Abstract
Roux-en-Y gastric bypass (RYGB) and laparoscopic adjustable gastric banding (LAGB) are anatomically different bariatric operations. RYGB achieves greater weight loss compared with LAGB. Changes in the gut microbiome have been documented after RYGB, but not LAGB, and the microbial contribution to sustainable surgical weight loss warrants further evaluation. We hypothesized that RYGB imposes greater changes on the microbiota and its metabolism than LAGB, and that the altered microbiota may contribute to greater weight loss. Using multi-omic approaches, we analyzed fecal microbial community structure and metabolites of pre-bariatric surgery morbidly obese (PreB-Ob), normal weight (NW), post-RYGB, and post-LAGB participants. RYGB microbiomes were significantly different from those from NW, LAGB and PreB-Ob. Microbiome differences between RYGB and PreB-Ob populations were mirrored in their metabolomes. Diversity was higher in RYGB compared with LAGB, possibly because of an increase in the abundance of facultative anaerobic, bile-tolerant and acid-sensible microorganisms in the former. Possibly because of lower gastric acid exposure, phylotypes from the oral cavity, such as Escherichia, Veillonella and Streptococcus, were in greater abundance in the RYGB group, and their abundances positively correlated with percent excess weight loss. Many of these post-RYGB microorganisms are capable of amino-acid fermentation. Amino-acid and carbohydrate fermentation products—isovalerate, isobutyrate, butyrate and propionate—were prevalent in RYGB participants, but not in LAGB participants. RYGB resulted in greater alteration of the gut microbiome and metabolome than LAGB, and RYGB group exhibited unique microbiome composed of many amino-acid fermenters, compared with nonsurgical controls.
Introduction
Rising obesity rates and ineffective weight management strategies have increased the prevalence of bariatric surgeries to treat morbid obesity (Korner et al., 2009). At present, laparoscopic adjustable gastric banding (LAGB) and Roux-en-Y gastric bypass (RYGB) are the most commonly performed surgeries worldwide (Yurcisin et al., 2009); however, RYGB achieves greater and more sustained weight loss, and better resolution of obesity-related comorbidities such as diabetes and hyperlipidemia (O'Brien, 2010).
Both bariatric operations limit food intake. Although LAGB surgery places an adjustable band around the proximal stomach to restrict the flow of ingested food, RYGB surgery creates a small gastric pouch by partitioning the stomach into proximal and distal parts (Korner et al., 2009). In addition, with RYGB, a section of jejunum (that is, the Roux limb) is then connected to the proximal gastric pouch, thereby excluding the majority of the stomach, duodenum and proximal jejunum from exposure to ingested food (Korner et al., 2009). RYGB may induce malabsorption of digested food, depending upon the length of the Roux limb, change hormonal response for appetite (Korner et al., 2009), increase intestinal transit and alter bile–acid metabolism (le Roux et al., 2006), hence enhance weight loss (Korner et al., 2009). Although an altered response of ghrelin has been observed after LAGB, the clinical significance of this finding remains uncertain (Shak et al., 2008).
A role of gut microbiota on host energy regulation and metabolism has been postulated. The microbiome structure has been shown to differ between lean and obese hosts (Ley et al., 2005) and altered microbial metabolism in obese hosts appears to contribute to weight gain (Turnbaugh et al., 2006). After RYGB, humans (Zhang et al., 2009; Furet et al., 2010) and animals (Li et al., 2011; Liou et al., 2013) exhibited significantly different microbiome structures than obese or lean comparators. Gut microbiome appears to change after two other bariatric operations: laparoscopic sleeve gastrectomy (Damms-Machado et al., 2015) and vertical banded gastroplasty (VBG) (Tremaroli et al., 2015). To our knowledge, changes in gut microbiota after LAGB surgery have not been studied.
Fecal transplants from post-RYGB mice/humans to germ-free mice induced weight loss (Liou et al., 2013) and decreased the size of adipose tissue (Tremaroli et al., 2015), independently from the diet. Although the microbiota-associated mechanism of weight loss remains unclear, several microbial metabolites, such as short chain fatty acids (SCFAs), have been suggested to have roles in host energy regulation (Lin et al., 2012). For instance, propionate and butyrate, which increase after RYGB surgery in mice (Liou et al., 2013), have been implicated as signaling molecules of the free fatty acid receptors and seem to protect against diet-induced obesity (Lin et al., 2012). In addition, branched chain fatty acids (BCFAs)—isobutyrate and isovalerate—also serve as signaling molecules and show potent activity against the G protein-coupled receptors (Le Poul et al., 2003) and free fatty acid receptors (Schmidt et al., 2011). By activating G protein-coupled receptors, microbiota can indirectly stimulate the release of appetite-regulating neuropeptides, such as peptide YY and glucagon-like peptide-1 (Psichas et al., 2015). Moreover, by producing BCFAs from branched chain amino acids (BCAAs), microbiota can reduce host BCAAs that were associated with metabolic syndrome (Yoon, 2016).
The objectives of our study were to: (i) determine differences in microbial community structure after RYGB and LAGB surgeries, (ii) identify microbe-produced metabolites that distinguish two different bariatric surgeries and (iii) reveal links between microbiome and weight loss-associated products after bariatric surgeries. To accomplish these objectives, we studied the microbial community structure and metabolic products in fecal samples from individuals who had previously undergone either RYGB or LAGB.
Materials and methods
Experimental design
The Institutional Review Boards of Mayo Clinic and Arizona State University (IRB# 10-008725 for both) approved our study and all participants provided signed informed consent. Our study compared gut microbial ecology in four groups: participants who had previously undergone RYGB (n=24) or LAGB (n=14) surgeries, healthy normal weight (NW) (n=10) and morbidly obese controls who were scheduled to undergo bariatric surgery (PreB-Ob) (n=15). Importantly, PreB-Ob participants were on a recommended weight loss diet. The participants were recruited between 2011 and 2014.
We excluded participants who had undergone other types of gastrointestinal surgery or had uncontrolled gastrointestinal or other chronic diseases. Prebiotic/probiotic agents were avoided 2 weeks before feces collection and antibiotic use was not allowed within 2 months of feces collection. Participants were asked to refrain from the use of medications that can alter gastrointestinal secretory or motor function (for example, proton pump inhibitors or prokinetic agents) for at least 2 weeks before feces collection; however, if they were unable to do so, this did not disqualify them from participation. Two post-LAGB participants were using metformin, whereas three post-LAGB and two post-RYGB participants were using a statin agent.
Our study focused on long-term changes in the gut microbiota; hence, we included only participants who had surgery >9 months before the sample collection. Fecal samples were collected at the Mayo Clinic, Scottsdale, AZ, USA, shipped to Arizona State University on dry ice and stored at –80 °C. Within 2 weeks before sample collection, we obtained 4-day prospective food diaries and food frequency questionnaires from the participants after they had been educated by a dietitian on how to record their food intake accurately. Dietary information was analyzed using DietOrganizer software (dietorganizer.com).
DNA extraction, 16S ribosomal RNA gene sequencing and genomic predictions
We extracted genomic DNA using the QIAamp DNA stool mini kit (Qiagen Inc., Valencia, CA, USA) as previously described by Kang et al. (2015). Sequencing was performed with an Illumina Miseq Instrument using V4-V6 primers (forward: 5′-GTGCCAGCMGCCGCGGTAA-3′ and reverse: 5′-ACAGCCATGCANCACCT-3′) at University of Minnesota Genomic Center. Sequences were submitted to NCBI Sequence Read Archive under BioProject PRJNA321731. Forward and reverse reads were paired using PANDAseq (Masella et al., 2012) and analyzed using the QIIME 1.9 suite (Caporaso et al., 2010). Details of the analysis can be found in the Supplementary Material S1. After quality filtering, we attained a total of 3 369 231 sequences and median number of sequences per sample was 50 035.
We calculated alpha diversity metrics: phylogenetic diversity whole tree (Faith, 1992) and equitability. We used the Unifrac metric (Lozupone et al., 2006) for beta diversity, and linear discriminant analysis effect size (LEFSe) (Segata et al., 2011) to identify microbial discriminants of surgeries. By overlaying class-level phylotypes on principal coordinate analysis plots, we identified phylotypes that separated groups on principal coordinate analysis. We predicted functional gene content from 16S ribosomal RNA gene data using Phylogenetic Investigation of the Communities by Reconstruction of Unobserved States (PICRUSt) (Langille et al., 2013). We analyzed wide-scale phenotypic properties of the microbiota with the BugBase tool (Caporaso et al., 2010; Langille et al., 2013).
Biochemical characterization of feces
For each fecal specimen, approximately 1 g of wet weight was diluted with 20 ml of 18 Ω water. The homogenate was vortexed for 10 min, centrifuged at 13 000 r.p.m. for 15 min and then filtered through 0.2-μm PVDF membranes (Pall Corporation, East Hills, NY, USA). A global metabolomics approach was used to obtain assignment and quantitation of metabolites via nuclear magnetic resonance (1H-NMR). The 1D 1H-NMR spectra of all samples were collected following standard Chenomx (Edmonton, Alberta, Canada) sample preparation and data collection guidelines (Weljie et al., 2006) (see Supplementary Material S1 for detailed protocol). The concentrations were normalized to grams of dry fecal weight. A subset of samples (n=44) were prepared in triplicate extractions and duplicates for 1H-NMR to verify intra-individual variation was smaller than inter-individual variation (Supplementary Table S1).
Statistics
Statistical analyses were conducted using the IBM SPSS for Macintosh, version 23.0 (Armonk, NY, USA). Group medians were compared using the Mann–Whitney U-test and were reported with median absolute deviation values. Spearman’s rank-order correlation coefficients were calculated between microbial phylotypes and percent excess weight loss (%EWL). False discovery rate corrected Padj values<0.05 using Monte Carlo simulations were considered statistically significant. Further statistical tests on distance matrixes: permutational analysis of variance, analysis of similarity and Adonis were performed (Supplementary Material S1).
Results and discussion
RYGB was more effective than LAGB
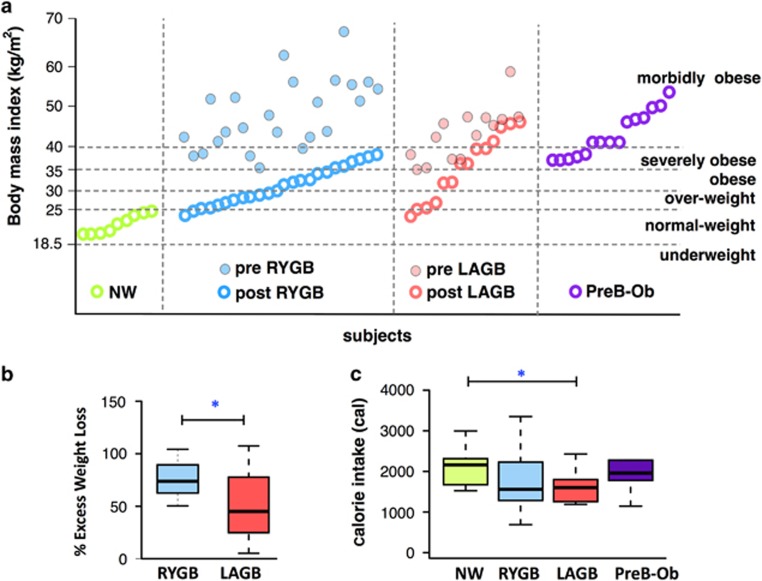
Table 1 shows number of participants and characteristics in each group. NW and RYGB participants had lower body mass index (BMI) than the LAGB and PreB-Ob participants, as shown in Figure 1a. The median BMI (kg m–2) of the populations were: NW: 22.32, RYGB: 30.8, LAGB: 36.6 and PreB-Ob: 43.48. Despite substantial weight loss, the majority of the participants from RYGB and LAGB groups remained in the overweight or obese categories based upon a BMI >25 kg m–2. Before surgery, the RYGB group was composed of mainly overweight and obese participants, whereas the LAGB group was mainly composed of obese and severely obese participants (BMI >30 kg m–2). Pre-RYGB and pre-LAGB BMIs were not statistically different (Mann–Whitney U-test P=0.104). Although post-LAGB BMIs were greater than post-RYGB BMIs, because of the large variation within the LAGB group, this difference was statistically insignificant (Mann–Whitney U-test P=0.118).
Table 1. Age, gender, time after surgery and medical condition summary for experimental groups.
| NW | RYGB | LAGB | PreB-Ob | |
|---|---|---|---|---|
| Number of participants | 10 | 24 | 14 | 15 |
| Age | 42±15 | 51±7 | 46±11 | 50±9 |
| Gender (F/M) | 7/3 | 16/8 | 14/2 | 8/7 |
| Time after surgery (months) | NA | 35± 8 | 34±12 | NA |
| Diabetes (# of positives) | 0 | 0 | 2 | 7 |
| Hyperlipidemia (# of positives) | 0 | 3 | 6 | 7 |
| Hypertension (# of positives) | 0 | 6 | 3 | 9 |
| Arthritis (# of positives) | 0 | 7 | 3 | 8 |
Abbreviations: F, female; LAGB, laparoscopic adjustable gastric banding; M, male; NA, not applicable; NW, normal weight, PreB-Ob, pre-bariatric surgery morbidly obese; RYGB, Roux-en-Y gastric bypass.
For age and time after surgery categories, the numbers represent median values with median absolute deviation.
Figure 1.
Subject’s BMI, %EWL and average daily calorie intake. (a) BMI distribution of the participants that participated in this study. Empty circles represent BMI at the time of sampling and full circles represent recorded BMI before surgeries retrospectively. The horizontal dashed lines indicate the BMI-based weight categories according to National Institutes of Health guidelines (nhlbi.nih.gov). (b) %EWL for the RYGB and LAGB groups. The RYGB group had significantly higher % EWL than the LAGB participants. (c) Median calorie intake per experimental group. *Denotes Mann–Whitney U-test P<0.05.
In order to assess surgical success, we calculated % EWL, a commonly used clinical outcome that defines the weight loss normalized to the EWL needed to reach an ideal body weight defined by a BMI of 25 kg m–2 (Boza et al., 2010). Figure 1b shows that %EWL was significantly higher for the RYGB group than the LAGB group (P<0.05), an observation consistent with previous long-term (Boza et al., 2010) and short-term findings (Tice et al., 2008). The higher success rate (%EWL) in the RYGB compared with the LAGB group was not related to a difference in the time of data collection with respect to the time from the surgery: the median times of data collection after the surgery were 35±8 and 34±12 months for RYGB and LAGB, respectively. In addition, Figure 1c shows that %EWL was independent of the average calories consumed based on 4-day food diaries, as LAGB and RYGB groups consumed similar amounts of calories that were less than the NW and PreB-Ob. Importantly, although LAGB and RYGB participants consumed similar amounts of calories per day (Supplementary Table S2), %EWL was significantly lower for LAGB group than RYGB group. Based on food frequency questionnaires (Supplementary Table S2), the RYGB group consumed fewer calories than the other groups, and PreB-Ob consumed more than the others, although this finding was not statistically significant.
Post-RYGB and PreB-Ob participants had distinctive fecal microbiomes and metabolomes
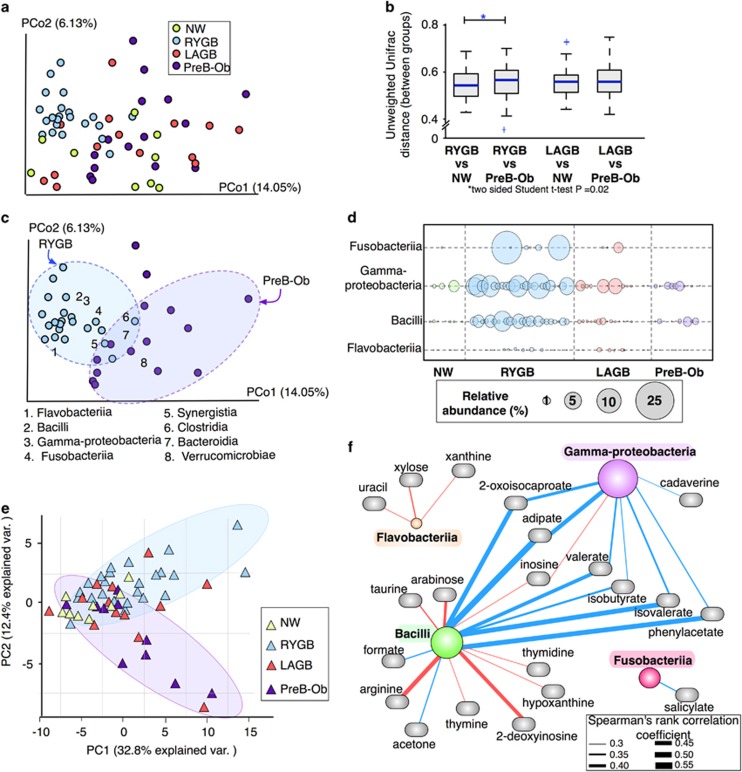
We evaluated community-level differences in the fecal microbiota of NW, RYGB, LAGB and PreB-Ob populations using principal coordinate analysis of unweighted and weighted Unifrac metrics (Lozupone et al., 2006). Weighted Unifrac analysis, which takes into account abundances of operational taxonomic units and reflects differences in the most dominant operational taxonomic units, did not show any specific clustering pattern (Supplementary Figure S1A), although NW group was significantly distant from the other groups on PCo1 (Supplementary Figure S1B). Unweighted Unifrac analysis accounts for the presence or absence of operational taxonomic units; hence, it reflects changes in less dominant phyla. In PCo1, the microbiomes of RYGB and PreB-Ob participants formed clusters having a degree of separation from the other groups based on unweighted Unifrac distances (Figure 2a). The microbiomes of LAGB participants were closer to PreB-Ob participants. Based on permutational analysis of variance and analysis of similarity analyses on unweighted Unifrac distances, RYGB and PreB-Ob groups (P=0.001 for both) and RYGB and LAGB groups (P=0.001 for both) were significantly different. Analysis of similarity R values indicate that bariatric surgery, in particular RYGB, had a significant effect on the microbiota variability (analysis of similarity: R between RYGB and PreB-Ob groups=0.51, and between RYGB and LAGB groups=0.42).
Figure 2.
Microbial community structure after RYGB and LAGB surgeries compared with NW and PreB-Ob controls. (a) Unweighted Unifrac distances between the samples visualized using principal coordinate analysis showed two distinctive clusters: RYGB and PreB-Ob. (b) Pairwise comparison of median unweighted Unifrac distances between the groups. (c) Class-level phylotypes on principal coordinates show the taxa that are responsible for clustering of RYGB participants. (d) Relative abundance of the four main class-level phylotypes that separated the RYGB group from NW, LAGB and PreB-Ob groups. (e) Principal component analysis (PCA) of fecal metabolites. (f) Spearman’s rho correlations between Gammaproteobacteria, Fusobacteriia, Flavobacteriia and Bacilli and fecal metabolites.
As shown in Supplementary Figure S2, microbiomes did not cluster based on other patient-specific data, such as BMI, diet composition, gender or age (permutational analysis of variance P>0.05). Therefore, RYGB surgery was the principal factor responsible for the alteration of gut microbial community structure.
As seen in Figure 2b, the unweighted Unifrac distance between the RYGB and PreB-Ob groups was greater than the distance between the RYGB and NW groups, evidence that RYGB community profiles were more similar to NW than PreB-Ob profiles. This trend was not valid for LAGB participants; the distance between LAGB and Pre-Ob were comparable to the distance between LAGB and NW.
As we observed the greatest difference between RYGB and PreB-Ob groups based on unweighted Unifrac distances, we visualized these groups with the class-level phylotypes that promoted their separation. Figure 2c illustrates that the presences of unique phylotypes from Bacilli, Gammaproteobacteria, Fusobacteriia and Flavobacteriia led the microbiome of RYGB participants to separate from the microbiome of PreB-Ob participants because of higher abundances in the RYGB (Figure 2d). They also were in greater abundance in the LAGB group, compared with nonsurgical groups. Based on LEFSe scores (Segata et al., 2011), as shown in Supplementary Figure S3, Bacilli, Gammaproteobacteria and Prevotellaceae were the microbial signatures discriminating RYGB microbiota from nonsurgical microbiome (NW and PreB-Ob). Gammaproteobacteria and Bacilli also discriminated RYGB from the LAGB group. Flavobacteriia and Porphyromonadacea discriminated LAGB participants from nonsurgical participants, and Bacteroidaceae discriminated the LAGB group from the RYGB group.
Our findings reinforce previous studies that Gammaproteobacteria and Fusobacteriia increased in relative abundance after RYGB (Zhang et al., 2009; Furet et al., 2010; Graessler et al. 2013). High levels of Gammaproteobacteria phylotypes have been associated with weight loss in post-RYGB participants (Furet et al., 2010), but Fusobacteriia have not been evaluated in the context of weight loss or adiposity. In addition, Bacilli and Flavobacteriia were enriched after RYGB surgery, a trend not previously reported. Interestingly, Bacilli include many species, such as Lactobacillus, which have been associated with weight loss (Santacruz et al., 2009). Flavobacteriia were found to be at lower abundance in diabetic population compared with non-diabetic population (Mrozinska et al., 2016).
We evaluated whether distinctive microbiomes of RYGB and PreB-Ob groups also resulted in distinctive fecal metabolomes by using principal component analysis. Figure 2e shows the distribution of fecal metabolomes of NW, RYGB, LAGB and PreB-Ob groups based on 71 metabolites that were detected with 1H-NMR. Although groups did not separate on principal component analysis based on their metabolomes, similar to the observations that are derived from microbiome data, RYGB and PreB-Ob groups showed the greatest separation. NW metabolomes showed narrow distribution, an indication that metabolomes of NW participants were similar to each other compared with participants in other groups. These findings suggest that obesity and RYGB surgery not only changed the microbial community structure, but also changed the microbial functions, reflected by the metabolome.
To probe the associations between fecal microbiome and metabolome, we calculated Spearman’s rho coefficients between the four phylotypes (Gammaproteobacteria, Mollicutes, Bacilli and Fusobacteriia) that separated RYGB from PreB-Ob and fecal metabolites; we considered coefficients greater than the critical value (for n=60, critical value=0.254) with a P-value <0.05 as significant. Figure 2f shows that Gammaproteobacteria and Bacilli positively correlated with amino-acid fermentation and degradation products: isobutyrate, isovalerate, valerate and o-cresol (see Supplementary Table S3 for concentrations). In addition, the abundance of Gammaproteobacteria positively correlated with cadaverine, another protein degradation product. Enrichment of the genes involved in protein degradation after RYGB was previously reported (Palleja et al., 2016), and our fecal metabolome data validate enhanced protein degradation after RYGB surgery. In addition, Bacilli and Flavobacteriia negatively correlated with nucleic acid degradation products such as xanthine, thymidine, uracil, hypoxanthine, thymine and 2-deoxyinosine.
In summary, RYGB and LAGB had different long-term effects on gut microbiomes and metabolomes. RYGB had a stringent clustering effect on microbial phylotypes, whereas LAGB did not impose any condition that distinguished its microbiome from nonsurgical groups. Differences in microbiomes and metabolomes between the RYGB and other groups may be explained by changes to the gastrointestinal environmental created by the surgery, which we discuss below.
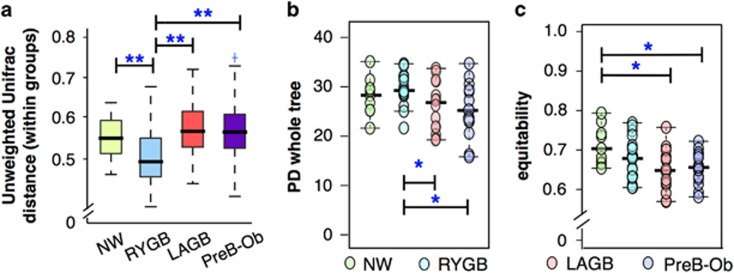
Bariatric surgeries change the microbial diversity in the gut
We evaluated variations within the experimental groups. As seen in Figure 3a, NW, LAGB and PreB-Ob groups had comparable within-group unweighted Unifrac distances. RYGB group had the smallest within-group distance (P<0.05) possibly because of restrictive diets or food intolerances after the surgery (Schweiger et al., 2010). Moreover, surgery-induced alterations in environmental conditions, such as an increased abundance of bile acids on gut microbiota after RYGB surgery (Bhutta et al., 2015), can reduce variation within the RYGB group.
Figure 3.
Within-community variation and alpha diversity indexes reveal differences in the community structure. (a) Within-group Unweighted Unifrac distance was the smallest for the RYGB group. Alpha diversity parameters: (b) PD whole tree and (c) equitability indexes for diversity and evenness.
The rearrangement of the gastrointestinal tract after RYGB alters gut pH, oxygen content and bile-acid concentrations, and nutrient exposure that are delivered to the colon (Li et al., 2011), these factors can affect microbial diversity. In order to understand the impact of anatomically distinct bariatric surgeries on microbial diversity, we compared post-RYGB and post-LAGB microbial diversity with non-matched nonsurgical NW and PreB-Ob participants. According to the alpha diversity indexes, the RYGB group had significantly higher diversity than LAGB and PreB-Ob groups (P<0.05), as seen in Figure 3b. The PreB-Ob group had the lowest microbial diversity, and our results are in agreement with previous reports that obese individuals have lower alpha diversity (Turnbaugh et al., 2006; Qin et al., 2010). NW and RYGB groups had higher equitability scores than LAGB and PreB-Ob (Figure 3c), indicating that phylotypes were more equally distributed in these groups. Lower diversity and evenness in PreB-Ob and LAGB groups may be attributed to their relatively higher dietary fat consumption and imbalanced diets (Supplementary Figure S4).
The high diversity in RYGB participants may be explained by the altered environmental conditions after RYGB (Li et al., 2011). For instance, a slight increase in the gut pH after RYGB can enhance the survival of acid-sensitive microorganisms (Chung et al., 2016), and increased oxygen in the gut can promote the growth of facultative anaerobes (Hartman et al., 2009). In addition, an altered bile flow after RYGB can stimulate the growth of bile-acid-transforming bacteria (Li et al., 2011). As anatomical rearrangement is not part of LAGB, similar changes would not be anticipated. Therefore, only RYGB creates the large environmental changes in the gut critical for colonization and succession of novel microbial species in the colon.
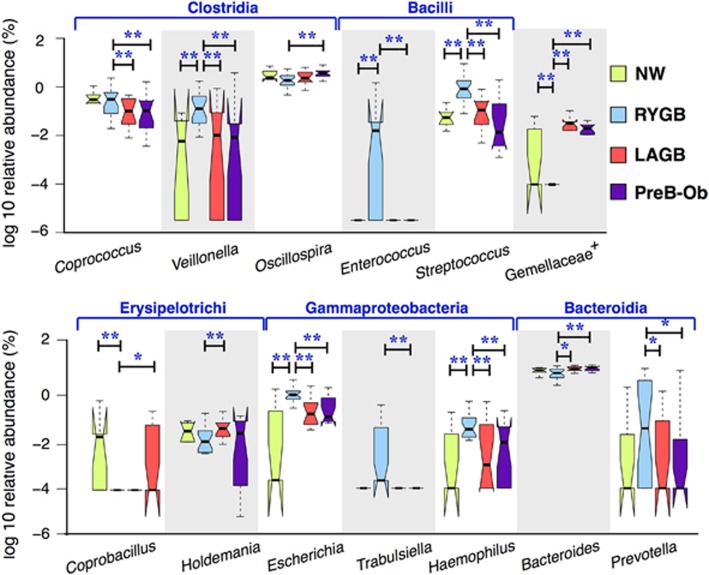
Weight loss-associated oral microbial phylotypes were enriched in the fecal microbiomes after RYGB
Supplementary Table S4 documents that the RYGB group had more facultative anaerobes and fewer anaerobes than the other groups, which supports a previous hypothesis that RYGB surgery increases the oxygen content of the gut (Zhang et al., 2009). As shown in Figure 4, RYGB significantly enriched or depleted microorganisms from five different classes: Clostridia, Bacilli, Erysipelotrichi, Gammaproteobacteria and Bacteroidia. Some of our findings confirm previous findings about the microbiome after RYGB surgery: Escherichia (Liou et al., 2013), Streptococcus (Palleja et al., 2016) and Veillonella (Palleja et al., 2016) were in greater abundance in post-RYGB individuals or animals. In addition, we observed greater abundances of Trabulsiella, Coprococcus and Prevotella and lower abundances of Oscillospira, Coprobacillus, Holdemania and Bacteroides in the RYGB group. The relative abundances of these microbial phylotypes were similar in LAGB and PreB-Ob groups.
Figure 4.
Significantly abundant genera in the RYGB participants compared with the other groups. * and ** denote P<0.01 and P<0.05, respectively, of Mann–Whitney U-test adjusted results.
Changes in microbial community composition after RYGB can be explained by the combination of multiple drastic alterations in the gut environment. For instance, bile-tolerant Streptococcus (Lim et al., 2004), Escherichia (Begley et al., 2005), Enterococcus (Begley et al., 2005) and Bacteroides (Begley et al., 2005); facultative anaerobic Escherichia (Turner et al., 2003) and Streptococcus (Turner et al., 2003); and acid-sensible Bacteroides (Duncan et al., 2009), Escherichia (Duncan et al., 2009), and Coprococcus (Duncan et al., 2009) species increased in relative abundance after RYGB.
Some of the phylotypes that were enriched in the RYGB, such as Veillonella, Enterococcus, Haemophilus, Prevotella and Streptococcus, are commonly found together in individuals with dental cavities or periodontitis (Liljemark and Bloomquist, 1996). Proton pump inhibitors, which reduce gastric acid secretions, increased the abundance of oral cavity associated microorganisms including Enterococcus, Streptococcus and Veillonella (Imhann et al., 2016). The great abundance of these phylotypes after RYGB suggests that the conditions in the gut after this surgery allowed higher transport and survival of oral microorganisms and promoted growth of the upper-gastrointestinal tract species in the distal bowel by a similar aggregation mechanism as oral biofilm formation. In addition, Prevotella and Escherichia proliferate in the large bowel of mice when gastric acid secretion is inhibited (Kanno et al., 2009). As RYGB reduces gastric acid contact with the nutrient stream (Rubin et al., 2004) and increases the distal excluded stomach pH to ~4 (Liou et al., 2013), these facultative anaerobes from the oral mucosa, which thrive at neutral pH levels, can bypass harsh stomach acid in transit to the colon (Palleja et al., 2016). In contrast, low abundance of these species after LAGB, where normal digestive physiology is retained, further supports that the observed microbiome changes were because of rearrangement of the gastrointestinal tract after RYGB surgery. We did not see a significant difference in fecal pH (NW=7.4, RYGB=7.2, LAGB=7.8 and PreB-Ob=7.4), probably because biochemical selection of species happens within the stomach and small intestine, before microorganisms reach the colon.
A majority of the genus-level phylotypes that were enriched in the RYGB group positively correlated with %EWL. Table 2 shows Spearman correlation coefficients of the relationships between genera and EWL% and BMI. When we performed Spearman’s correlation on the combined data set, we observed that Akkermansia, Escherichia, Veillonella, Enterococcus and Carnobacterium with unidentified phylotypes from Gemellaceae and Peptococcaeae, positively correlated with %EWL. In humans and animals, an increase in the relative abundance of Escherichia after RYGB surgery has been associated with weight reduction and adiposity independently from diet (Furet et al., 2010; Liou et al., 2013). Escherichia, Enterococcus, Streptococcus and Veillonella were signatures of RYGB surgery that discriminated RYGB from both nonsurgical and LAGB groups (Supplementary Figure S3). These phylotypes were not abundant in LAGB and PreB-Ob groups, although the participants within those groups were on negative energy balance. In addition, these phylotypes did not show any significant correlation with the dietary listed in Supplementary Table S2; hence, we conclude that changes in these phylotypes that correlated with %EWL were because of surgery and independent from diet.
Table 2. Spearman correlation coefficients between genus-level phylotypes in fecal samples and %EWL and BMI distribution of subjects.
|
%EWL |
BMI |
||||
|---|---|---|---|---|---|
| Spearman's rho | P-value | Spearman's rho | P-value | ||
| Escherichia | 0.34 | 0 | Ruminococcus | −0.316 | 0.012 |
| Akkermansia | 0.34 | 0 | Unidentified Rikenellaceae | −0.278 | 0.029 |
| Unidentified Peptococcaeae | 0.334 | 0 | Coprococcus | −0.273 | 0.032 |
| Streptococcus | 0.364 | 0 | Unidentified Ruminococcaceae | −0.271 | 0.033 |
| Veillonella | 0.367 | 0 | Bacteroides | 0.292 | 0.021 |
| Unidentified Gemellaceae | 0.405 | 0 | Sutterella | 0.314 | 0.013 |
| Carnobacterium | 0.458 | 0 | |||
| Bifidobacterium | −0.391 | 0 | |||
| Sutterella | −0.303 | 0 | |||
Abbreviations: BMI, body mass index; %EWL, % excess weight loss.
Critical values above 0.254 with P<0.05 were considered significant correlations.
We observed negative correlation between phylotypes including Bifidobacterium and Sutterella and %EWL (Table 2). Sutterella was observed in greater abundances in diet-induced obese mice compared with lean mice (Clarke et al., 2013) whereas Bifidobacterium is associated with lean metabolic types (Million et al., 2011). Sutterella and Bacteroides were positively correlated with BMI indicating that they were more abundant in the obese individuals. On the other hand, Ruminococcus, Coprococcus and unidentified phylotypes from Ruminococcaceae and Rikenellaceae negatively correlated with the BMI. Ruminococcus and Coprococcus (Kasai et al., 2015) have been associated with an obese metabolic type, but we speculate that these changes may reflect diverse diets and host physiology and, therefore, the relationship is ambiguous.
Butyrate, propionate and branched chain fatty acids were at great concentrations only in post-RYGB group
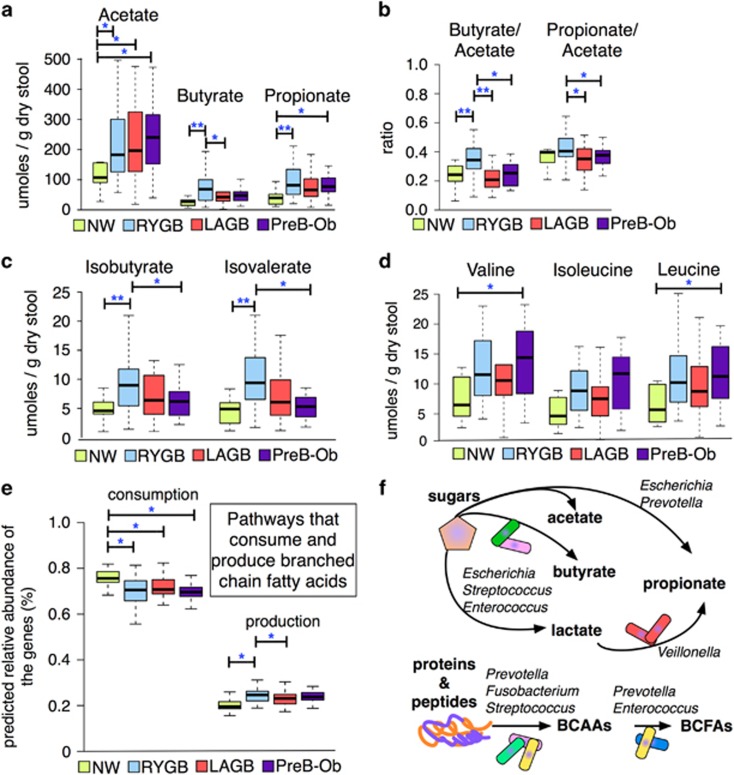
As seen in Figure 5a, the concentration of acetate, a major carbohydrate fermentation product, was higher in the PreB-Ob group, followed by the LAGB, RYGB and NW groups. The RYGB group had the highest concentrations of butyrate and propionate. In addition, LAGB and PreB-Ob groups had comparable levels of butyrate and propionate, and NW had the lowest concentrations of these two SCFAs. Liou et al. (2013) showed that fecal propionate concentration increases after RYGB surgery. In contrast to our findings, Tremaroli et al. (2015) reported that post-RYGB and post-VBG patients had lower levels of SCFAs including propionate. However, owing to minor anatomical differences between LAGB and VBG, variations between the populations, and differences in sample preparation methods, the results from Tremaroli et al. (2015) and our study are not comparable.
Figure 5.
Fecal fermentation products and substrates. (a) The most abundant fermentation products: acetate, propionate and butyrate showed different distribution among the groups. (b) Ratios of butyrate-to-acetate and propionate-to-acetate were greatest in the RYGB group compared with NW, LAGB and PreB-Ob groups. (c) Isobutyrate and isovalerate, which are fermentation products of BCAAs (isoleucine and valine), were at greater abundance in the RYGB group compared with the other groups. (d) BCAAs: valine, leucine and isoleucine concentrations did not significantly vary among the groups. (e) PICRUSt predicted relative abundance of the genes involved in the consumption and production of BCFAs. (f) Illustration of conversion of carbohydrates and proteins into short chain and branched chain fatty acids by significantly abundant taxa in RYGB participants. * and ** denote Mann–Whitney U-test Padj value <0.05 and <0.01, respectively.
Figure 5b shows the fecal butyrate-to-acetate and propionate-to-acetate ratios in the groups, which indicate changes in fermentative pathways of gut microbes after surgery. The butyrate-to-acetate ratio was significantly higher in the RYGB group, and the propionate-to-acetate ratio was significantly higher in RYGB compared with LAGB and PreB-Ob groups. Our findings suggest that RYGB-enriched microbiota or conditions that favor the accumulation of more reduced SCFAs, such as butyrate rather than acetate. Although we observed a greater concentration of butyrate in the RYGB group, relative abundances of many well-known butyrate producers, such as Eubacterium and Roseburia (Louis and Flint, 2009), were not significantly higher in the RYGB group compared with the other groups (P>0.05), with the exception of Coprococcus (Hold et al., 2003) (Figure 4). As relative abundances of phylotypes do not reflect their true quantities, we avoid making a connection between the relative abundances of butyrate producers and concentration of butyrate among the groups.
We also measured BCFAs, these are fermentation products of BCAA. Figure 5c shows that the RYGB samples had the highest concentrations of BCFAs: isobutyrate and isovalerate. Tremaroli et al. (2015) also observed greater concentrations of BCFAs in post-RYGB and post-VBG participants compared with BMI-matched controls. Our results are similar, however, we did not observe greater concentrations of these BCFAs in LAGB participants showing that VBG and LAGB affect the gut microbiome and metabolism very differently.
When we looked at the abundance of BCAAs, we observed that the concentrations were not statistically different among the groups, although NW participants had slightly lower concentrations of BCAAs (Figure 5d). Using PICRUSt (Supplementary Table S5) to analyze predicted genes involved in the BCFA production (leucine, isoleucine and valine degradation, KEGG pathway 00280) and consumption pathways (leucine, isoleucine and valine biosynthesis, KEGG pathway 00290), we found that genes associated with BCFA production were significantly more abundant in the NW group compared with the other groups, as shown in Figure 5e. The predicted genes involved in BCFA production, such as acyl-coA hydrolase and branched chain ketoacid dehydrogenases (Thierry et al., 2002), were in greater abundance in the RYGB group, compared with the other groups. Microorganisms can deaminate BCAAs to α-ketoisovaleric and α-keto-β-methylvaleric acids, the precursors of BCFAs (Thierry et al., 2002). Branched chain aminotransferase (BcaT) and branched chain-α-ketoacid dehydrogenase can yield α-ketoisocaproic acid and isovaleryl-CoA, respectively, which can be converted into BCFAs (Díaz-Pérez et al., 2016).
Tremaroli et al. (2015) showed that the microbial pathways in RYGB and VBG participants shifted from carbohydrate to protein fermentation. Our data on SCFAs, BCFAs, BCAAs, and the genes involved in amino-acid fermentation in the RYGB group strengthen the trend observed by Tremaroli et al. (2015). We did not note this kind of shift in post-LAGB participants although Tremaroli et al. (2015) observed it after VBG, a surgery similar to LAGB.
Phylotypes that produce SCFAs and BCFAs form a metabolic network that involves production of vitamins and fermentation of carbohydrates and proteins. For example, Streptococcus, Escherichia, Prevotella and Enterococcus ferment sugars and produce lactate and pyruvate for microorganisms that cannot utilize sugars, such as Veillonella (Hojo et al., 2009). Veillonella produces vitamin K, which stimulates the growth of Prevotella (Hojo et al., 2009). As illustrated in Figure 5f, microbial phylotypes such as Fusobacterium (Bachrach et al., 2004), Prevotella (Wallace et al., 1999), and Streptococcus (Wallace et al., 1999) can degrade proteins and peptides into amino acids. Microbial phylotypes that became enriched after RYGB, including Veillonella, Prevotella, Escherichia and Streptococcus can ferment amino acids and carbohydrates (Dai et al., 2011). Microbial phylotypes such as Prevotella and Enterococcus ferment BCAAs (valine and leucine) into BCFAs (isovalerate and isobutyrate) (Takahashi et al., 1997). Finally, Prevotella produce alkali to neutralize the pH because of acid production during amino-acid fermentation to BCFA (Takahashi et al., 1997).
SCFAs and BCFAs can have many effects on host energy metabolism. Acetate has been associated with fat depositions in the liver (Siddiqui et al., 2015), whereas butyrate and propionate have been associated with weight loss (Lin et al., 2012). Higher fecal propionate concentrations also were observed in mice that received fecal transplants from lean or post-RYGB mice, and they correlated with diet-independent weight loss (Liou et al., 2013; Ridaura et al., 2013). Propionate and, to a lesser extent, butyrate can regulate weight loss hormones, such as glucagon-like peptide-1 (Lin et al., 2012), and both were previously shown to protect against obesity (Lin et al., 2012). Besides glucagon-like peptide-1, propionate can induce secretion of appetite-reducing hormone peptide YY (Chambers et al., 2015). Therefore, propionate-to-acetate ratio is an important factor for the evaluation of microbiota’s contribution to host energy balance.
Conversion of BCAA to BCFA is optimal above neutral pHs (Thierry et al., 2002). The reduction in gastric acid secretion after RYGB (Rubin et al., 2004) may stimulate the production of BCFA by increasing the BCFA-producing bacteria or favor thermodynamics of these reactions. A previous study showed that leptin-deficient obese mice had lower isobutyrate levels than controls, indicating that the obese mice had lower leucine and valine fermentation (Won et al., 2013). Moreover, increased BCFAs and other SCFAs have been associated with increased satiety via increases in peptide YY (Haenen et al., 2013). An increase in satiety hormones, was previously observed in individuals who have had RYGB (Karra and Batterham, 2010). Our results of higher concentrations of BCFAs in the RYGB group, independent from diet, suggest that RYGB surgery enhanced the abundance of bacteria and pathways producing BCFAs. Limitations in carbohydrate intake may lead to a switch from fermentation of carbohydrates to amino acids by the gut microbiota in post-RYGB participants. In summary, RYGB had higher concentration of SCFAs and BCFAs that can interact with free fatty acid receptors and regulate the hunger/satiety response, suggesting a microbiota-associated role in sustained weight loss. These important changes were not observed after LAGB.
In conclusion, we observed major differences in the structure of the fecal microbiome and metabolome in post-RYGB and post-LAGB participants, compared with PreB-Ob and NW participants. The differences were more substantial in post-RYGB than post-LAGB and may be attributable to the very different environmental conditions caused by the radically altered gastrointestinal anatomy post-RYGB. This comparative bariatric surgery approach provides evidence that changes in human gastrointestinal anatomy are reflected in the microbiome and weight loss-associated microbial metabolites.
Acknowledgments
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under Award Number R01DK090379. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank Carole Flores for editing assistance, Irene Galasky and Susanna Murphy for recruitment efforts, and anonymous reviewers for their insightful comments. Portions of this research were supported by the PNNL Laboratory Directed Research and Development program and includes contribution from the Signatures Discovery Initiative. PNNL is a multi-program national laboratory operated by Battelle for the U.S. Department of Energy under Contract DE-AC05-76RL01830.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Bachrach G, Rosen G, Bellalou M, Naor R, Sela MN. (2004). Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol Immunol 19: 155–159. [DOI] [PubMed] [Google Scholar]
- Begley M, Gahan CGM, Hill C. (2005). The interaction between bacteria and bile. FEMS Microbiol Rev 29: 625–651. [DOI] [PubMed] [Google Scholar]
- Bhutta HY, Rajpal N, White W, Freudenberg JM, Liu Y, Way J et al. (2015). Effect of Roux-en-Y gastric bypass surgery on bile acid metabolism in normal and obese diabetic rats. PLoS One 10: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boza C, Gamboa C, Awruch D, Perez G, Escalona A, Ibanez L. (2010). Laparoscopic Roux-en-Y gastric bypass versus laparoscopic adjustable gastric banding: five years of follow-up. Surg Obes Relat Dis 6: 470–475. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SEK et al. (2015). Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64: 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WSF, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D et al. (2016). Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol 14: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, O’Sullivan O, Ross RP, O’Toole PW, Shanahan F et al. (2013). Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS One 8: e65790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z-L, Wu G, Zhu W-Y. (2011). Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci 16: 1768–1786. [DOI] [PubMed] [Google Scholar]
- Damms-Machado A, Mitra S, Schollenberger AE, Kramer KM, Meile T, Koenigsrainer A et al. (2015). Effects of surgical and dietary weight loss therapy for obesity on gut microbiota composition and nutrient absorption. Biomed Res Int 2015: 806248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Pérez AL, Díaz-Pérez C, Campos-García J. (2016). Bacterial l-leucine catabolism as a source of secondary metabolites. Rev Environ Sci Biotechnol 15: 1–29. [Google Scholar]
- Faith DP. (1992). Conservation evaluation and phylogenetic diversity. Biol Conserv 61: 1–10. [Google Scholar]
- Furet J-P, Kong L-C, Tap J, Poitou C, Basdevant A, Bouillot J-L et al. (2010). Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss links with metabolic and low-grade inflammation markers. Diabetes 59: 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong ML et al. (2013). Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J 13: 514–522. [DOI] [PubMed] [Google Scholar]
- Haenen D, Zhang J, da Silva CS, Bosch G, van der Meer IM, van Arkel J et al. (2013). A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. J Nutr 143: 274–283. [DOI] [PubMed] [Google Scholar]
- Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M et al. (2009). Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci USA 106: 17187–17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo K, Nagaoka S, Ohshima T, Maeda N. (2009). Bacterial interactions in dental biofilm development. J Dent Res 88: 982–990. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Thomson JM, Flint HJ. (2009). The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11: 2112–2122. [DOI] [PubMed] [Google Scholar]
- Hold GL, Schwiertz A, Aminov RI, Blaut M, Flint HJ. (2003). Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl Environ Microbiol 69: e-pub ahead of print; doi: 10.1128/aem.69.7.4320-4324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhann F, Bonder MJ, Vila AV, Fu J, Mujagic Z, Vork L et al. (2016). Proton pump inhibitors affect the gut microbiome. Gut 65: 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D-W, DiBaise JK, Ilhan ZE, Crowell MD, Rideout JR, Caporaso JG et al. (2015). Gut microbial and short-chain fatty acid profiles in adults with chronic constipation before and after treatment with lubiprostone. Anaerobe 33: 33–41. [DOI] [PubMed] [Google Scholar]
- Kanno T, Matsuki T, Oka M, Utsunomiya H, Inada K, Magari H et al. (2009). Gastric acid reduction leads to an alteration in lower intestinal microflora. Biochem Biophys Res Commun 381: 666–670. [DOI] [PubMed] [Google Scholar]
- Karra E, Batterham RL. (2010). The role of gut hormones in the regulation of body weight and energy homeostasis. Mol Cell Endocrinol 316: 120–128. [DOI] [PubMed] [Google Scholar]
- Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H et al. (2015). Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol 15: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner J, Inabet W, Fabres G, Conwell IM, McMahon DJ, Salas R et al. (2009). Prospective study of gut hormone and metabolic changes after adjustable gastric banding and Roux-en-Y gastric bypass. Int J Obes 33: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME et al. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 278: 25481–25489. [DOI] [PubMed] [Google Scholar]
- le Roux CW, Aylwin SJB, Batterham RL, Borg CM, Coyle F, Prasad V et al. (2006). Gut hormone profiles following bariatric surgery favor an anorectic state, facilitate weight loss, and improve metabolic parameters. Ann Surg 243: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. (2005). Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JV, Ashrafian H, Bueter M, Kinross J, Sands C, le Roux CW et al. (2011). Metabolic surgery profoundly influences gut microbial-host metabolic cross-talk. Gut 60: 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljemark WF, Bloomquist C. (1996). Human oral microbial ecology and dental caries and periodontal diseases. Crit Rev Oral Biol Med 7: 180–198. [DOI] [PubMed] [Google Scholar]
- Lim HJ, Kim SY, Lee WK. (2004). Isolation of cholesterol-lowering lactic acid bacteria from human intestine for probiotic use. J Vet Sci 5: 391–395. [PubMed] [Google Scholar]
- Lin HV, Frassetto A, Kowalik EJ Jr, Nawrocki AR, Lu MM, Kosinski JR et al. (2012). Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. Plos One 7: e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AP, Paziuk M, Luevano J-M Jr, Machineni S, Turnbaugh PJ, Kaplan LM. (2013). Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 5: 178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294: 1–8. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. (2006). UniFrac - An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform 7: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. (2012). PANDAseq: Paired-end assembler for Illumina sequences. Bmc Bioinform 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P et al. (2011). Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes 36: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mrozinska S, Radkowski P, Gosiewski T, Szopa M, Bulanda M, Ludwig-galezowska AH et al. (2016). Qualitative parameters of the colonic flora in patients with HNF1A-MODY are different from those observed in type 2 diabetes mellitus. J Diabetes Res 2016: 3876764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien PE. (2010). Bariatric surgery: mechanisms, indications and outcomes. J Gastroenterol Hepatol 25: 1358–1365. [DOI] [PubMed] [Google Scholar]
- Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–U70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleja A, Kashani A, Allin KH, Nielsen T, Zhang C, Li Y et al. (2016). Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual microbiota. Genome Med 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psichas A, Sleeth ML, Murphy KG, Brooks L, Bewick GA, Hanyaloglu AC et al. (2015). The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int J Obes 39: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1079–U1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin JP, Nguyen V, Schwentker A. (2004). Perioperative management of the post-gastric-bypass patient presenting for body contour surgery. Clin Plast Surg 31: 601–610. [DOI] [PubMed] [Google Scholar]
- Santacruz A, Marcos A, Warnberg J, Marti A, Martin-Matillas M, Campoy C et al. (2009). Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity 17: 1906–1915. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Smith NJ, Christiansen E, Tikhonova IG, Grundmann M, Hudson BD et al. (2011). Selective orthosteric free fatty acid receptor 2 (FFA2) agonists identification of the structural and chemical requirements for selective activation of FFA2 versus FFA3. J Biol Chem 286: 10628–10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger C, Weiss R, Keidar A. (2010). Effect of different bariatric operations on food tolerance and quality of eating. Obes Surg 20: 1393–1399. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shak JR, Roper J, Perez-Perez GI, Tseng C-h, Francois F, Gamagaris Z et al. (2008). The effect of laparoscopic gastric banding surgery on plasma levels of appetite-control, insulinotropic, and digestive hormones. Obes Surg 18: 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui MS, Fuchs M, Idowu MO, Luketic VA, Boyett S, Sargeant C et al. (2015). Severity of nonalcoholic fatty liver disease and progression to cirrhosis are associated with atherogenic lipoprotein profile. Clin Gastroenterol Hepatol 13: 1000–U1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Saito K, Schachtele CF, Yamada T. (1997). Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalisPrevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol 12: 323–328. [DOI] [PubMed] [Google Scholar]
- Thierry A, Maillard MB, Yvon M. (2002). Conversion of L-leucine to isovaleric acid by Propionibacterium freudenreichii TL 34 and ITGP23. Appl Environ Microbiol 68: 608–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice JA, Karliner L, Walsh J, Petersen AJ, Feldman MD. (2008). Gastric banding or bypass? A systematic review comparing the two most popular bariatric procedures. Am J Med 121: 885–893. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Karlsson F, Werling M, Stahlman M, Kovatcheva-Datchary P, Olbers T et al. (2015). Roux-en-Y gastric bypass and vertical banded gastroplasty inducelong-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab 22: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Turner NJ, Thomson BM, Shaw IC. (2003). Bioactive isoflavones in functional foods: the importance of gut microflora on bioavailability. Nutr Rev 61: 204–213. [DOI] [PubMed] [Google Scholar]
- Wallace RJ, Atasoglu C, Newbold CJ. (1999). Role of peptides in rumen microbial metabolism. Asian-Australasian J Anim Sci 12: 139–147. [Google Scholar]
- Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. (2006). Targeted profiling: quantitative analysis of H-1NMR metabolomics data. Anal Chem 78: 4430–4442. [DOI] [PubMed] [Google Scholar]
- Won E-Y, Yoon M-K, Kim S-W, Jung Y, Bae H-W, Lee D et al. (2013). Gender-specific metabolomic profiling of obesity in leptin-deficient ob/ob mice by H-1NMR spectroscopy. Plos One 8: e75998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M-S. (2016). The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients 8: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurcisin BM, Gaddor MM, DeMaria EJ. (2009). Obesity and bariatric surgery. Clin Chest Med 30: 539–553. [DOI] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y et al. (2009). Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 106: 2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.