Abstract
The biological significance of Archaea in the human gut microbiota is largely unclear. We recently reported genomic and biochemical analyses of the Methanomassiliicoccales, a novel order of methanogenic Archaea dwelling in soil and the animal digestive tract. We now show that these Methanomassiliicoccales are present in published microbiome data sets from eight countries. They are represented by five Operational Taxonomic Units present in at least four cohorts and phylogenetically distributed into two clades. Genes for utilizing trimethylamine (TMA), a bacterial precursor to an atherosclerogenic human metabolite, were present in four of the six novel Methanomassiliicoccales genomes assembled from ELDERMET metagenomes. In addition to increased microbiota TMA production capacity in long-term residential care subjects, abundance of TMA-utilizing Methanomassiliicoccales correlated positively with bacterial gene count for TMA production and negatively with fecal TMA concentrations. The two large Methanomassiliicoccales clades have opposite correlations with host health status in the ELDERMET cohort and putative distinct genomic signatures for gut adaptation.
Introduction
The human microbiome is now recognized as an environmental modifier of disease risk (Grice and Segre, 2012). Definition of the catalog of taxa and gene functions in multiple large-scale projects, with a particular emphasis on the gut, has allowed the identification of altered microbiota compositions or functional states that correlate with disease states (Qin et al., 2010; Human Microbiome Project Consortium, 2012; Le Chatelier et al., 2013; Forslund et al., 2015; Falony et al., 2016). Most large-scale microbiome studies have focussed on the Bacteria, with relatively little attention being paid to the viral, micro-eukaryotic and archaeal components. It is notable, however, that the domain Archaea is the only domain of life that includes no pathogenic members. Their association with human health have been questioned for many years since the pioneering study by the Levitt group (Bond et al., 1971) and the isolation of the first two methanogen species consistently found in the human microbiota, Methanobrevibacter smithii (Miller et al., 1982) and Methanosphaera stadtmanae (Miller and Wolin, 1985). These two taxa belong to the Methanobacteriales order. There is one clear case of health association evident in the higher occurrence rate of Methanobrevibacter oralis in periodontal diseases, whereby this methanogen could promote the growth of pathogenic bacteria by lowering the redox level (Lepp et al., 2004). Independent of health condition, another commonly reported trend is the increase of methanogen occurrence and diversity among older adults (Mihajlovski et al., 2010; Dridi et al., 2012a; Fernandes et al., 2013; Polag et al., 2014). However, beyond those two associations, few links between methanogen occurrence/abundance and health conditions have been consistently reported over multiple studies. Several recent findings and mechanistic hypotheses have offered new perspectives on potential implications of methanogens for human health (as reviewed in Gaci et al., 2014; Bang and Schmitz, 2015; Horz, 2015; Lurie-Weinberger and Gophna, 2015). Among these, three studies have reported that Msp. stadtmanae, but not Mbr. smithii, could have an important pro-inflammatory effect in the lung (Blais Lecours et al., 2011) and in the gut (Bang et al., 2014; Blais Lecours et al., 2014). Moreover, occurrence of Msp. stadtmanae is more frequent in subjects with inflammatory bowel disease (Blais Lecours et al., 2014), raising questions about its potential role in the development of allergic and autoimmune diseases (Bang and Schmitz, 2015).
During the same time frame, our understanding of the diversity of human-associated methanogens was extended by the description of three novel methanogen species, Methanomassiliicoccus luminyensis (Gorlas et al., 2012; Dridi et al., 2012b), ‘Candidatus Methanomassiliicoccus intestinalis Mx1’ (Borrel et al., 2013) and ‘Candidatus Methanomethylophilus alvus Mx1201’ (Borrel et al., 2012), constituting the first cultured and sequenced representatives of a novel order of Archaea, the Methanomassiliicoccales. The characterization of these methanogens led us to propose that human-associated Archaea might even have unexpected positive effects on health (Brugère et al., 2014; Gaci et al., 2014). Specifically, these species have the genetic potential to deplete a deleterious compound, trimethylamine (TMA), through methanogenesis based on the H2-dependent reduction of methyl compounds (Borrel et al., 2014). This predicted metabolic activity was subsequently confirmed by biochemical characterization of Mmc. luminyensis (Brugère et al., 2014). In the human body, TMA is produced exclusively by the activity of gut bacteria (Koeth et al., 2013). After diffusion into the blood, TMA is oxidized by the liver enzyme FMO3 monooxygenase into trimethylamine-N-oxide (TMAO), a molecule increasingly recognized as an important factor for the development of cardiovascular disease (Wang et al., 2011; Zhu et al., 2016) and chronic kidney diseases (Tang et al., 2015). Also, the physiological deficiency of the liver FMO3 monooxygenase prevents the oxidation of TMA to TMAO and causes trimethylaminuria, a condition that affects 0.5%–11% of people across distinct ethnic populations (Mitchell et al., 1997; Mackay et al., 2011). By removing TMA directly from its source, naturally occurring Methanomassiliicoccales representatives could help to prevent cardiovascular disease, chronic kidney diseases or trimethylaminuria. Thus we proposed that human-associated Methanomassiliicoccales strains could be used as probiotics, namely, ‘Archaebiotics’, particularly for mitigating trimethylaminuria (Brugère et al., 2014). However, the functional diversity of human-associated Methanomassiliicoccales is still poorly understood and many aspects need further exploration in order to clarify their role in TMA bioremediation (naturally occurring or deliberately instigated) and other yet undefined connections with human health.
In the present study, we identified the predominant Methanomassiliicoccales representatives associated with the human gut microbiome based on a meta-analysis of published data sets and novel data from the ELDERMET cohort of older subjects (>64 years of age) in Ireland (Claesson et al., 2011, 2012; O’Toole and Jeffery, 2015). The capacity of predominant Methanomassiliicoccales species to utilize TMA was then assessed by analyzing six genomes assembled de novo from ELDERMET metagenomics data and by the biochemical characterization of ‘Ca. Mmp. alvus’. Additionally, we determined Methanomassiliicoccales levels in ELDERMET gut samples and compared them with fecal TMA concentrations. Functional links between TMA-producing bacteria and Methanomassiliicoccales were further explored by evaluating the effect of the redundancy and level of four distinct bacterial TMA-producing pathways on Methanomassiliicoccales distribution. Previous analyses of the ELDERMET cohort have focussed on diet–microbiota–health interactions and reported an accelerated health decline linked with diet modification and a strong shift in the bacterial microbiota composition after subjects entered long-term residential care (Claesson et al., 2011, 2012; O’Toole and Jeffery, 2015). We investigated how Methanomassiliicoccales distribution is affected by this important lifestyle modification. Putative genomic signatures accounting for differential abundance of these Archaea were identified.
Experimental Procedures
ELDERMET cohort data
The subjects are Irish people aged >64 years. They were recruited as previously described (Claesson et al., 2011). Ethical approval was obtained from the Cork Clinical Research Ethics Committee. Subjects provided an informed consent. In case of cognitive impairment, consent was given by the next-of-kin to the subject. None of the subjects has an advanced organic disease, alcohol abuse or participates to a drug intervention. The average age of the 371 ELDERMET subjects is 78 (±8) years and range from 64 to 102 years. Frailty of the subjects was previously assessed with Functional Independence Measure index and Barthel Score (Jeffery et al., 2016).
Molecular methods and bioinformatics
DNA extraction and bacterial 16S rRNA amplicon sequencing was previously performed on 732 stool samples from the 371 ELDERMET subjects as described in Jeffery et al., (2016). Shotgun sequencing of the total fecal DNA of a subset of 190 ELDERMET samples was performed using the Illumina HiSeq 2000 (Illumina Inc., San Diego, CA, USA) to generate 100 bp paired-end read libraries following the manufacturer’s instructions. Reads were assembled de novo using IDBA_UD v1.1.1 (Peng et al., 2012).
Methanomassiliicoccales 16S rRNA sequences from publically available cohorts/data sets were obtained from NCBI, EMBL or were provided by the authors of published studies and used to query (with blastn) the ELDERMET metagenomes to identify Methanomassiliicoccales 16S rRNA within this cohort. Operational Taxonomic Units (OTUs) were defined using the USEARH suite of algorithms on publically available and newly identified ELDERMET 16S rRNA gene sequences (Edgar, 2010). The cluster_fast script was implemented with the length sorting option and a 97% identity cutoff.
Six ELDERMET samples having Methanomassiliicoccales 16S rRNA genes on contigs >10 kb in their associated metagenomes were resequenced in order to improve the length and coverage of Methanomassiliicoccales sequences. Newly obtained reads were pooled and assembled with reads from the first sequencing round. Draft genomes were constructed from six metagenomic samples with Methanomassiliicoccales contigs identified based on binning by read coverage, guanine–cytosine content and tetranucleotide frequency using Emergent Self-Organizing Maps (Dick et al., 2009; Supplementary Figure S1) and comparisons to publically available reference genomes. Tetranucleotide frequency calculations were performed on 5–10 kb fragments of scaffolds >5 kb. Completeness and contamination of the draft genomes were estimated based on a presence/absence analysis of 199 single-copy marker genes shared by complete genomes belonging to the Diaforarchaea, the super-class of Archaea containing the Methanomassiliicoccales (Supplementary Table S1). KEGG (Kyoto Encyclopedia of Genes and Genomes) orthology (KO) annotation was carried out with BlastKOALA (Kanehisa et al., 2016). The core, variable and pan-genome were computed with SiLiX (Miele et al., 2011), using the online Microscope platform of Genoscope (http://www.genoscope.cns.fr) with threshold cutoffs of 50% identity and 80% alignment length. Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline and results were manually curated. Proteins containing the Flg_New (pfam09479) and Sel1_repeat (pfam09479) domains were identified with IMG/ER database (November 2015) against 13 871 draft and complete prokaryotic and eukaryotic genomes (Markowitz et al., 2012).
Quantification of the ‘Host-associated clade’ Methanomassiliicoccales and ‘Free-living clade’ Methanomassiliicoccales, as well as Mbr. smithii and Msp. stadtmanae were performed by quantitative PCR (qPCR) using newly designed primers (Supplementary Table S2). The detection level and specificity of these primers was evaluated based on comparisons with metagenomic reads mapping to the corresponding 16S rRNA genes as described in Supplementary Experimental Procedures and Supplementary Figure S2. For absolute quantifications, a standard was added to all qPCR. It was prepared using a plasmid containing a cloned 16S rRNA or mcrA amplicon, which was serially 10 times diluted from 108 to 102 copies per μl. Bacteria were quantified using the F_Bact1369/R_Prok1492 primer set (Sokol et al., 2009) and were used to determine relative abundance of the four quantified archaeal lineages. Quantifications were performed on 342 ELDERMET subjects. The potential association of the four lineages of Archaea with the four bacterial microbiota modules defined by Jeffery et al. (2016), grouping OTUs as Core, Diversity-Associated, Longstay-Associated and Reduced Core, was determined by correlating their abundance with the sum of logged abundances of rarefied OTUs from each module for each subject. The correlation test performed was Pearson’s product moment correlation. Differences in prevalence or relative abundance of methanogens (species or clades) were determined by generalized linear models of either the binomial (prevalence) or quasibinomial (relative abundance) family, in order to adjust for the age of the subjects. The stability of carriage of the four archaeal lineages was evaluated by quantifications performed on 107 stool samples collected 3 months and/or 6 months after the first sampling time. Four additional species specific primer sets were designed to independently target the 16S rRNA gene and the mcrA gene of ‘Ca. Mmc. intestinalis’ and Mmc. luminyensis (Supplementary Table S2).
Samples containing Methanomassiliicoccales representatives that likely have the capacity to deplete TMA were identified based on metagenomic analysis and qPCR, as described in Supplementary Experimental Procedures. Identification of bacterial TMA production markers (cntA/yeaW, cutC, grdH, torA) in the ELDERMET metagenomes is described in Supplementary Experimental Procedures.
Measurement of fecal TMA levels
TMA concentrations were measured by proton nuclear magnetic resonance spectroscopy in fecal water samples from 189 subjects, corresponding to the same samples used for molecular biology (amplicon sequencing, shotgun sequencing, qPCR) analysis. The measurements were carried out by The Metabolomic Innovation Center (TMIC, Edmonton, Canada). Sample preparation, nuclear magnetic resonance spectroscopy and compound identification are described in Supplementary Experimental Procedures. The limit of detection was 1 μM and no values were below this limit.
Characterization of ‘Ca. Mmp. alvus’ enrichment culture
The enrichment culture of ‘Ca. Mmp. alvus’ corresponds to the culture used for the sequencing of ‘Ca. Mmp. alvus Mx1201’ (Borrel et al., 2012). Growth experiments were performed in a modified DSMZ medium 119 under a N2/H2/CO2 (55:35:10) gas phase. Modifications consist of the addition of 5 ml l−1 of trace element solution and vitamin solution from the DSMZ medium 141 and replacement of sludge fluid by rumen fluid. Methanol and TMA (both Sigma-Aldrich, Munich, Germany) were added at a final concentration of 20 mM. Growth of ‘Ca. Mmp. alvus’ and bacteria in the presence of TMA+H2, methanol+H2 or with H2 in the absence of methyl compound was measured by qPCR with primers used for quantifying the ‘Host-associated clade’ Methanomassiliicoccales ELDERMET cohort (Supplementary Table S2). Methane and hydrogen present in the headspace were measured with a gas chromatograph after 5 days of incubation.
Results and Discussion
Five candidate species from the order Methanomassiliicoccales were most frequently found across 10 cohort data sets analyzed
To date, only two studies specifically investigated Methanomassiliicoccales diversity and distribution in multiple human subjects. Our previous study by Mihajlovski et al., (2010) reported five different Methanomassiliicoccales OTUs designated Mx-01 to Mx-05, while the study of Vanderhaeghen et al. (2015) identified only one OTU that showed high sequence similarity to Mx-02. Mx-01 and Mx-05 Methanomassiliicoccales have now been cultured and correspond to ‘Ca. Mmc. intestinalis’ and ‘Ca. Mmp. alvus’, respectively (Borrel et al., 2012, 2013). In order to determine the prevalence and abundance of Methanomassiliicoccales representatives in the human microbiome, we made a census of Methanomassiliicoccales sequences present in 10 studies on the human gut microbiota (Mihajlovski et al., 2008; Scanlan et al., 2008; Mihajlovski et al., 2010; Hoffmann et al., 2013; Goodrich et al., 2014; Nielsen et al., 2014; Clemente et al., 2015; Pozuelo et al., 2015; Vanderhaeghen et al., 2015), extending these data sets with 16S rRNA gene sequences extracted from 190 previously unpublished shotgun metagenome data sets from the ELDERMET cohort.
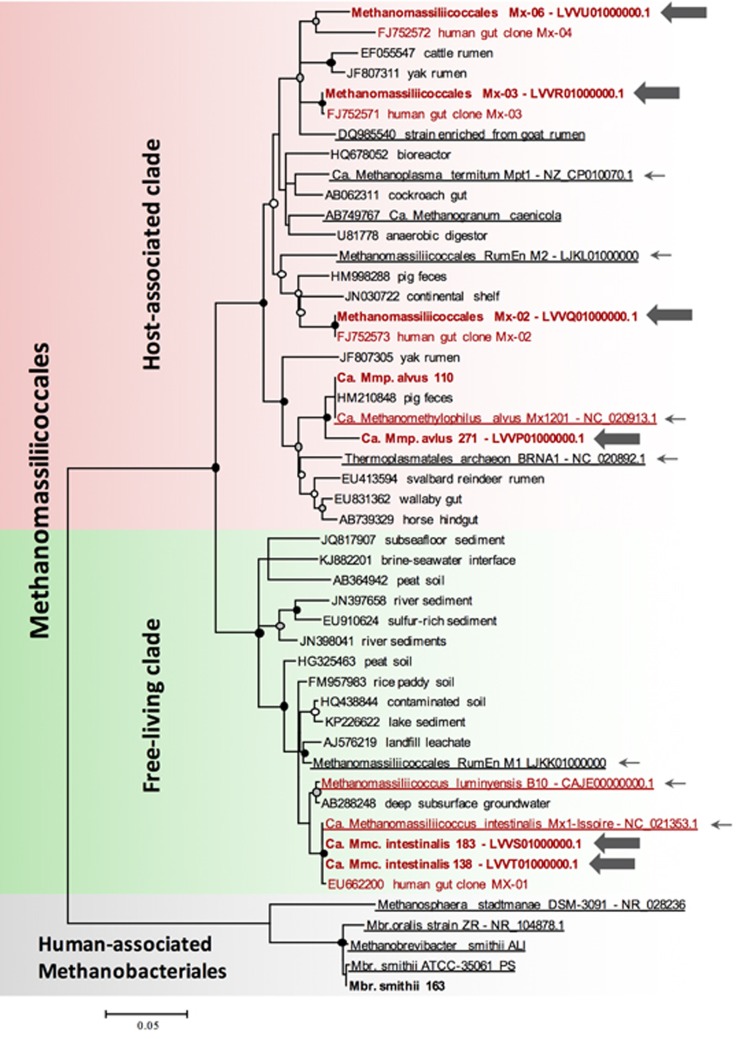
A total of nine Methanomassiliicoccales OTUs (97% identity cutoff) were identified across this combined survey of the human gut microbiome and these likely represent distinct species. Among them, five were detected in at least four cohorts from four different countries and could be considered as the Methanomassiliicoccales representatives most predominantly associated with the human gut microbiome (Table 1). The most widely distributed OTU, found in 8 out of the 11 cohorts, clusters with ‘Ca. Mmc. intestinalis’ and Mmc. luminyensis sequences. However, in all cohorts analyzed, sequences of this OTU were 100% identical to ‘Ca. Mmc. intestinalis’ or closer to this species than to Mmc. luminyensis. This suggests that ‘Ca. Mmc. intestinalis’ is more often associated with the human gut microbiome than Mmc. luminyensis. One of the five predominant OTUs has never been reported previously and we now designate it as Mx-06 in concordance with our previous nomenclature in Mihajlovski et al. (2010). The three others correspond to ‘Ca. Mmp. alvus’ and to the uncultured Mx-02 and Mx-03 sequences identified by Mihajlovski et al. (2010). Almost all the predominant human-associated Methanomassiliicoccales OTUs belong to a clade formed by sequences found in the digestive tract of animals (‘Host-associated clade’ Figure 1). The only exception is the ‘Ca. Mmc. intestinalis’/Mmc. luminyensis OTU, which belongs to a clade of sequences that are mainly associated with soil and sediment environment (‘Free-living clade’ Figure 1). We previously suggested that this phylogenetic and environmental distribution indicates at least two independent adaptation events to the gut microbiome during Methanomassiliicoccales evolution, an ancient one in the last common ancestor of the ‘Host-associated clade’ and a more recent one for ‘Ca. Mmc. intestinalis’ and for Mmc. luminyensis (Borrel et al., 2014). Differential adaptations as well as health and microbiota associations of the members of these two clades are investigated below to substantiate that hypothesis.
Table 1. Methanomassiliicoccales OTUs predominantly associated with the human gut microbiome across multiple cohorts.
| Reference | Method | Target gene | Cohort origin | Number of subjects | Age range | Number of OTUs | Occurrence of OTUs found across >4 studies |
|---|---|---|---|---|---|---|---|
| Mihajlovski et al. (2008) | PCR cloning | 16S/mcrA | France | 6 | 25–69 | 1 | Minta/Mlum |
| Scanlan et al. (2008) | PCR cloning | mcrA | Ireland | 207 | 19–74 | 1 | Mint/Mlum |
| Mihajlovski et al. (2010) | PCR cloning | 16S/mcrA | France | 60 | 0–90 | 5 | Mint/Mlum, Malvb, Mx-02, Mx-03, Mx-04 |
| Dridi et al. (2012a) | qPCR | 16S | France | 500 | 0–100 | — | Mint/Mlum?c |
| Hoffman et al. (2013) | NGS on amplicons | 16S | USA | 96 | 18–40 | 3 | Mint/Mlum, Mx-06 |
| Nielsen et al. (2014) | Metagenomic | — | Denmark/Spain | 396 | 18–70 | 1 | Malv |
| Goodrich et al. (2014) | NGS on amplicons | 16S | UK | 977 | 23–86 | 5 | Mint/Mlum, Mx-02, Mx-03, Mx-06 |
| Clemente et al. (2015) | NGS on amplicons | 16S | Amerindian | 12 | 7–48 | 3 | Mx-02, Mx-06 |
| Vanderhaeghen et al. (2015) | PCR cloning | 16S | Switzerland | 44 | 8–78 | 1 | Mx-02 |
| Pozuelo et al. (2015) | NGS on amplicons | 16S | Spain | 179 | 18–86 | 5 | Mint/Mlum, Malv, Mx-06 |
| This study | Metagenomic | — | Ireland | 190 | 64–102 | 5 | Mint/Mlum, Malv, Mx-02, Mx-03, Mx-06 |
Abbreviations: NGS, next-generation sequencing; OTU, Operational Taxonomic Unit; qPCR, quantitative PCR. For the ‘Ca. Mmc. intestinalis’/Mmc. luminyensis OTU, ‘Mint’ was underlined when the sequences were identical or closer to ‘Ca. Mmc. intestinalis’ than Mmc. luminyensis.
Named Mx-01.
Mx-05 in this early study.
Presence of Mmc. luminyensis was determined with primers and probe having only one mismatch with ‘Ca. Mmc. intestinalis’. Mint, ‘Ca. Mmc. intestinalis’ Mlum, Mmc. luminyensis; Malv, ‘Ca. Mmp. alvus’. Presence of ‘Ca. Mmc. intestinalis’ in Scanlan et al. (2008) and Mihajlovski et al. (2010) was inferred by identification of mcrA sequences having >99% nucleotide identity with the mcrA sequence of this species.
Figure 1.
Maximum likelihood phylogenetic tree of the Methanomassiliicoccales showing the diversity of human-associated Methanomassiliicoccales. Sequences in red are from human-associated Methanomassiliicoccales and sequences in bold are obtained in this study; large arrows indicate draft genomes obtained in this study and small arrows indicate sequences corresponding to complete or draft genomes obtained in previous studies. Underlined sequences correspond to cultured representatives. Sequences of human-associated Methanobacteriales were used to root the tree. Mx-07 to Mx-09 sequences are not included due to too short sequences available for correct assignment but are given in Supplementary Information. Bootstrapping using 200 iterations was performed, black-filled dots indicate nodes with bootstrap supports >90%, gray-filled dot >70% and white-filled dot >50%.
Within the 190 stool metagenome data sets from the ELDERMET cohort, all Methanomassiliicoccales sequences correspond to the five predominant human gut Methanomassiliicoccales OTUs (Figure 1). All other archaeal sequences from the ELDERMET cohort were assigned to the Methanobacteriales, with 99–100% identity to Mbr. smithii. No Msp. stadtmanae sequences were detected among assembled genes of the ELDERMET metagenomes (but they were detected by qPCR that has a lower detection limit in our current study; see following sections), suggesting that this species occurs at lower abundance than the five Methanomassiliicoccales OTUs and Mbr. smithii in these elderly subjects. Because this shotgun sequence data are not subject to PCR biases, this finding indicates that Methanobacteriales and Methanomassiliicoccales are the two most abundant archaeal orders in the ELDERMET cohort. The dominance of these two orders is in line with our previous census of the archaeal lineages detected across multiple cohorts (Gaci et al., 2014). Among the predominant human-associated Archaea currently known, only Methanomassiliicoccales Mx-02, Mx-03 and Mx-06 still lack genomic and physiological characterization.
The pan-genome of predominant human-associated Methanomassiliicoccales
We successfully assembled six draft genome sequences of Methanomassiliicoccales from ELDERMET shotgun sequence data (Table 2, Figure 1, Supplementary Figure S1). Each draft genome was obtained from a single metagenome. Three draft genomes correspond to the uncultured Methanomassiliicoccales Mx-02, Mx-03 and Mx-06. The three others were closely related to previously sequenced Methanomassiliicoccales genomes. Two were very similar to ‘Ca. Mmc. intestinalis Mx1’ and were named ‘Ca. Mmc. intestinalis 138’ and ‘Ca. Mmc. intestinalis 183’. The sixth assembled genome was related to ‘Ca. Mmp. alvus Mx1201’ and was named ‘Ca. Mmp. alvus 271’. The draft genomes of Mx-02, Mx-03, ‘Ca. Mmc. intestinalis 138’, ‘Ca. Mmc. intestinalis 183’ and ‘Ca. Mmp. alvus 271’ were estimated to be complete and not contaminated (Table 2). The draft genome of Mx-06 was estimated to be 87% complete and was thus not used for the pan-genome analysis.
Table 2. Human-associated Methanomassiliicoccales genome characteristics.
| Mx-02 | Mx-03 | Mx-06 | ‘Ca. Mmp. alvus 271’ | ‘Ca. Mmc. intestinalis 138’ | ‘Ca. Mmc. intestinalis 183’ | ‘Ca. Mmp. alvus Mx1201’ | ‘Ca. Mmc. intestinalis Mx1’ | Mmc. luminyensis | |
|---|---|---|---|---|---|---|---|---|---|
| Genome length (Mb) | 1.42 | 1.91 | 1.48/1.70a | 1.68 | 1.84 | 1.89 | 1.67 | 1.93 | 2.64 |
| Number of scaffolds | 2 | 4 | 269 | 23 | 29 | 24 | 1 | 1 | 26 |
| Number of contigs | 2 | 5 | 366 | 23 | 33 | 24 | 1 | 1 | 26 |
| Scaffold N50 (Mb) | 1.06 | 1.49 | 0.005 | 0.1 | 0.17 | 0.17 | NA | NA | NA |
| Estimated completeness (%)b | 100 | 100 | 87 | 100 | 100 | 100 | NA | NA | NA |
| Estimated contamination (%)b | 0 | 0 | 0 | 0c | 0 | 0 | NA | NA | NA |
| Coverage | 17× | 13× | 4× | 17× | 12× | 12× | 37× | 43× | ND |
| G+C content (%) | 47.6 | 59.6 | 58.3 | 60.3 | 41.2 | 41.4 | 55.6 | 41.3 | 60.5 |
| rRNA genes (5S-16S-23S) | 3 (1-1-1) | 3 (1-1-1) | 3 (1-1-1) | 4 (2-1-1) | 4 (2-1-1) | 4 (2-1-1) | 4 (2-1-1) | 4 (2-1-1) | 4 (2-1-1) |
| tRNA genes | 46 | 46 | 36 | 45 | 45 | 45 | 45 | 45 | 48 |
| Number of protein-coding genes | 1430 | 1861 | 1240 | 1594 | 1780 | 1814 | 1653 | 1832 | 2661 |
| Accession number | LVVQ01000000 | LVVR01000000 | LVVU01000000 | LVVP01000000 | LVVS01000000 | LVVT01000000 | CP004049 | CP005934 | CAJE0100000 |
| Reference | This study | This study | This study | This study | This study | This study | Borrel et al. (2012) | Borrel et al. (2013) | Gorlas et al. (2012) |
Abbreviations: NA, not applicable; ND, not displayed.
Complete genome length estimated from current genome completeness.
Based on the single occurrence of 199 genes that are present in single copy in all previously published complete genomes of Methanomassiliicoccales and 3 other Diaforarchaea, Ferroplasma acidarmanus (CP004145.1), Aciduliprofundum boonei (CP001941.1) and Uncultured marine group II (CM001443.1).
One of the single-copy genes is present twice in the ‘Ca. Mmp. alvus 271’ genome, one copy is closely similar to the Methanomassiliicoccales homologs and the other is distantly related to bacterial homologs but is surrounded by genes with high identity to ‘Ca. Mmp. alvus Mx1201’ genes, suggesting an horizontal transfer rather than a contamination.
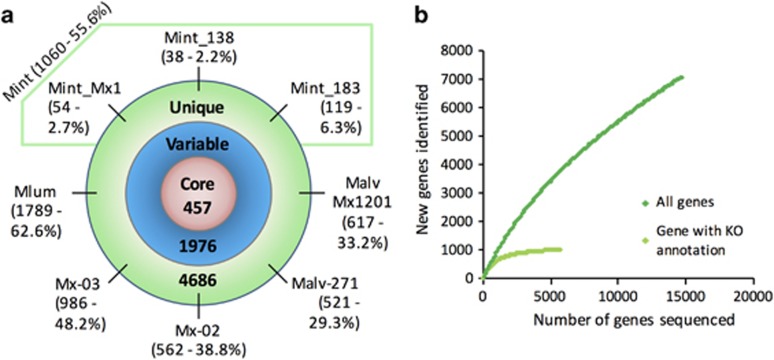
The pan-genome of the five new Methanomassiliicoccales draft genomes analyzed in combination with the ‘Ca. Mmp. alvus Mx1201’, ‘Ca. Mmc. intestinalis Mx1’ and Mmc. luminyensis genomes was predicted to contain 7119 different gene families. Among these, 457 were shared by all 8 genomes, 1976 were shared by 2–7 genomes and 4686 were unique to 1 genome (Figure 2a). The Mmc. luminyensis genome revealed the highest proportion of unique genes (62%). Interestingly, while ‘Ca. Mmp. alvus Mx1201’ and ‘Ca. Mmp. alvus 271’ were phylogenetically close (Figure 1), they harbored a relatively high proportion of unique gene families (around 30% of their genome), which suggests recent gene losses and acquisitions. This highlights the potential importance and scale of genome plasticity in Methanomassiliicoccales in the gut. The variability of the number of genes with a defined KEGG function is much lower (Supplementary Figure S3). These KO annotated genes mainly correspond to core metabolism. Rarefaction analysis on all genes revealed that the rate of discovery of new gene families is far from reaching a plateau based on the eigth genomes available, which is not the case for the KO annotated genes (Figure 2b). Thus a large proportion of the genes that are unique or variably present had only a general predicted function or an unknown function. Completing the current genomic data on the most common archaeal species found in the human gut microbiota (Fricke et al., 2006; Samuel et al., 2007; Hansen et al., 2011; Borrel et al., 2012, 2013), those genomes are useful to better understand the biology of Archaea in the human gut. We next investigated them to predict their TMA-utilizing capacity and the gut adaptations of these Archaea.
Figure 2.
Pan-genome of human-associated Methanomassiliicoccales. (a) Core, variable and unique gene families in the pan-genome of the human-associated Methanomassiliicoccales. Between brackets, the number of unique gene families and the percentage they represent within each genome. The number of unique genes and its percentage are also given for the pan-genome of ‘Ca. Mmc. intestinalis’ as the three genomes of this species are very close. Mint, ‘Ca. Mmc. intestinalis’ Malv, ‘Ca. Mmp. alvus’ Mlum, Mmc. luminyensis. (b) Rarefaction analysis of gene family discovery in the pan-genome of human-associated Methanomassiliicoccales, determined on the basis of identity percentage (dark green) and KO annotation (light green).
Variation in TMA utilization potential in human gut Methanomassiliicoccales
The five Methanomassiliicoccales representatives with an estimated complete genome are very likely to be methanogens as they harbored genes coding for the methyl coenzyme M reductase complex as well as the 17 other marker genes that are universal and unique to methanogens (Borrel et al., 2014) (Supplementary Table S3). The same assumptions can be made for Mx-06, which was only missing 1 of the 20 methanogenesis marker genes. In line with previously characterized Methanomassiliicoccales genomes, the six novel genomes lacked the genes for reduction of CO2 by H2 or methyl group oxidation, and they contained homologs of mtaA/mtbA involved in methylated substrate utilization. This genomic composition indicated that they likely operate a methanogenesis based on the reduction of methyl compound with H2, a characteristic shared by all Methanomassiliicoccales so far analyzed. Presence of the genes involved in the utilization of specific methyl compounds (methanol, methanethiol, mono-, di- and TMA) as substrates for methanogenesis was variable among the six newly assembled genomes (Supplementary Table S3). The genes for TMA utilization (mttBC) were found in the genomes of ‘Ca. Mmp. alvus 271’, Mx-06, ‘Ca. Mmc. intestinalis 138’ and ‘Ca. Mmc. intestinalis 183’. Moreover, there was a congruent occurrence of the 16S rRNA and mttB genes of these three Methanomassiliicoccales across ELDERMET metagenomes and a strong correlation between the read counts of the ‘Ca. Mmc. intestinalis’ 16S rRNA and mttB genes (Pearson linear correlation: r2=0.74; P=10−54). This indicates that the different strains of these three Methanomassiliicoccales consistently share the predicted capacity to deplete TMA. In contrast, this capacity might be less regularly shared by other predominant human gut Methanomassiliicoccales. Indeed, Mx-03 mttB gene is split into two genes coding for N- and C-terminal parts of the canonical mttB gene (Supplementary Figure S4), and among the four metagenomes where Mx-03 genome is enriched, no read was mapped on Mx-03 mttB. Regarding Mx-02, no mttB homolog was found in its draft genome and within the three metagenomes enriched in this methanogen. Supporting this lack in Mx-02, the genes encoding pyrrolysine synthesis and use (pylSBCD), an unusual amino acid conserved in the catalytic site of all characterized MttB proteins, were not detected in the Mx-02 genome or respective metagenomes. Interestingly, a similar absence of the genes for methylated compound utilization as well as pylSBCD genes was also observed in the Methanomassiliicoccales RumEn M2, and for the moment, the electron acceptors of these methanogens remain unknown (Söllinger et al., 2016). The capacity to deplete TMA is thus potentially not shared by all the Methanomassiliicoccales that are commonly present in the human microbiota.
Intestinal Methanomassiliicoccales prevalence/abundance correlated with microbiota potential for modulating TMA
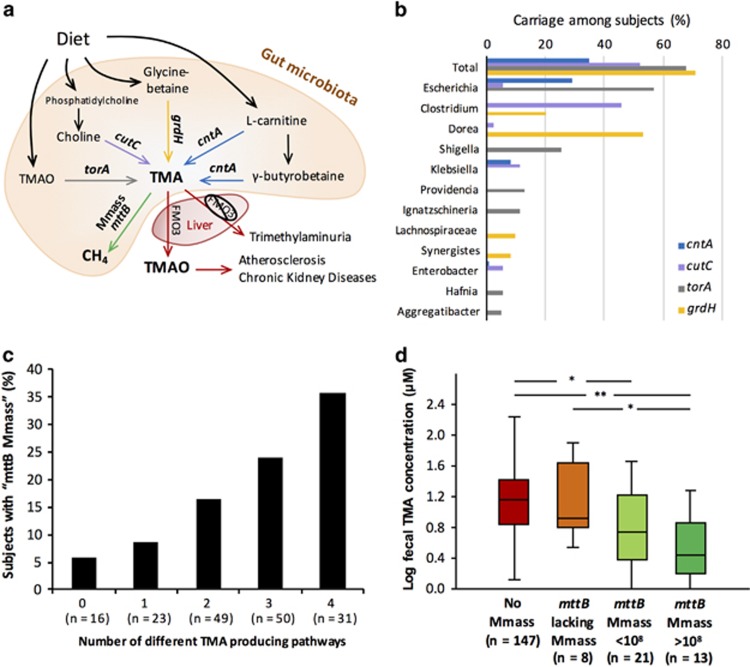
We hypothesized that the occurrence of Methanomassiliicoccales representatives with predicted ability to utilize TMA for growth (‘mttB Methanomassiliicoccales’) would be influenced by the TMA production potential of the gut microbiota. Several pathways converting distinct dietary compounds into TMA have been characterized and found in many gut bacteria through genomic analyses (Falony et al., 2015; summarized in Figure 3a). However, the genes encoding these pathways could be variably present between strains of the same species or genera, as recently exemplified for choline conversion to TMA (Martínez-del Campo et al., 2015). For this reason, we did not assess TMA production potential on the basis of 16S rRNA gene composition but based on functional markers responsible for the final step of TMA production within the four pathways (Figure 3a). Those proteins are encoded by torA for TMAO conversion (Méjean et al., 1994), cutC for choline conversion (Craciun and Balskus, 2012), grdH for betaine conversion (Meyer et al., 1995; Wagner et al., 1999) and cntA/yeaW for L-carnitine conversion (Koeth et al., 2014; Zhu et al., 2014). At the threshold of detection allowed by the sequencing coverage of the current study, marker genes were not detected in every subject but in 70.8% of the subjects for grdH, 67.7% for torA, 52.1% for cutC and 34.9% for cntA (Figure 3b). Affiliation of those functional markers and mapping of metagenomic reads on gene sequences (including complete marker genes from HMP genomes) revealed that only a few genera of TMA producers were widely distributed among ELDERMET subjects (Figure 3b, Supplementary Table S4). Among these, Escherichia coli strains could be important players in TMA production, being variably involved in TMA production from choline, L-carnitine and TMAO (Figure 3b). Clostridium spp., potentially converting choline and betaine to TMA, are phylogenetically diverse and partially comprise uncharacterized species (or not yet sequenced) but collectively could represent important agents of TMA production in the human gut (Figure 3b, Supplementary Table S4). Dorea formicigenerans with a potential for conversion of betaine to TMA is widely distributed among subjects but never occurs at high relative abundance.
Figure 3.
Microbial metabolisms modulating TMA concentrations in the human gut microbiome. (a) Schematic representation of the pathways and associated marker genes modulating TMA concentrations in the gut microbiome and health output related to high TMA productions. (b) Composition and carriage (%) of the potential TMA producers in the ELDERMET cohort. ‘Total’ corresponds to the percentage of carriage of each marker gene among the subjects, regardless of the affiliation of the marker. (c) Percentage of carriage of Methanomassiliicoccales representatives in subgroups of subjects having no TMA production pathway or one to four different TMA production pathways detected in their microbiota. (d) Concentrations of fecal TMA (logged) among subjects without or with Methanomassiliicoccales detected at different concentrations. No Mmass, no Methanomassiliicoccales detected; Mmass without mttB, Methanomassiliicoccales potentially lacking the capacity to deplete TMA; mttB Mmass <108 and mttB Mmass >108, Methanomassiliicoccales potentially depleting TMA having a concentration lower and higher than 108 cell per g stool, respectively. Significant differences between groups are determined with Mann–Whitney test, *P<0.05, **P<0.001.
Samples containing ‘mttB Methanomassiliicoccales’ were identified by a combination of qPCR and metagenomic analyses as described in Supplementary Experimental Procedures. Subjects with a higher number of different TMA production pathways encoded by their gut microbiota are more likely to harbor Methanomassiliicoccales representatives that are able to use TMA as substrate. Indeed, ‘mttB Methanomassiliicoccales’ were detected in 6% of the ELDERMET subjects with no TMA production pathway detected in their gut microbiota and this proportion progressively increased to 36% among the subjects carrying a microbiota able to produce TMA from four different precursors (Figure 3c). More diversified TMA production pathways in a single subject could allow the conversion of a larger number of different food ingredients into TMA. A more regular production of this molecule might help Methanomassiliicoccales to colonize and persist in the microbiota. The influence of TMA-producing bacteria in the presence of Methanomassiliicoccales is also suggested by a higher abundance of TMA production pathways, and more specifically those for carnitine (cntA) and choline (cutC) conversion, in subjects with ‘mttB Methanomassiliicoccales’ than without Methanomassiliicoccales (Supplementary Figure S5). In addition to pathway redundancy, a higher TMA production capacity of the microbiota would thus help the Methanomassiliicoccales to develop in the human gut.
The predicted ability of human-associated Methanomassiliicoccales to deplete TMA in the gut was supported by comparison of experimentally determined fecal TMA concentrations in subjects harboring ‘mttB Methanomassiliicoccales’, ‘mttB lacking Methanomassiliicoccales’ and subjects not carrying Methanomassiliicoccales (Figure 3d). Lower TMA concentrations were found in fecal samples of subjects carrying ‘mttB Methanomassiliicoccales’ than in subjects without Methanomassiliicoccales. The difference in fecal TMA concentrations between subjects carrying ‘mttB Methanomassiliicoccales’ and subjects without Methanomassiliicoccales was more significant when the abundance of ‘mttB Methanomassiliicoccales’ was >108 cells per gram of stool than below (Figure 3d). Moreover, no significant difference in fecal TMA concentration was found between subjects carrying ‘mttB lacking Methanomassiliicoccales’ and subject without Methanomassiliicoccales (Figure 3d). Those results parallel the negative correlation between Methanomassiliicoccales abundance and urinary TMAO level recently reported in a study of rumen communities in lambs (Morgavi et al., 2015). In the current study, while the presence of ‘mttB Methanomassiliicoccales’ was associated with lower fecal TMA concentrations, an increased number of different TMA production pathways was not significantly associated with higher fecal TMA concentrations. This could be due to variable level of expression of those pathways depending on diet and potentially through leveling of high TMA values by its assimilation. In situ depletion of TMA by Methanomassiliicoccales representatives awaits experimental confirmation by following blood TMAO concentration in an animal model with defined diet and microbiota composition. These biomanipulations will require isolated strains of the ‘mttB Methanomassiliicoccales’ predominantly found in the human microbiota, that is, ‘Ca. Mmp. alvus’, ‘Ca. Mmc. intestinalis’ or Mx-06. Using an enrichment culture of ‘Ca. Mmp. alvus’ (Borrel et al., 2012), we confirmed the capacity of this species to use TMA as a substrate by growth and metabolite analysis (Supplementary Figure S6), making it promising for future experiments on controlled gut microbiota systems for TMA production/depletion. This in vivo validation of genome annotation further supports the notion that the predominant ‘mttB Methanomassiliicoccales’ could deplete TMA within the human gut. Beyond the interesting potential for the removal of deleterious methylated compounds by Methanomassiliicoccales in the human gut, it is important to better understand the general biology and ecology of these methanogens in the human gut and their overall interaction with the bacterial microbiome.
Representatives of the two main Methanomassiliicoccales clades have different interactions with the gut microbiome and clinical metadata
Subjects of the ELDERMET cohort are living in the community or in residential care (either long term or rehabilitation). Previous studies on this cohort have revealed a progressive shift in the gut microbiota composition of subjects entering long-term residential care (‘longstay subjects’) (Claesson et al., 2012; O’Toole and Jeffery, 2015; Jeffery et al., 2016). Detailed analysis of these changes has shown a gradual loss of health-associated bacteria (gathered under the Diversity-Associated module) and their replacement by bacteria (Longstay-Associated module) associated with increased frailty (Jeffery et al., 2016). This recent analysis showed that frailty and frailty-related microbiota alterations may occur independently of age and residence location (Jeffery et al., 2016). This alteration of the bacterial microbiota is likely driven by diet and could contribute to accelerated age-related health decline (Claesson et al., 2012). We now report that the composition of human gut Archaea, albeit a much less diverse than bacteria, is also gradually changing after the subjects enter residential care. (For simplicity, one intermediate residence location, day hospital, was omitted). This was investigated by using qPCR to determine the prevalence of members of the two main clades of Methanomassiliicoccales, the ‘Host-associated clade’ and the ‘Free-living clade’ (Figure 1), as well as Mbr. smithii and Msp. stadtmanae. Three hundred and forty-two of these samples had been collected at one time point (T0) and 107 subjects sampled at one or two additional time points separated by 3 months (T3 and T6).
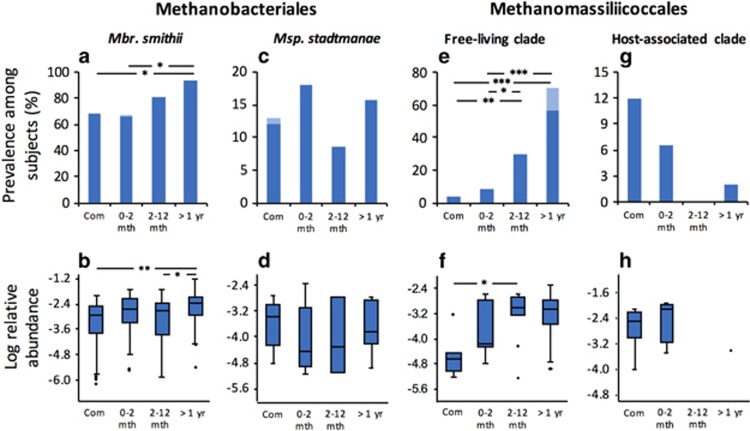
Of the two Methanobacteriales species targeted, only Mbr. smithii carriage rates interacted with time spent in residential care (Figure 4a). Being present in almost 70% of community-dwelling subjects, it became nearly ubiquitous (94%) among residents of >1 year and occurred at a higher relative abundance than in community-dwelling subjects (Figure 4b). Accordingly, Mbr. smithii positively correlates with the Longstay-Associated module of intestinal bacterial taxa (ρ=0.46; 10−21). Despite this association, Mbr. smithii has been previously reported as a common component of the human gut microbiota without particular distribution according to health status (Scanlan et al., 2008; Blais Lecours et al., 2014). Moreover, Mbr. smithii has been reported to have a low inflammatory potential (Bang and Schmitz, 2015), suggesting that it is not directly contributing to increased inflammation associated with health decline among longstay subjects (Claesson et al., 2012). The carriage rate and abundance of Msp. stadtmanae was not significantly different between community and residential care dwellers (Figures 4c and d). This suggests that, in the present case, Msp. stadtmanae is not associated with increased inflammation levels of longstay subjects (Claesson et al., 2012) and contrasts with the previously suggested link between this methanogen species and inflammation (Blais Lecours et al., 2014).
Figure 4.
Prevalence and relative abundance of methanogens among elderly subjects living in community and according to the time spent in residential care. Prevalence and relative abundance are presented for Methanobrevibacter smithii (a and b), Methanosphaera stadtmanae (c and d), ‘Free-living clade’ Methanomassiliicoccales (e and f) and the ‘Host-associated clade’ Methanomassiliicoccales (g and h). In panels (a), (c), (e) and (g), the dark blue bars represent prevalence among subjects at T0, while the clear blue bars take into account subjects positive for at least one time point. For prevalence, Community (Com), n=125; Residential care 0–2 months (mth), n=61; 2–12 months, n=47; >1 year, n=51. Statistical analysis and representation of the relative abundance of methanogens were carried out on subjects with a positive detection of methanogens. *P<0.05, **P<0.01, ***P<0.001. Day hospital category is not presented for clarity but is never different from Community.
In comparison to Methanobacteriales, more significant prevalence differences were noted for the Methanomassiliicoccales. Indeed, the carriage rate of ‘Free-living clade’ Methanomassiliicoccales was low among community-dwelling subjects (4%) but progressively increased with time spent in residential care to 55% among subjects being resident for longer than a year (Figure 4e). Additionally, six longstay subjects with no ‘Free-living clade’ Methanomassiliicoccales detected at T0 became positive for these methanogens at either T6 or T3+T6 and were thus potentially newly colonized during this 6-month period. Taking into account those subjects, the maximum prevalence of this clade could reach up to 70% (Figure 4e). Furthermore, this higher prevalence was coupled with a >10-fold increase in the average relative abundance of ‘Free-living clade’ Methanomassiliicoccales after 2 months living in residential care (Figure 4f), at which they stabilized (Supplementary Figures S7A and B). In addition, subjects harboring ‘Free-living clade’ had a higher frailty level, when considering all subjects or only residential care subjects (Supplementary Figure S8). This prompted us to identify which members of this clade are associated with this strong trend. Detailed investigation using species-specific primers and metagenomic analysis revealed that 90% of the subjects positive for this clade were carrying ‘Ca. Mmc. intestinalis’, whereas only 5% carried Mmc. luminyensis. This supports our analysis of other cohorts that suggests a generally higher proportion of ‘Ca. Mmc. intestinalis’ (Table 1). The frequency of carriage of the ‘Host-associated clade’ Methanomassiliicoccales followed an opposite trend as they gradually disappeared with time spent in residential care (Figure 4g). This clade is more often present among subjects in better health condition according to frailty indices (Supplementary Figure S8). Despite the progressive shift with time spent in residential care, carriage and relative abundance of Methanomassiliicoccales representatives was mostly stable over a 6-month period (Supplementary Figure S7C). The occurrence of the two clades tended to increase with the age of the subjects but only among community-dwellers for the ‘Host-associated clade’ and only among residential care subjects for the ‘Free-living clade’ (Supplementary Figure S9). Moreover, for the same age range, ‘Host-associated clade’ tended to be higher among community and residential care dwellers and inversely for ‘Ca. Mmc. intestinalis’ (Supplementary Figure S9). In agreement with the opposite distribution of the two clades, the ‘Free-living clade’ Methanomassiliicoccales was positively correlated with the Longstay-Associated module (ρ=0.39; P<10−14) while the ‘Host-associated clade’ was positively correlated to the Diversity-Associated module (ρ=0.21; P<0.001).
Two questions arise from these contrasting distributions and module associations among Methanomassiliicoccales. Is ‘Ca. Mmc. intestinalis’ a deleterious invader while ‘Host-associated clade’ representatives are normal members of the human gut microbiome? What are the factors leading to the increased colonization of one and the loss of the other among longstay subjects? A part of the answer to these two questions may lie in the relationships between Methanomassiliicoccales and TMA-producing bacteria. Indeed, the prevalence of the different TMA-producing pathways and their redundancy increased with the time subjects spent in residential care (Supplementary Figure S10), potentially raising the level of plasma TMAO and the development of associated cardiac and kidney diseases. The greater level of ‘Ca. Mmc. intestinalis’ among longstay subjects could reduce the adverse effect of this molecule on health, and in this way, ‘Ca. Mmc. intestinalis’ could be seen more as a beneficial microorganism than a pathobiont. However, the immunogenic potential of this methanogen has not yet been evaluated and should not be neglected when considering its overall impact on health. Although the potentially higher availability of TMA could be a factor promoting the colonization of ‘Ca. Mmc. intestinalis’, it does not explain why the TMA using representatives of the ‘Host-associated clade’ (‘Ca. Mmp. alvus’ and Mx-06) are not favored but instead tend to be lost. Despite multiple attempts to isolate members of the ‘Host-associated clade’, including those from the human gut, no pure culture of these Archaea has been obtained to date (Borrel et al., 2012; Paul et al., 2012; Borrel et al., 2013; Iino et al., 2013; Jin et al., 2014; Lang et al., 2015; Söllinger et al., 2016). From this observation, it has been previously suggested that members of this clade could be dependent on unrecognized metabolite(s) produced by co-occurring bacteria (Lang et al., 2015). This aspect will need to be further studied to determine whether members of the ‘Host-associated clade’ present in the human gut could be dependent on some specific bacteria for such metabolites, and whether those bacteria make part of the Diversity-Associated module that tend to be lost among residential care subjects (Jeffery et al., 2016).
As previously mentioned, specialization to the gut microbiome could have occurred several times during the evolution of the Methanomassiliicoccales (Figure 1) (Borrel et al., 2014). In an evolutionary context, it is interesting to observe that the Methanomassiliicoccales representatives that potentially co-evolved with the host for a longer evolutionary period (those of the ‘Host-associated clade’) were preferentially found in community-dwelling subjects having a health-associated microbiota, while ‘Ca. Mmc. intestinalis’, which adapted more recently to this environment, became more prevalent with the progressive alteration of the gut microbiota composition among residential care subjects. Four of the six subjects newly colonized by ‘Ca. Mmc. intestinalis’ during the 6 months monitoring had an unstable microbiota as defined by our previous analyses (Jeffery et al., 2016) and one was taking antibiotics. This suggests that microbiota instability could be one of the factors that contribute to helping ‘Ca. Mmc. intestinalis’ to colonize the gut among longstay subjects. High prevalence of ‘Host-associated clade’ has been observed in several cohorts of healthy subjects, such as an Amerindian cohort (Clemente et al., 2015) in which data we found Mx-06 to be present among 80% of the subjects, in a Swiss cohort where Mx-02 was detected in 50% of the subjects (Vanderhaeghen et al., 2015) and in a French cohort of older adults (Mihajlovski et al., 2010) where members of this clade were detected among 40% of the subjects. A high prevalence of ‘Ca. Mmc. intestinalis’ in a cohort of healthy subjects has never been observed to date.
Genomic signatures suggesting different archaeal strategies to colonize and persist in the gut microbiome
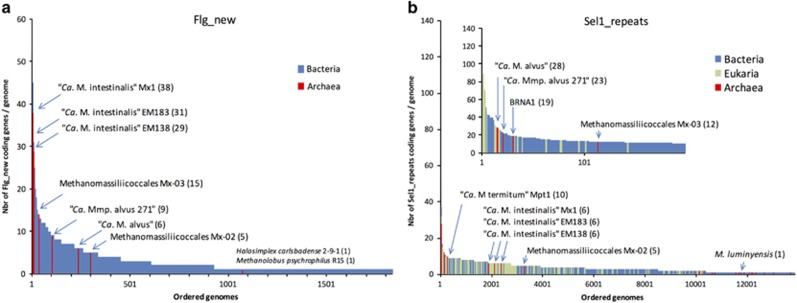
Previously sequenced and de novo assembled genomes of Methanomassiliicoccales were screened for traits that might account for the different distribution of the two clades and potentially explain why ‘Ca. Mmc. intestinalis’ is more commonly detected than Mmc. luminyensis in the human gut microbiome. Sequences encompassing two conserved domains, Sel1_repeat (pfam08238) and Flg_new (pfam09479, also called Listeria-Bacteroides repeats or B-repeats), were highly enriched in several Methanomassiliicoccales genomes. Comparison to all genomes from the IMG/ER database revealed that the three ‘Ca. Mmc. intestinalis’ genomes were among those with the highest number of genes coding for Flg_new-containing proteins while the two ‘Ca. Mmp. alvus’ and Mx-03 were among genomes with the highest number of genes coding for Sel1_repeat-containing proteins (Figure 5). Interestingly, Flg_new proteins are absent from all other Archaea (with the exception of two Archaea, Halosimplex carlsbadense and Methanolobus psychrophilus that have one Flg_new gene each). Our investigation of the lifestyle of the bacteria with high number of genes coding for Flg_new and Sel1_repeat proteins revealed that most of them have been isolated from various animal hosts and body sites (Supplementary Table S5), which is in line with previous functional characterizations and hypotheses on the role of these repeats in host–microbe interactions (Mittl and Schneider-Brachert, 2007; Ebbes et al., 2011). The Flg_new domain was characterized in the internalin InlB of Listeria monocytogenes, a protein required for host cell invasion, but this domain is also found in many other membrane-bound proteins with no given function (Ebbes et al., 2011). This domain have structural similarities with β-grasp fold and mucin-binding domains (Ebbes et al., 2011). Most of the Methanomassiliicoccales Flg_new repeat proteins have a transmembrane domain at either one or both terminuses and are thus predicted to be almost entirely outside of the cell (Supplementary Table S6). These proteins were very variable in length (ranging from 174 to 5834 amino acids) and in number of Flg_new domains (ranging from 1 to 25) and could have different accessory domains that are redundant across the Methanomassiliicoccales genomes (Supplementary Table S6). This is reminiscent of the asn/thr-rich and adhesin-like proteins that are also associated with the membrane, variable in length and in number of repeat domains and encoded by a high number of genes in Msp. stadtmanae and Mbr. smithii, respectively (Fricke et al., 2006; Samuel et al., 2007; Hansen et al., 2011). The high heterogeneity and cell surface location of Flg_new proteins suggest that they could be rapidly evolving proteins that underwent repeated recombination events and duplications in order to interact with a wider range of sites in the gut microbiome, as previously suggested for the asn/thr-rich large protein of Msp. stadtmanae (Fricke et al., 2006) and the adhesin-like proteins of Mbr. smithii (Samuel et al., 2007; Hansen et al., 2011). The Sel1_repeat domain is an eukaryotic-like domain found in proteins that regulate cell processes and that are involved in interactions between bacteria and eukaryotes (Mittl and Schneider-Brachert, 2007). It has been previously reported that a high level of eukaryotic-like proteins is a characteristic feature of bacteria that have a long history of co-evolution with eukaryotic hosts (Cazalet et al., 2004; Hentschel et al., 2012). Interestingly, genes coding for Sel1_repeat proteins tended to be more abundant in genomes of the ‘Host-associated clade’ Methanomassiliicoccales while genes coding for Flg_new proteins occurred more frequently in ‘Ca. Mmc. intestinalis’ genomes (Figure 5). Biochemical characterization of these proteins will help to determine whether they could account for different colonization strategies between members of the ‘Host-associated clade’ and ‘Ca. Mmc. intestinalis’. Mmc. luminyensis was distinguished from all other human-associated Methanomassiliicoccales representatives by having only one gene coding for a Sel1_repeat protein and none for a Flg_new protein (Figure 5). The lack of a Flg_new protein in Mmc. luminyensis is particularly striking considering that the closely related ‘Ca. Mmc. intestinalis’ have up to 38 genes coding for these proteins. It could thus be wondered whether this distinctive gene composition in Mmc. luminyensis could participate to the low prevalence of this species in human cohorts compared with other Methanomassiliicoccales representatives (Table 1). We previously suggested that Mmc. luminyensis could have a wider environmental distribution than the human gut, harboring genomic adaptations to thrive in soils such as an number of important genes for oxidative stress resistance or diazotrophy (Borrel et al., 2014). To this can also be added the high-affinity K+ transport system (Kdp), which was proposed as an adaptation to low K+ concentration in oligotrophic peat lands (Bräuer et al., 2015). The other human gut-associated Methanomassiliicoccales have a lower number of genes for oxidative stress-resistance proteins and none of the other genes reflecting adaptation to soils present in Mmc. luminyensis, highlighting their stronger specialization to gut microbiome membership. Additionally, Mmc. luminyensis harbors genes for mercury methylation (hgcAB) whose activity was experimentally validated (Podar et al., 2015). These genes were widely detected in anaerobic environments but never in the mammalian gut (Podar et al., 2015), suggesting that they were not retained during the evolution of species composing the commensal microbiota due to the high toxicity of methylated mercury for the host. Their presence in the Mmc. luminyensis genome supports that this methanogen is not a common gut commensal. The hgcAB genes are not present in any of the six new draft genomes reported in the current study and so mercury methylation by these methanogens is not a consideration for their role in health or disease.
Figure 5.
Occurrence of genes encoding for Flg_new and Sel1_repeat domains in Methanomassiliicoccales and other genomes from the three domains of life. Flg_new domains (a) and Sel1 repeat domains (b). Genomes are ordered from the highest occurrence to the lowest occurrence of the genes coding those domains.
Conclusion
This study highlights that human intestinal Archaea should not be investigated as a uniform group when exploring their potential relationships with health, in line with previous reports of uneven immunogenic potential of human-associated Methanobacteriales species (Blais Lecours et al., 2014; Bang and Schmitz, 2015). Our data provide evidence that some archaeal species among the Methanomassiliicoccales could reduce the adverse effect of TMA through their specific activity and higher occurrence among subjects with high TMA production potential. Our results also indicate that the archaeal component of the gut microbiome is clearly affected after the subjects of this cohort enter residential care. As previously reported for bacteria, this alteration could be related to the significant physiological and lifestyle changes that are features of long-term residential care (Claesson et al., 2011, 2012; O’Toole and Jeffery, 2015). The intriguing high prevalence of ‘Ca. Mmc. intestinalis’ among subjects with higher frailty will need further exploration to define whether this species may be favored merely by factors including altered gut transit, instability of the microbiota and increased availability of TMA or if by itself it could trigger inflammation and contribute to frailty. Evaluation of the immunogenic potential of the dominant human gut Methanomassiliicoccales, and more particularly those with a predicted ability to utilize TMA, is needed to determine whether their role could be mostly positive through TMA removal or if a more nuanced role is being played. A better knowledge of the factors influencing their prevalence will be of particular interest as Methanomassiliicoccales representatives are unevenly distributed in human populations and some cohorts/subjects could be dominated by Methanomassiliicoccales that potentially lack the capacity to deplete TMA (Vanderhaeghen et al., 2015). Although Mmc. luminyensis is currently the only Methanomassiliicoccales species isolated from the human gut (or even in general) (Dridi et al., 2012b), it might not be the best model to study the interactions of the representatives of this archaeal order with the gut microbiome and the host. Isolated strains of the Methanomassiliicoccales predominantly associated with the human microbiome will be very valuable to better define the role of Methanomassiliicoccales in health and disease.
Acknowledgments
We thank JK Goodrich, RE Ley, JC Clemente and MG Domingo-Bello for sharing their data to identify human-associated Methanomassiliicoccales, as well as J-F Mangot for his early advice with binning. PWO’T was supported by Science Foundation Ireland through a Principal Investigator award, by a CSET award to the APC Microbiome Institute and by an FHRI award to the ELDERMET project by the Department of Agriculture, Fisheries and Marine of the Government of Ireland.
Author contributions
Conceptualization: GB, J-FB, and PWO’T; methodology: GB and PWO’T; investigation: GB, JD, and MCN; formal analysis: GB, AMC, and DBL; writing—original draft: GB; writing—review and editing: AMC, J-FB, and PWO’T.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Bang C, Schmitz RA. (2015). Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol Rev 39: 631–648. [DOI] [PubMed] [Google Scholar]
- Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA. (2014). The intestinal Archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS ONE 9: e99411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais Lecours P, Duchaine C, Taillefer M, Tremblay C, Veillette M, Cormier Y et al. (2011). Immunogenic properties of archaeal species found in bioaerosols. PLoS ONE 6: e23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais Lecours P, Marsolais D, Cormier Y, Berberi M, Haché C, Bourdages R et al. (2014). Increased prevalence of Methanosphaera stadtmanae in inflammatory bowel diseases. PLoS ONE 9: e87734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond Jr JH, Engel RR, Levitt MD. (1971). Factors influencing pulmonary methane excretion in man. An indirect method of studying the in situ metabolism of the methane-producing colonic bacteria. J Exp Med 133: 572–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Harris HM, Tottey W, Mihajlovski A, Parisot N, Peyretaillade E et al. (2012). Genome sequence of "Candidatus Methanomethylophilus alvus" Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol 194: 6944–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Parisot N, Harris HM, Peyretaillade E, Gaci N, Tottey W et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic Archaea that encodes pyrrolysine. BMC Genomics 15: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Harris HM, Parisot N, Gaci N, Tottey W, Mihajlovski A et al. (2013). Genome sequence of "Candidatus Methanomassiliicoccus intestinalis" Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1: e00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer S, Cadillo-Quiroz H, Kyrpides N, Woyke T, Goodwin L, Detter C et al. (2015). Genome of Methanoregula boonei 6A8 reveals adaptations to oligotrophic peatland environments. Microbiology 161: 1572–1581. [DOI] [PubMed] [Google Scholar]
- Brugère JF, Borrel G, Gaci N, Tottey W, O'Toole PW, Malpuech-Brugère C. (2014). Archaebiotics: proposed therapeutic use of Archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes 5: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Brüggemann H, Zidane N, Magnier A, Ma L et al. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36: 1165–1173. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E et al. (2011). Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 108(Suppl 1): 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488: 178–184. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B et al. (2015). The microbiome of uncontacted Amerindians. Sci Adv 1: e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun S, Balskus EP. (2012). Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA 109: 21307–21312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Henry M, Richet H, Raoult D, Drancourt M. (2012. a). Age-related prevalence of Methanomassiliicoccus luminyensis in the human gut microbiome. APMIS 120: 773–777. [DOI] [PubMed] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. (2012. b). Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62: 1902–1907. [DOI] [PubMed] [Google Scholar]
- Ebbes M, Bleymüller WM, Cernescu M, Nölker R, Brutschy B, Niemann HH. (2011). Fold and function of the InlB B-repeat. J Biol Chem 286: 15496–15506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Falony G, Vieira-Silva S, Raes J. (2015). Microbiology meets big data: the case of gut microbiota-derived trimethylamine. Annu Rev Microbiol 69: 305–321. [DOI] [PubMed] [Google Scholar]
- Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K et al. (2016). Population-level analysis of gut microbiome variation. Science 352: 560–564. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Wang A, Su W, Rozenbloom SR, Taibi A, Comelli EM et al. (2013). Age, dietary fiber, breath methane, and fecal short chain fatty acids are interrelated in Archaea-positive humans. J Nutr 143: 1269–1275. [DOI] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WF, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol 188: 642–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaci N, Borrel G, Tottey W, O’Toole PW, Brugère J-F. (2014). Archaea and the human gut: new beginning of an old story. World J Gastroenterol 20: 16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R et al. (2014). Human genetics shape the gut microbiome. Cell 159: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlas A, Robert C, Gimenez G, Drancourt M, Raoult D. (2012). Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated Archaea species. J Bacteriol 194: 4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. (2012). The human microbiome: our second genome. Annu Rev Genomics Hum Genet 13: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A et al. (2011). Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA 108: 4599–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Piel J, Degnan SM, Taylor MW. (2012). Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10: 641–654. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD et al. (2013). Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE 8: e66019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz H-P. (2015). Archaeal lineages within the human microbiome: absent, rare or elusive? Life 5: 1333–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K et al. (2013). Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ 28: 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery IB, Lynch DB, O'Toole PW. (2016). Composition and temporal stability of the gut microbiota in older persons. ISME J 10: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Cheng Y, Sheng Y, Zhu W. (2014). Discovery of a novel rumen methanogen in the anaerobic fungal culture and its distribution in the rumen as revealed by real-time PCR. BMC Genomics 14: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428: 726–731. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Levison BS, Culley MK, Buffa JA, Wang Z, Gregory JC et al. (2014). γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab 20: 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K, Schuldes J, Klingl A, Poehlein A, Daniel R, Brune A. (2015). New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of “Candidatus Methanoplasma termitum”. Appl Environ Microbiol 81: 1338–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. (2004). Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci USA 101: 6176–6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie-Weinberger MN, Gophna U. (2015). Archaea in and on the human body: health implications and future directions. PLoS Pathog 11: e1004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay RJ, McEntyre CJ, Henderson C, Lever M, George PM. (2011). Trimethylaminuria: causes and diagnosis of a socially distressing condition. Clin Biochem Rev 32: 33–43. [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40: D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-del Campo A, Bodea S, Hamer HA, Marks JA, Haiser HJ, Turnbaugh PJ et al. (2015). Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6: e00042-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjean V, Lobbi‐Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal MC. (1994). TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol 11: 1169–1179. [DOI] [PubMed] [Google Scholar]
- Meyer M, Granderath K, Andreesen JR. (1995). Purification and characterization of protein PB of betaine reductase and its relationship to the corresponding proteins glycine reductase and sarcosine reductase from Eubacterium acidaminophilum. Eur J Biochem 234: 184–191. [DOI] [PubMed] [Google Scholar]
- Miele V, Penel S, Duret L. (2011). Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlovski A, Alric M, Brugère JF. (2008). A putative new order of methanogenic Archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene. Res Microbiol 159: 516–521. [DOI] [PubMed] [Google Scholar]
- Mihajlovski A, Dore J, Levenez F, Alric M, Brugère JF. (2010). Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity. Environ Microbiol Rep 2: 272–280. [DOI] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ. (1985). Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol 141: 116–122. [DOI] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ, Conway de Macario E, Macario AJ. (1982). Isolation of Methanobrevibacter smithii from human feces. Appl Environ Microbiol 43: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S, Zhang A, Barrett T, Ayesh R, Smith R. (1997). Studies on the discontinuous N-oxidation of trimethylamine among Jordanian, Ecuadorian and New Guinean populations. Pharmacogenet Genomics 7: 45–50. [DOI] [PubMed] [Google Scholar]
- Mittl PR, Schneider-Brachert W. (2007). Sel1-like repeat proteins in signal transduction. Cell Signal 19: 20–31. [DOI] [PubMed] [Google Scholar]
- Morgavi DP, Rahahao-Paris E, Popova M, Boccard J, Nielsen KF, Boudra H. (2015). Rumen microbial communities influence metabolic phenotypes in lambs. Front Microbiol 6: 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S et al. (2014). Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32: 822–828. [DOI] [PubMed] [Google Scholar]
- O’Toole PW, Jeffery IB. (2015). Gut microbiota and aging. Science 350: 1214–1215. [DOI] [PubMed] [Google Scholar]
- Paul K, Nonoh JO, Mikulski L, Brune A. (2012). "Methanoplasmatales," Thermoplasmatales-related Archaea in termite guts and other environments, are the seventh order of methanogens. Appl Environ Microbiol 78: 8245–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu S-M, Chin FYL. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR et al. (2015). Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv 1: e1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polag D, Leiß O, Keppler F. (2014). Age dependent breath methane in the German population. Sci Total Environ 481: 582–587. [DOI] [PubMed] [Google Scholar]
- Pozuelo M, Panda S, Santiago A, Mendez S, Accarino A, Santos J et al. (2015). Reduction of butyrate-and methane-producing microorganisms in patients with irritable bowel syndrome. Sci Rep 5: 12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R et al. (2007). Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA 104: 10643–10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, Marchesi JR. (2008). Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol 8: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion‐Larmurier I, Beaugerie L et al. (2009). Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189. [DOI] [PubMed] [Google Scholar]
- Söllinger A, Schwab C, Weinmaier T, Loy A, Tveit AT, Schleper C et al. (2016). Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol Ecol 92: fiv149. [DOI] [PubMed] [Google Scholar]
- Tang WW, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B et al. (2015). Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res 116: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen S, Lacroix C, Schwab C. (2015). Methanogen communities in stool of humans of different age and health status and co-occurrence with bacteria. FEMS Microbiol Lett 362: fnv092. [DOI] [PubMed] [Google Scholar]
- Wagner M, Sonntag D, Grimm R, Pich A, Eckerskorn C, Söhling B et al. (1999). Substrate‐specific selenoprotein B of glycine reductase from Eubacterium acidaminophilum. Eur J Biochem 260: 38–49. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z et al. (2016). Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165: 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Jameson E, Crosatti M, Schäfer H, Rajakumar K, Bugg TD et al. (2014). Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci USA 111: 4268–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.