTUs (Figure 3b), suggesting that the dominant phyllosphere taxa may not be recruiting from this core seed bank.
Figure 3.
Contribution of reactive OTUs across communities. Contribution to total bacterial sequences of the OTUs from the ‘reactive’ pool that were detected (a) or not (b) in the three deep samples across the different types of ecosystems. Letters refer to results of an ANOVA with a Tukey’s post hoc test. Different letters indicate significant differences (P<0.05) between types of ecosystems.
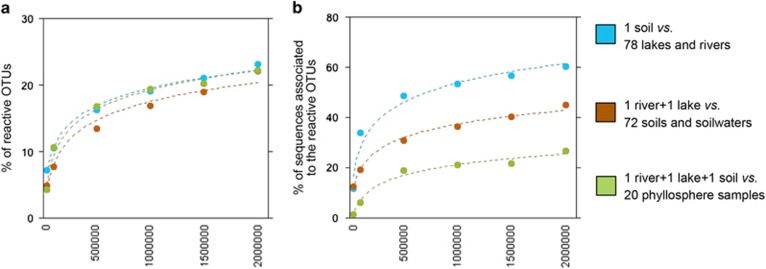
We then assessed the extent to which the ‘reactive’ pool of bacteria from one type of ecosystem could be detected in another type of ecosystem within the metacommunity, by comparing (1) the fraction of ‘reactive’ taxa from aquatic ecosystems (all lakes and rivers) that was detected in the deep soil sample; (2) the fraction of ‘reactive’ taxa found in all terrestrial samples that was found in the two deep aquatic samples (lake and river), and (3) the fraction of phyllosphere ‘reactive’ OTUs that were detected in the three deep samples (soil, river, and lake) pooled together. Moreover, to assess whether the sequencing depth influences this recovery of reactive OTUs, we randomly rarefied the OTU table generated by each of the 3 deep samples to an increasing number of reads, from 100 000 to 2 000 000 sequences per sample, which allowed us to recover increasingly rare OTUs. In all three comparisons, increasing the sequencing depth led to the detection of additional ‘reactive’ OTUs from a different ecosystem. Beyond 500 000 reads per sample, however, very few additional ‘reactive’ OTUs were recovered (Figure 4a) and the linear relationship observed when plotting the sequencing depth on a logarithmic axis suggests we may need a much greater sequencing effort to recover the bulk of the ‘reactive’ pool in adjacent ecosystems (Supplementary Figure S5). Interestingly, this pattern was remarkably similar for the three cases (Figure 4a) regardless of the number of OTUs considered in each comparison (Supplementary Table S3). We further explored whether the recruiting potential of these recovered ‘reactive’ OTUs varied as a function of their relative abundance in neighboring ecosystems. In all 3 comparisons described above, we found that ‘reactive’ OTUs detected at a shallower rarefaction (that is, not very rare OTUs) in one type of system accounted for a large portion of sequences in neighboring systems, but that the cumulative contribution of OTUs recovered at deeper rarefaction to total sequences stabilized at 500 000 reads per sample. This suggests that the probability of these seed taxa of recruiting in a given system declined as a function of their degree of rareness in the surrounding systems (Figure 4b). Interestingly, whereas ‘reactive’ aquatic OTUs found in the deeply sequenced soil accounted for ca. 60% of the sequences from all aquatic systems (Figure 4b), ‘reactive’ terrestrial bacteria recovered in the combined deep river and lake samples (Figure 4a) represented only 45% of the sequences detected across all soils (Figure 4b). Finally, the phyllosphere ‘reactive’ taxa detected in the three deeply sequenced samples accounted for a much smaller fraction of the sequences associated to all leaf communities (Figure 4b), suggesting a much weaker coupling between this component of the landscape and soils and water.
Figure 4.
Recruitment across ecosystem types as a function of sequencing depth. (a) Proportion of the ‘reactive’ OTUs from one type of habitat (aquatic—blue-, terrestrial—brown-, and phyllosphere—green-) detected in a different type of ecosystem as a function of the depth of sequencing for the three comparisons (see legend). (b) Proportion of sequences associated to those detected ‘reactive’ OTUs (of the total sequences considered in each comparison) as a function of the depth of sequencing. For these analyses, the OTU table with the 3 deeply sequenced samples was randomly subsampled to 100 000, 500 000, 1 000 000, 1 500 000 and 2 000 000 sequences per sample. Even though number of OTUs considered in each comparison differed (Supplementary Table S3), these differences did not influence the observed patterns because the percentage of recovered reactive OTUs shown in Figure 4a does not change among comparisons.
Discussion
The existence of a bacterial seed bank that underlies the compositional changes observed across aquatic and terrestrial microbial communities has become one of the paradigms of microbial ecology (Jones and Lennon, 2010; Lennon and Jones, 2011; Gibbons et al., 2013), but one that has been challenging to define quantitatively. The greatest difficulty is to determine which taxa comprise this seed bank, something that requires defining ecologically relevant criteria to discriminate between seed and non-seed taxa. Here we identified the seed bank of a bacterial metacommunity using two basic premises to discriminate seed from non-seed taxa: (1) bacterial seeds must have the potential to grow locally somewhere within the metacommunity (that is, shift from rare/dormant to abundant), and (2) bacterial seeds must be ubiquitous within the metacommunity, persisting across space. We used OTUs spatial distribution throughout the network to identify taxa that met the above criteria, by selecting those that transitioned from rare to abundant across an array of terrestrial and aquatic bacterial assemblages known to make part of this network metacommunity (Ruiz-González et al., 2015). We further identified, within this ‘reactive’ pool of taxa, those that were widely distributed within the metacommunity (‘ubiquitous reactive’ taxa). Since detection, and therefore the perceived occurrence of OTUs, is largely influenced by the sequencing depth, we assessed the ubiquity of taxa by checking their presence in three deeply sequenced samples from the metacommunity, testing the assumption that bacteria belonging to the core seed bank should be found in any random sample if sequenced deeply enough.
In this interconnected and environmentally heterogeneous network (Supplementary Figure S1,Supplementary Table S1), no OTU was consistently abundant and dominant across all sites, and thus all ‘reactive’ OTUs represented taxa that dominated at certain sites but became rare elsewhere within the metacommunity. Within them, the ‘ubiquitous reactive’ OTUs were the most abundant on average, representing only 13% of all OTUs but >75% of all metacommunity sequences (Figures 1a and b), and most sequences within any given terrestrial or aquatic assemblage (Figure 3a). These taxa were also the most widely distributed not only across sites (Figure 1d), but also across different habitats, since most of them were present in four to six different ecosystem types within the network (Figure 1g). This agrees with previous studies that have suggested that higher local abundance facilitates regional dispersal (Nemergut et al., 2011; Caporaso et al., 2012a; Martiny, 2015; Salazar et al., 2015). However, abundance alone did not explain the greater ubiquity of this group, since we observed a large overlap in mean relative abundances between the ‘ubiquitous’ and ‘restricted’ reactive OTUs (Figure 1c), and yet these two pools differed greatly in their cross-system distribution patterns (Figures 1e and g). This would suggest that the ‘restricted reactive’ group, despite recruiting within the metacommunity and being locally abundant, may comprise taxa either adapted to a very narrow range of conditions and infrequent disturbances, or which may originate from very localized sources. In contrast, the ‘ubiquitous reactive’ taxa are widespread across the network, and likely make up the core metacommunity seed bank from which all these terrestrial and aquatic communities recruit. This idea was supported by the fact that the taxa dominating the ‘ubiquitous reactive’ pools largely differed between the different ecosystem types (see examples in Supplementary Figure S4), suggesting that, although they are widely distributed, they recruit under specific conditions.
We expected to find stronger environmental responses among the ‘reactive’ compared to the ‘non-reactive’ taxa, and yet both groups showed strikingly similar spatial distribution patterns (Supplementary Figure S3). This agrees with studies reporting that dominant and rare components of bacterial communities show similar biogeographic patterns (Galand et al., 2009; Campbell et al., 2011; Logares et al., 2013; Vergin et al., 2013; Liu et al., 2015). When we explored the changes in taxonomic composition associated to gradients in pH within each type of ecosystem, however, we found clear differences in the response of the ‘reactive’ and ‘unreactive’ pools (Figure 2a). Overall, the stronger differences occurred in lakes, where the longer residence time leads to a more intense local sorting of species, compared to lotic systems (Lindström et al., 2006; Crump et al., 2007; Ruiz-González et al., 2015; Niño-García et al., 2016a). Although in lakes the pH-driven responses were stronger for the ‘ubiquitous’ OTUs than the ‘restricted’ ones, the magnitude of these responses was surprisingly similar between the ‘reactive’ and ‘non-reactive’ pools of taxa (Figure 2a). The underlying mechanisms, however, appear to be fundamentally different: as a group, the ‘ubiquitous reactive’ OTUs had the lowest turnover rates, suggesting that whereas their pH-driven responses implied changes in the relative abundances of taxa across communities, changes in the other three pools involved an almost complete substitution of OTUs between pairs of sites (Figure 2b). This supports the notion that the ‘ubiquitous reactive’ category comprises taxa that are strongly reacting to environmental conditions within the metacommunity, while the spatial structuring of the other categories may reflect other processes, such as the dispersal of taxa from spatially structured communities (Niño-García et al., 2016b).
In agreement with other studies showing that most rare taxa are permanently rare within a particular ecosystem type (for example, Galand et al., 2009), we found that >80% of the taxa making up this complex metacommunity never surpassed the local abundance threshold fixed here (0.01%) in spite of the large environmental gradients and different physical structuring covered by our sampled sites. We acknowledge, however, that our approach has limitations, since it does not allow assessing the physiological state or activity of rare taxa, so we cannot discriminate between rare taxa that were metabolically active at the time of sampling, from those that were dormant but reactive, or simply inactive or dead. However, quantifying dormancy at the scale of this study would be extremely difficult and, as pointed out before, it is not a sufficient criterion for a taxon to be part of the seed bank. In any case, in previous studies in these boreal networks (Niño-García et al., 2016b), we identified a group of consistently rare bacteria that appear to be actively selected in lakes, yet it contained very few taxa (<400) relative to the thousands of rare OTUs whose presence seemed largely accidental and associated to hydrologic transport. Thus, although within the lake ‘rare biosphere’ there may be taxa for which rareness is not accidental but rather adaptive, as has been hypothesized before (Logares et al., 2014; Lynch and Neufeld, 2015), this suggests that this group of constitutively rare bacteria would not comprise a large fraction of our ‘non-reactive’ pools. Even though the abundance threshold we used is operational and the proportions may change if applying a more or less stringent limit, it seems that the overwhelming majority of taxa in these assemblages is most likely not part of the seed bank from which this metacommunity recruits.
Our study provides only a seasonal snapshot and thus the actual size and composition of the reactive and unreactive pools might also vary temporally, since rare ‘non-reactive’ taxa may shift in abundance at other times of the year (Campbell et al., 2011; Shade et al., 2014; Neuenschwander et al., 2015; Saunders et al., 2016; Niño-García et al., 2017). To address this issue, we repeated the exercise of identifying the ‘reactive’ pool for a different data set of 46 rivers and lakes from 3 boreal regions of Québec, each sampled on 3 occasions (spring, summer and fall, n=138, details not shown), and we found that the proportion of ‘reactive’ OTUs increased from 10%, when only the summer samples were considered, to 15% using the whole data set. Thus, it does not seem that including the temporal dimension, at least at this coarse level of resolution, would fundamentally alter the contribution of the different pools to the metacommunity structure presented here. It is possible, however, that some of these apparently non-reactive OTUs recruit in connected microbial ecosystems not considered here (for example, lake sediments, biofilms and so on), and thus a more intensive characterization of the surrounding habitats, or a deeper sequencing of all communities, could lead to changes in the size of the different pools.
Regardless of the uncertainties associated to the actual size of these pools, our analysis suggests that the taxa potentially belonging to the core metacommunity seed bank represent a very small proportion of the vast bacterial richness found in these boreal metacommunities. Although a handful studies have attempted to explore aspects related to the notion of a seed bank, either by determining the activity or dormancy of rare bacteria (Jones and Lennon, 2010; Lennon and Jones, 2011; Hugoni et al., 2013; Aanderud et al., 2015) or assessing the presence of cosmopolitan taxa between samples within or among habitats (Caporaso et al., 2012a; Gibbons et al., 2013, Valter de Oliveira and Margis, 2015), so far no study had attempted to determine the size, composition and spatial boundaries of the seed bank of complex bacterial metacommunities. Moreover, most seed bank studies have focused on single ecosystem types (Jones and Lennon, 2010; Lennon and Jones, 2011; Caporaso et al., 2012a; Valter de Oliveira and Margis, 2015), yet there is unambiguous evidence that the taxa pool from which a local community may recruit most likely transcends the limits of a given ecosystem type (Crump et al., 2012; Sjöstedt et al., 2012; Lee et al., 2013; Comte et al., 2014; Ruiz-González et al., 2015).
We observed that OTUs populating the phyllosphere were greatly underrepresented within our ‘ubiquitous reactive’ pool (Figures 2c and d), and that >60% of the sequences detected in tree leaves were exclusively found in the phyllosphere, as opposed to the rest of ecosystems where the percentage of ecosystem-endemic OTUs was <5% (Table 1 in Ruiz-González et al., 2015). Most phyllosphere OTUs thus do not appear to recruit in the soil-aquatic metacommunity, and viceversa, in spite of the fact that we sampled the leaves exactly at the same place were we collected soil, soil water and stream samples. Although a deeper sequencing might result in greater taxonomic overlap between phyllosphere and other communities, this suggests that there are spatial boundaries to the potential core seed bank related to the dispersal of microbes and the interactions between local assemblages within the metacommunity. Thus, not all the taxa that inhabit a certain landscape automatically become part of its microbial seed bank.
We also found evidence for a spatial structuring of the seed bank. Whereas most dominant aquatic OTUs could be found in a single deeply sequenced soil sample, a significantly smaller fraction of the dominant soil OTUs could be found in the two deeply sequenced aquatic samples (Figure 4), even though these integrated very large catchments (see Materials and methods section). Besides supporting previous evidence that freshwater bacterial communities are largely dominated by terrestrially-derived taxa that have the potential to grow in inland waters (Crump et al., 2012; Ruiz-González et al., 2015; Niño-García et al., 2016a), this further suggests a directionality in the seed bank driven by the flow of water in the network. Finally, the observation that the 3 deep samples together only recovered 27% of phyllosphere sequences confirms that there are physical dispersal-related limits that constrain the dimensions of the seed bank independently of the sequencing depth used.
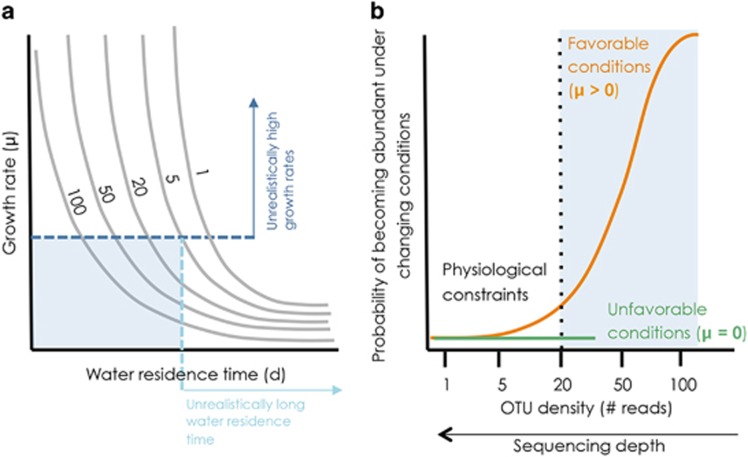
In addition to the physical boundaries and structure of the seed bank, the degree of rareness itself may impose constraints on whether taxa can effectively recruit and therefore be part of the seed bank. For example, even if a dormant taxon could potentially grow when exposed to favorable conditions, it is the interplay between the maximum growth rate that this taxon can express and the physical structure of the environment (for example, the water residence time) what will determine whether it can actually attain the density levels required to become abundant in a particular environment (Figure 5a). This implies that there may be thresholds of rareness below which a taxon will not be able to recruit within a set of plausible combinations of growth rate and ecosystem water retention time (shaded area in Figure 5a and b), which may further limit the size of the effective metacommunity seed bank. Although this hypothesis is difficult to test, our results do provide some preliminary support. For example, we have shown that the detection of increasingly rare taxa in soils did not result in a proportional increase of their associated sequences in the receiving waters, implying that these extremely rare soil taxa could not attain high abundances downstream (Figure 4b), and therefore did not contribute to the metacommunity seed bank. This suggests that although there may be effective recruitment at even lower cell densities than those reached by our sequencing effort, there appears to be an abundance threshold required for a taxon to be effectively recruited (Figure 5b). This applies in particular to the aquatic portion of the metacommunity, where the recruitment and growth of terrestrial bacteria seems to be strongly modulated by the water retention time within the network (Ruiz-González et al., 2015; Niño-García et al., 2016a,2016b).
Figure 5.
Physiological constraints may limit the size of the effective seed bank. The degree of rareness of a given bacterial taxon may itself impose constrains on whether it can effectively recruit and become part of the effective seed bank (i.e., the seed bank that we can identify as such because it is expressed at some point within the network). (a) This may be particularly relevant in the aquatic portion of these complex boreal networks, where, depending on the water residence time of the ecosystem (horizontal axis) and the initial cell densities of potentially recruiting taxa (different lines), the bacterial growth rates (vertical axis) required for a taxon to attain a certain dominance in a community will vary largely: for example, taxa that are extremely rare (initial theoretical density of 1) would need to have unrealistically high growth rates to overcome extreme rarity and attain high abundances within the ecosystem water residence time. This implies that a taxon will only be able to behave as a seed within a limited set of combinations of realistic growth rates and water residence times (shaded area in a), and that its probability to recruit will thus be dependant on its initial degree of rareness (orange line in b). Even if the conditions for its activation and growth are favourable, a taxon will only be able to recruit above a threshold initial abundance (shaded area in b). Below this threshold, unreactive taxa (green line in b) and potentially reactive taxa (orange line in b) behave essentially as unreactive and do not make part of the seed bank (non-shaded area), at least within the time frame dictated by the movement of water in the landscape.
Taken together, our results indicate the existence of a core seed bank that underlies the patterns in composition of these bacterial metacommunities, which is shared between the terrestrial and aquatic portions of complex boreal networks. We show that bacterial communities inhabiting other less connected habitats within the same landscape, such as the phyllosphere, are largely decoupled from the rest of the metacommunity and poorly represented in the metacommunity seed bank, suggesting that there may be distinct seed banks coexisting within a landscape. Our results indicate that the core seed bank is composed of a small fraction of the total bacterial richness found in these networks, and that most rare bacteria do not appear to recruit, at least within the spatial confines of the metacommunity. We further hypothesize that the degree of rarity may impose limits on the capacity of taxa to recruit, since slower growing taxa may not be able to overcome extreme rareness within the constraints of their residence time within the network, and thus this may impose additional physiological limits to the potential metacommunity seed bank.
Acknowledgments
We thank Terhi Rasilo, Annick St-Pierre, Alice Parkes, Isabelle Laforest-Lapointe, Travis Dawson, and the whole CarBBAS team for their contribution to the field and laboratory components of this research. This study is part of the program of the Carbon Biogeochemistry in Boreal Aquatic Systems (CarBBAS) Industrial Research Chair, co-funded by the Natural Science and Engineering Research Council of Canada (NSERC) and Hydro-Quebec. We acknowledge funding from FRQNT, NSERC, and the Canada Research Chairs program. CRG benefited of a Beatriu de Pinós post-doctoral fellowship (2014 BP_B 00116) from the Marie-Curie COFUND programme and the Catalan Government.
Author contributions
PdG, CRG designed the sampling, JPNG, CRG collected the data, CRG, JPNG and SK analyzed the data, CRG, JPNG, SK and CRG discussed and interpreted the results and CRG, PdG and SK wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aanderud ZT, Jones SE, Fierer N, Lennon JF. (2015). Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front Microbiol 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aanderud ZT, Vert JC, Lennon JT, Magnusson TW, Breakwell DP, Harker AR. (2016). Bacterial dormancy is more prevalent in freshwater than in hypersaline lakes. Front Microbiol 7: 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga A, Orme CDL. (2012). Betapart: an R package for the study of beta diversity. Methods Ecol Evol 3: 808–812. [Google Scholar]
- Campbell BJ, Yua L, Heidelberg JF, Kirchman DL. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012. a). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. (2012. b). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR. (1993). Non-parametric multivariate analysis of changes in community structure. Aust J Ecol 18: 117–143. [Google Scholar]
- Comte J, Lindström ES, Eiler A, Langenheder S. (2014). Can marine bacteria be recruited from freshwater sources and the air? ISME J 8: 2423–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Adams HE, Hobbie JE, Kling GW. (2007). Biogeography of bacterioplankton in lakes and streams of an Arctic tundra catchment. Ecology 88: 1365–1378. [DOI] [PubMed] [Google Scholar]
- Crump BC, Amaral-Zettler LA, Kling GW. (2012). Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J 6: 1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Giorgio PA, Gasol JM. (2008) Physiological structure and single-cell activity in marine bacterioplankton. In:Kirchman D (ed), Microbial Ecology of the Ocean. John Wiley Sons, Inc: Hoboken, NJ, USA, pp 243–298. [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA 106: 22427–22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. (2013). Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci USA 110: 4651–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugoni M, Taiba N, Debroas D, Domaizon I, Doufornel IJ, Bronner G et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA 110: 6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA 107: 5881–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, O'Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL. (2014). Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci USA 111: 13715–13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Comte J, Langenheder S, Comte J, Zha Y, Samad MS et al. (2016). Remnants of marine bacterial communities can be retrieved from deep sediments in lakes of marine origin. Environ Microbiol Rep 8: 479–485. [DOI] [PubMed] [Google Scholar]
- Lee JE, Buckley HL, Etienne RS, Lear G. (2013). Both species sorting and neutral processes drive assembly of bacterial communities in aquatic microcosms. Fems Microbiol Ecol 86: 288–302. [DOI] [PubMed] [Google Scholar]
- Lennon JT, Jones SE. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 119: 119–130. [DOI] [PubMed] [Google Scholar]
- Lindh MV, Sjöstedt J, Andersson AF, Baltar F, Hugerth LW, Lundin D et al. (2015). Disentangling seasonal bacterioplankton population dynamics by high-frequency sampling. Environ Microbiol 17: 2459–2476. [DOI] [PubMed] [Google Scholar]
- Lindström ES, Forslund M, Algesten G, Bergström A. (2006). External control of bacterial community structure in lakes. Limnol Oceanogr 51: 339–342. [Google Scholar]
- Liu L, Yang J, Yu Z, Wilkinson DM. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J 9: 2068–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logares R, Audic S, Bass D, Bittner L, Boutte C, Christen R et al. (2014). Patterns of rare and abundant marine microbial eukaryotes. Curr Biol 24: 813–821. [DOI] [PubMed] [Google Scholar]
- Logares R, Lindström ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J et al. (2013). Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J 7: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MDJ, Neufeld JD. (2015). Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13: 217–229. [DOI] [PubMed] [Google Scholar]
- Magoc T, Salzberg S. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. (2014). Swarm: robust and fast clustering method for amplicon-based studies. Peer J 2: e593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH. (2015). Dispersal and the Microbiome: Learning how fast and how far microorganisms move will help us better understand the diversity of microbial communities. Microbe 10: 191–196. [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander SM, Pernthaler J, Posch T, Salcher MM. (2015). Seasonal growth potential of rare lake water bacteria suggest their disproportional contribution to carbon fluxes. Environ Microbiol 17: 781–795. [DOI] [PubMed] [Google Scholar]
- Niño-García JP, Ruiz-González C, del Giorgio PA. (2016. a). Interactions between hydrology and water chemistry shape bacterioplankton biogeography across boreal freshwater networks. ISME J 10: 1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño-García JP, Ruiz-González C, del Giorgio PA. (2016. b). Landscape-scale spatial abundance distributions discriminate core from random components of lake bacterioplankton. Ecol Lett 19: 1506–1515. [DOI] [PubMed] [Google Scholar]
- Niño-García JP, Ruiz-González C, del Giorgio PA. (2017). Exploring the ecological coherence between the spatial and temporal patterns of bacterioplankton in boreal Lakes. Front Microbiol 8: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen JF, Blanchet G, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2015). Vegan: community ecology package. R Package Version 2: 3–0. [Google Scholar]
- Pedrós-Alió C. (2012). The rare bacterial biosphere. Ann Rev Mar Sci 4: 449–466. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-González C, Niño-García JP, del Giorgio PA. (2015). Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecol Lett 18: 1198–1206. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2013) R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing Vienna, Austria, Available from http://www.R-project.org/. [Google Scholar]
- Salazar G, Cornejo-Castillo FM, Benitez-Barrios V, Fraile-Nuez E, Álvarez-SalgadonXA, Duarte CM et al. (2015). Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J 10: 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Albertsen M, Vollertsen J, Nielsen PH. (2016). The activated sludge ecosystem contains a core community of abundant organisms. ISME J 10: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5: e01371–01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstedt J, Koch-Schmidt P, Pontarp M, Canbäck B, Tunlid A, Lundberg P et al. (2012). Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl Environ Microbiol 78: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C, Unno T, Gould TJ, Jarvis B, Phillips JB, Cotner JB et al. (2013). Application of Illumina next-generation sequencing to characterize the bacterial community of the Upper Mississippi River. J Appl Microbiol 115: 1147–1158. [DOI] [PubMed] [Google Scholar]
- Valter de Oliveira LF, Margis R. (2015). The source of the river as a nursery for microbial diversity. PLoS One 10: e0120608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergin KL, Done B, Carlson CA, Giovannoni SJ. (2013). Spatiotemporal distributions of rare bacterioplankton populations indicate adaptive strategies in the oligotrophic ocean. ISME J 71: 1–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.