Abstract
Methane-oxidizing bacteria represent a major biological sink for methane and are thus Earth’s natural protection against this potent greenhouse gas. Here we show that in two stratified freshwater lakes a substantial part of upward-diffusing methane was oxidized by filamentous gamma-proteobacteria related to Crenothrix polyspora. These filamentous bacteria have been known as contaminants of drinking water supplies since 1870, but their role in the environmental methane removal has remained unclear. While oxidizing methane, these organisms were assigned an ‘unusual’ methane monooxygenase (MMO), which was only distantly related to ‘classical’ MMO of gamma-proteobacterial methanotrophs. We now correct this assignment and show that Crenothrix encode a typical gamma-proteobacterial PmoA. Stable isotope labeling in combination swith single-cell imaging mass spectrometry revealed methane-dependent growth of the lacustrine Crenothrix with oxygen as well as under oxygen-deficient conditions. Crenothrix genomes encoded pathways for the respiration of oxygen as well as for the reduction of nitrate to N2O. The observed abundance and planktonic growth of Crenothrix suggest that these methanotrophs can act as a relevant biological sink for methane in stratified lakes and should be considered in the context of environmental removal of methane.
Introduction
Freshwater lakes represent large natural sources of methane and contribute more to methane emissions than the oceans despite their comparably smaller area (Bastviken et al., 2004). Highest rates of methane removal are usually measured at the oxyclines, either in the water column or in the sediment. Lake Rotsee and Lake Zug in Central Switzerland are typical examples of temperate lake systems with methane fluxes across the oxycline of 13±3 mmol and 10±3 mmol m−2 d−1, respectively (Oswald et al., 2015, 2016). Both lakes are stratified, with methane-rich hypolimnia, but whereas the shallow Lake Rotsee overturns annually, the deep Lake Zug remains stratified throughout the year. In both lakes, the vast majority of the upward-diffusing methane is removed at the base of the oxycline at in situ oxygen concentrations in the low micromolar range (Oswald et al., 2015, 2016). Methane oxidation at the oxycline was shown to be coupled to the reduction of residual or in situ-produced oxygen, but there were also indications for methane-oxidizing activity under oxygen-deficient conditions (Oswald et al., 2015, 2016).
Abundant gamma-proteobacterial methane-oxidizing bacteria (gamma-MOB) were shown to be involved in methane removal in both lakes (Oswald et al., 2015, 2016). Gamma-MOB are considered aerobes requiring oxygen for methane activation, even though some cultured representatives can perform methane oxidation under denitrifying conditions (Kits et al., 2015a, 2015b). Environmentally relevant representatives of gamma-MOB in lakes and other freshwater habitats belong to the ‘classical’ genera of Methylobacter, Methylomonas, Methylosarcina and Methylomicrobium (Boschker et al., 1998; Bodelier et al., 2013; Oshkin et al., 2015), and all possess particulate methane monooxygenase (pMMO) as the key methane-oxidizing enzyme (Bowman, 2005). In Lake Rotsee and Lake Zug, unicellular gamma-MOB represented a stable community at the oxycline. The bacteria showed rapid growth on methane as evidenced by the increase in cell abundances and the uptake of 13C-methane into their biomass (Oswald et al., 2015, 2016).
In these studies, gamma-MOB were identified by fluorescence in situ hybridization using the 16S rRNA-targeted oligonucleotide probes Mgamma84+705. Interestingly however, these probes do not bind to members of a potentially important subgroup of gamma-proteobacterial MOB, the putative family Crenothrichaceae. Contrary to ‘classical’ MOB, these gamma-MOB are multicellular and filamentous. So far, only two of these bacteria have been documented in literature, Crenothrix polyspora and Clonothrix fusca, and both were retrieved from groundwater (Stoecker et al., 2006; Vigliotta et al., 2007). Sporadically, environmental occurrence of Crenothrix is reported in literature based on retrieved 16S rRNA or pmoA sequences (Dörr et al., 2010; Drewniak et al., 2012), but its role in methane cycling has remained unclear.
The metabolism of Crenothrix has been a matter of debate since its first description as ‘Brunnenfaden’ (‘a well thread’ Cohn, 1870). Initially, Crenothrix/Clonothrix filaments were considered to belong to the ‘iron bacteria’ due to the presence of metal particles in their sheaths (Roze, 1896; Jackson, 1902; Molisch, 1910). This belief was challenged by studies that failed to observe iron encrustation in Crenothrix/Clonothrix filaments (Kolk, 1938; Wolfe, 1960), and the later discovery of membrane invaginations has prompted suggestions for a methanotrophic lifestyle (Völker et al., 1977). Eventually, the capacity to oxidize methane was experimentally confirmed on filaments retrieved from man-made habitats (Stoecker et al., 2006; Vigliotta et al., 2007). Interestingly, C. polyspora was reported to possess an ‘unusual’ pMMO, which was only distantly related to ‘classical’ MMO of gamma-proteobacterial methanotrophs (Stoecker et al., 2006), and has now been recognized to cluster together with the ammonium monooxygenases of completely nitrifying ‘comammox’ bacteria (Daims et al., 2015; van Kessel et al., 2015).
Here we investigated the occurrence and involvement of these filamentous bacteria in methane oxidation at and below the oxyclines of Lake Rotsee and Lake Zug. We performed stable isotope labeling experiments followed by single-cell imaging to explore the role of these microorganisms in environmental methane cycling, and metagenomic analyses to investigate their metabolic potential with respect to aerobic and anaerobic respiration. For comparison, we also performed metagenomic analysis of a sample from Wolfenbüttel waterworks sand filter reportedly containing high proportions of C. polyspora.
Materials and methods
Geochemical profiling in Lake Rotsee
Profiling was done in October 2014 at the deepest point (16 m depth, 47°04.259‘N, 8°18.989‘E). A multi-parameter probe was used to measure photosynthetically active radiation (PAR; LI-193 Spherical Underwater Quantum Sensor, LI-COR, Lincoln, NE, USA) along with conductivity, turbidity, depth (pressure), temperature and pH (XRX 620, RBR, Ottawa, ON, Canada). Dissolved oxygen was simultaneously monitored online with normal and trace micro-optodes (types PSt1 and TOS7, Presens, Regensburg, Germany) with detection limits of 125 and 20 nm, respectively, and a response time of 7 s (Kirf et al., 2014).
Water samples for dissolved methane analysis were retrieved from distinct depths with a Niskin bottle. Serum bottles (120 ml) were filled completely without bubbles or headspace through a gas-tight outlet tubing allowing water to overflow. Solid copper chloride [Cu(I)Cl] was immediately added in excess to the water samples and the bottles were crimped. Before analysis, a 30 ml headspace was set with N2 and after overnight equilibration methane concentrations were measured in the headspace with a gas chromatograph (GC; Agilent 6890 N, Agilent Technologies, Santa Clara, CA, USA) equipped with a Carboxen 1010 column (30 m × 0.53 mm, Supelco, Bellefonte, PA, USA) and a flame ionization detector. Methane concentrations in the water phase were back-calculated according to (Wiesenburg and Guinasso, 1979). Stable carbon isotopes of methane were determined in the same headspace by isotope ratio mass spectrometry with a trace gas instrument (T/GAS PRE CON, Micromass UK Ltd., Wilmslow, UK) coupled to a mass spectrometer (GV Instruments, Manchester, UK; Isoprime, Stockport, UK). Isotopic ratios are given in δ-notation relative to the Vienna Pee Dee Belemnite reference standard.
Oxygen, PAR, methane concentration and methane isotope profiles for the sampling campaign in October 2014 are shown in Supplementary Figure 1. Geochemical profiles from other Lake Rotsee campaigns are reported in Oswald et al. (2015).
Lake Rotsee methane oxidation rates
Methane oxidation rates were measured in incubations set up in October 2014, with water from the 7 m depth (oxycline), and from 8 m depth (with no detectable oxygen). Water was collected with a Niskin bottle and filled into sterile 1 l Schott bottles without a headspace, closed with butyl stoppers and kept cold and dark until further handling. In the laboratory, 120 ml was distributed into 160 ml serum bottles in an anoxic (N2-containing) glove box (Iner Tec, Grenchen, Switzerland), closed with butyl stoppers and crimped. Each incubation was supplemented with 13C-labeled methane (99 at%, Campro Scientific, Berlin, Germany) and 12C-methane to reach 2 bar overpressure, resulting in ~1.8 mmol l−1 CH4 in the water phase and 50 at% 13C labeling percentage. For comparison, in situ methane concentrations at 7 and 8 m depth were ca. 15 and 35 μmol l−1 (Supplementary Figure 1). Duplicate bottles were incubated at 6 °C under dark and light conditions along with a control (sterile filtered lake water). Methane oxidation was monitored during an incubation period of 7 days as production of 13CO2. Anoxically withdrawn water samples (2 ml) were transferred into 6 ml Exetainers (Labco, Lampeter, UK), fixed with 200 μl zinc chloride (50% w/v) and acidified with concentrated H3PO4 (100 μl). Isotopic ratios of CO2 were determined in the headspace with a preparation system (MultiFlow, Isoprime) coupled to an isotope ratio mass spectrometry (Micromass, Isoprime). Subsequently, methane oxidation rates were calculated as described previously (Oswald et al., 2015). These rates are shown in Supplementary Figure 1. As these incubations were unamended (apart from methane addition), aerobic methane oxidation in these incubations was presumably sustained solely by oxygenic photosynthesis (Milucka et al., 2015; Oswald et al., 2015). At selected time points, sub-samples were also taken for catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) analysis. These data are shown in Supplementary Figure 3. Nanometer-scale secondary ion mass spectrometry (NanoSIMS) and metagenome analyses reported for Lake Rotsee (shown in Figure 1 and Supplementary Figure 2) were performed on samples collected on a previous sampling campaign in August 2013 (rates and other data from this campaign are reported in Oswald et al. (2015)).
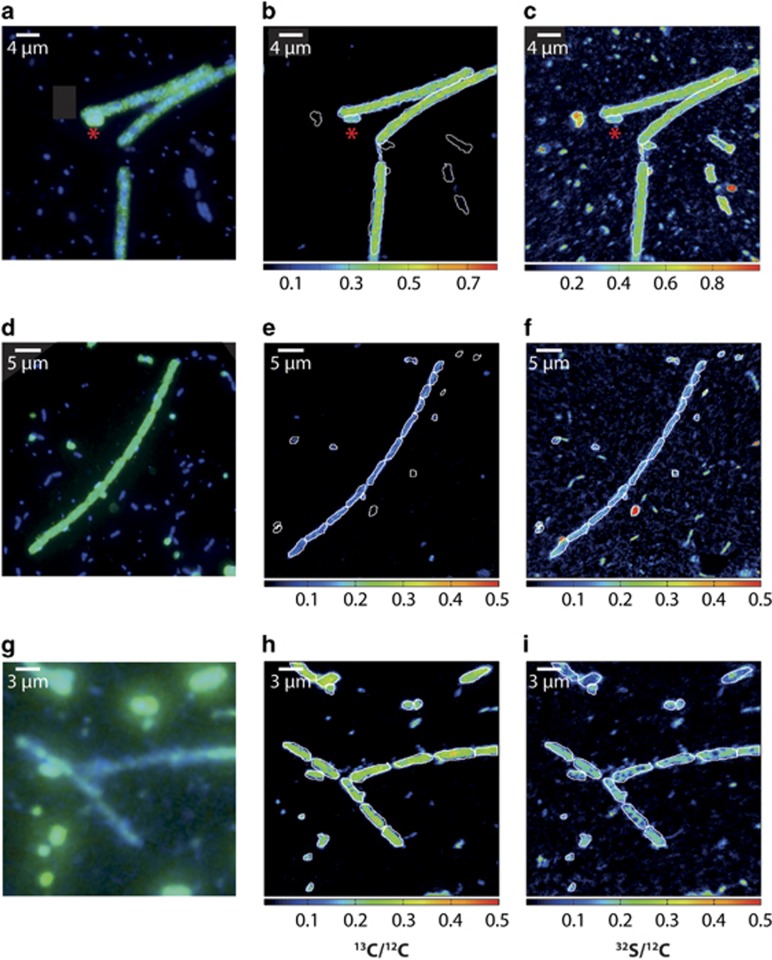
Figure 1.
Methane-dependent growth of Crenothrix in Lake Rotsee and Lake Zug. (a) Crenothrix in the Lake Rotsee oxic incubation visualized by CARD-FISH (green; counterstained by DAPI in blue) with a specific probe Creno445 (Stoecker et al., 2006). A small coccoid cell targeted by the probe (marked by the asterisk) might represent a gonidial cell, which Crenothrix is reportedly capable of producing (Völker et al., 1977). (b) The corresponding 13C/12C nanoSIMS image shows homogeneous 13C enrichment throughout the cell filament. The small coccoid cell is also significantly enriched, albeit less. (c) The corresponding 32S/12C nanoSIMS image showing distribution of organic material on the filter. (d) Putative Crenothrix filaments in the Lake Zug oxic incubation visualized by DAPI (blue) and CARD-FISH (green) with probe Mgamma669. (e) Corresponding 13C/12C and (f) 32S/12C nanoSIMS images. Note the fragmented nature of the Crenothrix filaments and the attached small (unidentified) bacteria. (g) Putative Crenothrix filaments in the Lake Zug anoxic incubation visualized by DAPI (blue) and CARD-FISH (green) with probe Mgamma669. (h) Corresponding 13C/12C and (i) 32S/12C nanoSIMS images.
Lake Zug nitrate addition experiment
The sampling campaign was carried out in October 2013. Water samples from the anoxic 160 m depth were collected with a Niskin bottle, filled into sterile Schott bottles, closed with a stopper and stored as described above. The water was distributed into sterile 160 ml serum bottles (a 120 ml) in an N2 glove box (Mecaplex, Grenchen, Switzerland) as described in detail in Oswald et al. (2016). 13C-labeled methane (99 at%, Campro Scientific) was supplied at a ~20% labeling percentage. A 2 bar methane overpressure was set using 12C-methane. One set of duplicate bottles received no further addition and served as a control and one set of duplicate bottles was amended with 15NO3− (from a sterile anoxic 100 mmol l−1 stock solution) to a final concentration of 50 μmol l−1. Bottles were incubated in the dark under in situ temperatures (~5 °C) for 16 days. At regular intervals, bottles were subsampled for 13CO2 measurements in order to determine methane oxidation rates. For this, anoxically withdrawn water samples (2 ml) were transferred into 6 ml Exetainers, fixed with zinc chloride and acidified with concentrated H3PO4. Isotopic ratios of CO2 were determined in the headspace using a Finnigan GasBench II attached to an isotope ratio mass spectrometer (IRMS; Finnigan Delta Plus, Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, methane oxidation rates were calculated as described previously (Oswald et al., 2015). At selected time points, sub-samples were also taken for CARD-FISH and nanoSIMS analyses. An early time point (T= 2d) was analyzed by nanoSIMS to obtain data for the calculation of methane uptake rates reported in Table 1. FISH and nanoSIMS images from Lake Zug nitrate incubation (Figure 1; Supplementary Figure 6) originate from the last time point of the incubation (T=16 d). The sample for metagenome analysis (sample Z3) was also taken at this time point. Additionally, an in situ water sample from 160 m was also used for metagenome analysis (sample Z1). During this sampling campaign, no incubations with added oxygen were performed.
Table 1. Overview of methane carbon uptake rates by Crenothrix and unicellular gamma-MOB in Lake Rotsee and Lake Zug.
| 13C at %a (n) | Avg biovolume (CARD-FISH-based)b (μm3;n) | Methane-C uptake per cellc (fmol cellavg−1 d−1) | Cell countd (cell ml−1) | Total population biovolumee(μm3 ml−1) | Methane-C uptake per populationf (μmol l−1 d−1) | |
|---|---|---|---|---|---|---|
| Lake Rotsee (oxic) | ||||||
| Crenothrix (Mgamma669) | NA | 85±8.3 (59) | 147.7±26.3g | 1.2E+04 | 1.0E+06 | 1.73g |
| Crenothrix (Creno445) | 22.00±4.8 (17) | 73.7±8.4 (51) | 128.0±22.8 | 9.2E+03 | 6.8E+05 | 1.18 |
| Other gamma-MOBh | 28.77±4.1 | 4.2 | 10.6±0.9 | 2.6E+04 | 1.1E+05 | 0.27 |
| Lake Zug (oxic) | ||||||
| Crenothrix (Mgamma669; low O2) | 9.26±1.7 (19) | 32.5±5.5 (20) | 38.1±6.9 | 1.1E+03 | 3.5E+04 | 0.041 |
| Crenothrix (Mgamma669; high O2) | 8.68±1.9 (10)i | 32.5±5.5 (20) | 35.3±7.8 | 1.1E+03 | 3.5E+04 | 0.038 |
| Other gamma-MOB (low O2)j | 10.39±3.1 | 4.2 | 5.7±1.2 | 6.8E+04 | 2.9E+05 | 0.39 |
| Other gamma-MOB (high O2)j | 12.13±3.75 | 4.2 | 6.9±1.6 | 6.8E+04 | 2.9E+05 | 0.47 |
| Lake Zug (anoxic) | ||||||
| Crenothrix (Mgamma669) | 13.27±4.9 (6) | 49.7±20.3 (15) | 74.2±26.6 (6) | 0.4E+03 | 2.0E+04 | 0.03 |
| Other gamma-MOB | NA | NA | NA | NA | NA | NA |
Abbreviations: CARD-FISH, catalyzed reporter deposition fluorescence in situ hybridization; MOB,
methane-oxidizing bacteria; n, number of analyzed cells, NA, not analyzed.
Calculated as an average (± s.d.) of the 13C/12C ratios of individual regions of interest (i.e., cells) determined by nanoSIMS.
Calculated from CARD-FISH data as an average biovolume (± s.d.) using Mgamma669 or Creno445 probe (Crenothrix) and Mgamma84+705 probes (other gamma-MOB).
Calculated as follows: data from column a were converted into 13C excess in fmol per cell (of a given average biovolume; cellavg) using the avg cell biovolume reported in column b and a conversion factor of 6.4 fmol C μm−3 (Musat et al., 2008). The numbers were corrected for labeling percentage and incubation time.
Counted from the same filters from which avg biovolumes (column b) were obtained. As the boundaries between individual cells within the filament were often not recognizable, only hybridized filaments were counted. Cell counts refer to cell abundances at the start of each incubation and thus do not account for increase of cell abundances during the incubation period.
Calculated as follows: data from column b were upscaled using data in column d.
Calculated as follows: data from column c were upscaled using data from column d.
Assuming the same 13C enrichment as determined with the probe Creno445 on the same sample.
According to Oswald et al., 2015.
In this sample, three analyzed filaments had 13C/12C<0.015 and were not included in the analysis.
According to Oswald et al., 2016.
Calculations are based on incubations from Lake Rotsee (oxic, 2013) and Lake Zug (oxic and anoxic, 2013, 2014; see Supplementary Table 4 for sample details).
O2-supplemented incubations referred to in this manuscript were only performed during a sampling campaign in June 2014 and are described in detail in Oswald et al. (2016), where also the corresponding geochemical profiles and methane oxidation rates from relevant depths and incubations are reported. Briefly, O2-supplemented incubations were set up as described above, with the difference that instead of nitrate, sterile air was injected to the incubations to reach final O2 concentrations of ca. 80 μmol l−1 (‘low O2’) and ca. 200 μmol l−1 (‘high O2’), respectively. Incubations were subsampled at regular intervals for methane oxidation rates, CARD-FISH and nanoSIMS analyses. The CARD-FISH and nanoSIMS analyses shown in Figure 1 were performed on samples taken from 160 m incubation after T=2 d. The sample for metagenome analysis was taken at the last time point of the ‘low O2’ 160 m incubation (T=11 d).
Catalyzed reporter deposition fluorescence in situ hybridization
Formaldehyde- (2% (v/v) final concentration) fixed water samples were incubated for 30 min at room temperature before being filtered onto polycarbonate GTTP filters (0.2 μm pore size; Merck Millipore, Darmstadt, Germany). For nanoSIMS analysis, samples were filtered onto Au or Au/Pd-coated GTTP filters (0.2 μm pore size). Permeabilization with lysozyme, peroxidase inactivation, hybridization with specific oligonucleotide probes labeled with horseradish peroxidase in combination with tyramide signal amplification (Oregon Green 488) and DAPI counter staining was performed as described previously (Pernthaler et al., 2002). An overview of probes used (Biomers, Ulm, Germany) is included in Supplementary Table 2. For cell counts and biovolume determinations, one filter was analyzed for each sample. Hybridized filaments (using probe Mgamma669) were enumerated in randomly selected fields of view with a confocal laser scanning microscope (SP5 DMI 6000, Leica, Wetzlar, Germany). For biovolume calculations, length and width of >15 filaments in >10 fields of view were then measured directly in confocal micrographs using LAS AF Lite software (Leica). Values for the cell counts and methane uptake rates of unicellular gamma-MOB cells were taken from Oswald et al. (2015) and Oswald et al. (2016).
Nanometer-scale secondary ion mass spectrometry
Areas of interest containing positive CARD-FISH hybridization signals were marked with a laser micro-dissection microscope (DM 6000, Leica Microsystems, Mannheim, Germany). Laser-marked areas were analyzed by nanoSIMS (NanoSIMS 50 l, Cameca, Paris, France) at the MPI Bremen as described previously. For Lake Rotsee (light incubation, 9 m depth), 12 and 26 filaments were analyzed in five fields of view after 2 and 7 days of incubation, respectively. For the Lake Zug low and high O2 addition experiments, 19 and 13 filaments were measured in 9 and 7 fields of view, respectively, after 2 days of incubation. For the Lake Zug nitrate addition incubation, 6 filaments were measured in 5 fields of view after 2 days of incubation and 7 filaments were measured in 5 fields of view after 16 days of incubation. Obtained secondary ion images were drift corrected, accumulated and processed with Look@NanoSIMS (Polerecky et al., 2012).
Biovolume and carbon assimilation rates
The biovolume of individual Crenothrix filaments was calculated from their measured length and width by assuming a cylindrical shape. The length and the width of filaments were determined from the CARD-FISH images that were used for cell counting. Due to the varying length of filaments, an average biovolume of Crenothrix was calculated and is reported in Table 1. The ‘average biovolume determined from CARD FISH’ was calculated as an average of biovolumes of individual filaments hybridized with a Crenothrix-targeting probe (Mgamma669 or Creno445) at the start of the respective incubation and is reported with the s.d. ‘Total’ Crenothrix biovolume reported in Table 1 and Supplementary Figure 4 was obtained by multiplying the average filament biovolume by the number of filaments per ml of water. For comparison, the biovolume of unicellular gamma-MOB cells was calculated from total cell counts and by assuming an average spherical cellular diameter of 2 μm.
Cellular 13C at % were calculated from 13C/12C values of individual ROIs (regions of interest). Regions of interests were drawn to outline single Crenothrix cells (for example, Figures 1h and i), whole filaments (Figures 1e and f) or parts of filaments (Figures 1b and c). Both the background (cell-free polycarbonate filter in the same field of view) and the 13C enrichment of all cells in every field of view was evaluated and compared for all measurements. Rates of methane carbon uptake (fmol C cellavg−1 d−1) of Crenothrix and unicellular gamma-MOB were calculated from the 13C excess of the measured cells using a conversion factor of 6.4 fmol C μm−3 reported in Musat et al. (2008). These uptake rates were corrected for the labeling percentage and the incubation time. The methane uptake rates were calculated only for filamentous cells, which were stained with the Creno445 or Mgamma669 probe. Hybridized single cells (such as in Figures 1a–c) were not considered in the calculation.
DNA extraction, 16S rRNA gene amplicon sequencing and analysis
Two in situ water samples from Lake Rotsee were used for 16S rRNA gene amplicon sequencing. One was collected from the oxycline (9 m depth) during a campaign in August 2013 and the other from anoxic water (8 m depth) during a campaign in October 2014 (Supplementary Table 4). Volumes of ca. 250 ml were filtered onto polycarbonate Nuclepore Track-Etched Membrane filters (0.2 μm pore size; Whatman, Maidstone, UK). Filters were stored at −80 °C until DNA was extracted with the UltraClean Soil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA). Extraction procedure was performed according to manufacturer’s instructions with the following adjustment: vortexing with the Bead Solution was reduced to 30 s with subsequent incubation on ice (30 s), and this cycle was repeated four times.
The V3–V4 regions of the 16S rRNA gene were targeted with primer pair 341 F (5′-CCTACGGGNGGCWGCAG-3′) and 805 R (5′-GACTACCAGGGTATCTAATC-3′). The forward primers contained unique identifier sequences at the 5’-end for each sample to allow for multiplex sequencing. Ten separate PCR reactions (25 μl volume) were set up for each sample including both forward and reverse primers (500 nm each), deoxyribose nucleotide triphosphates (dNTPs; 800 μm), 1 × Taq reaction buffer, Taq DNA polymerase (0.25 U) and DNA extracts of the respective samples (0.5–1 μl). The reactions proceeded as follows: initial denaturation (3 min at 95 °C), 25 cycles of denaturation (30 s at 95 °C), annealing (30 s at 54 °C) and elongation (90 s at 72 °C); and final elongation (10 min at 72 °C). Parallel reactions were combined and purified with the QIA quick PCR Purification Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions, with a final elution in 1 × TE buffer (30 μl; 10 mm Tris-HCl (pH 8.0)+1 mm EDTA). The DNA was further purified with a gel using SYBR Green I Nucleic Acid Gel Stain (Invitrogen, Carlsbad, CA, USA) followed by gel extraction with QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s protocol. Extract concentrations were measured fluorometrically using the Qubit dsDNA HS Assay Kit and the Qubit 2.0 Fluorometer (Invitrogen). Illumina sequencing was performed on the amplicons at the Max Planck-Genome Centre (Cologne, Germany).
16S rRNA gene amplicon paired-end reads were trimmed (right end only, trim quality threshold=10) and merged (20 bases minimum overlap) using BBmap software version 35.43 (sourceforge.net/projects/bbmap). Reads were then separated by barcode and trimmed (minimum length=300, maximum homopolymer length=8, maximum number of ambiguous bases=0, minimum average quality score allowed over 50 bp window=20) using mothur v.1.36.1 (Schloss et al., 2009). The separated reads were processed using SILVAngs and standard parameters (Quast et al., 2013).
Lake metagenome sequencing and assembly
Two in situ water samples (Lake Rotsee, 9 m depth, August 2013 (sample R1) and Lake Zug, 160 m depth, October 2013 (sample Z1)) and four end time points of incubations (Lake Rotsee, O2-supplemented (sample R2), Lake Rotsee, light (sample R3), Lake Zug, low O2-supplemented (sample Z2), Lake Zug, anoxic, nitrate-supplemented (sample Z3); see Supplementary Tables 3 and 4 for additional sample information) were analyzed by Illumina sequencing. The following water volumes were filtered onto polycarbonate Nucleopore Tracked-Etched membrane filters (0.2 μm pore size; Whatman) and stored at −80 °C: 250 ml for in situ samples (R1 and Z1), 50 ml for Lake Rotsee incubations (R2 and R3) and 40 ml for Lake Zug incubations (Z2 and Z3). DNA was extracted from cut-up filters using the PowerSoil DNA isolation kit according to manufacturer’s instructions (MoBio Laboratories). DNA from lake Zug was fragmented by sonication (MiSeq: 600–700 bp; HiSeq2500: 300 bp) using a Covaris S2 sonicator (Covaris, Woburn, MA, USA). The library was prepared using Ovation Ultra Low Library Systems V1 (for MiSeq) or V2 (for HiSeq2500) kits (NuGEN Technologies, San Carlos, CA, USA) and paired-end sequencing (2 × 300 or 2 × 150 bp) was performed using the Illumina MiSeq (2 × 300 bp) or HiSeq2500 (2 × 150 bp) platform (Illumina Inc., San Diego, CA, USA). DNA from Lake Rotsee was fragmented by sonication (350 bp) using a Covaris S2 sonicator (Covaris), the library was prepared using NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) and paired-end sequencing (2 × 150 or 2 × 100 bp) was performed using the Illumina HiSeq2500 or 3000 platform (Illumina Inc.). Both MiSeq and HiSeq sequencing was performed by the Max Planck-Genome-centre, Cologne, Germany (http://mpgc.mpipz.mpg.de/home/; Supplementary Table 3).
Sequences were quality checked using FastQC (Andrews, 2010) and trimming, as well as adapter removal was done using Trimmomatic 0.32 and parameters MINLEN:20 ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:50 (Bolger et al., 2014). Metagenome assembly of sequences from the Lake Zug incubation (anoxic, nitrate-supplemented (Z3; Supplementary Tables 3, 4)) was performed using SPAdes 3.5.0 (Bankevich et al., 2012) with mismatch corrector enabled and default parameters.
Sand filter Crenothrix metagenome sequencing and assembly
Samples containing high proportions of C. polyspora filaments were taken from the backwash water of rapid sand filters of the Wolfenbüttel waterworks (Germany), which treats a mixture of oxic and anoxic groundwater. During sampling, Crenothrix filaments were retained from 600 to 850 liters of backwash water by either sedimentation or filtration through a fine-mesh sieve (200 or 400 μm). One sample was collected in 2004 (on 21 June; sample C) and was incubated with 500 μmol l−1 ammonium for 212 h. The second sample was collected in 2005 (10 October, sample B) and was incubated at different methane concentrations for 24 h. It should also be noted that earlier we deposited one additional partial and unpublished Crenothrix genome from a sand filter sample from the Wolfenbüttel waterworks at IMG (genome ID 3300005627). We did not analyze that older genome sequence in the course of the present study, because it originated from the same site but had been sequenced less deeply than the two sand filter Crenothrix genomes described here.
After the incubations, samples B and C were frozen at −20 °C and DNA was extracted in 2016 using a phenol chloroform protocol (Zhou et al., 1996) including two bead-beating steps. Paired-end sample libraries were prepared using Illumina Nextera DNA Library Preparation Kit (Illumina Inc.) and sequenced at Aalborg University (Denmark) using an Illumina MiSeq with MiSeq Reagent Kit v3 (2 × 301 bp; Supplementary Table 3). Paired-end reads were imported to CLC Genomics Workbench v. 8.0 (CLCBio, Aarhus, Denmark) and trimmed using a minimum phred score of 20, a minimum length of 50 bp, allowing no ambiguous nucleotides and trimming off Illumina sequencing adaptors if found. All trimmed paired-end metagenome reads were assembled using CLC’s de novo assembly algorithm, using a kmer of 63 and a minimum scaffold length of 1 kbp.
Metagenome binning, reassembly and annotation
Binning of contigs of the Lake Zug metagenomic assembly (sample Z3, Supplementary Table 3) was performed by exploiting differential contig coverage from three sequenced metagenomic data sets: Z1 (Lake Zug, in situ), Z2 (Lake Zug, O2-supplemented incubation) and Z3 (Lake Zug, anoxic, nitrate-supplemented incubation) as described previously (Albertsen et al., 2013) and implemented in the mmgenome R package (http://madsalbertsen.github.io/mmgenome/; Karst et al., 2016). Only contigs longer than 500 bp were used and the average coverage of each contig was computed directly using BBmap 35.43 (http://sourceforge.net/projects/bbmap/) with default parameters. Prodigal 2.60 (Hyatt et al., 2010) in metagenomic mode (-p meta) and standard parameters was used to predict open reading frames, which were translated to amino-acid sequences and subsequently searched for using HMMER 3.1b (Eddy et al., 2013) against a set of 107 hidden markov models of essential single-copy genes (Dupont et al., 2012) using default settings and trusted cutoff (-cut_tc) enabled. Protein sequences coding for essential single copy genes were searched against NCBI non-redundant database (retrieved in August 2015) using BLASTP (Camacho et al., 2009) and an e-value cutoff of 10−6. The taxonomy (class level) of each essential single-copy gene was assigned using MEGAN5 (Huson et al., 2011; with the previously generated BLASTP xml file as input) and the mmgenome script ‘hmm.majority.vote.pl’. Bowtie2 (Langmead and Salzberg, 2012) with standard settings was used to map reads to contigs and the number of paired-end connections between separate contigs was calculated from the SAM file using the mmgenome script ‘network.pl’.
Differential coverage of contigs between the two sand filter Crenothrix metagenomes (Supplementary Figure 8) and between the Lake Zug metagenomes (Supplementary Figure 7), as well as paired-end connections between separate contigs were used to extract genomic bins from the metagenome using the mmgenome R package (http://madsalbertsen.github.io/mmgenome/; Karst et al., 2016). Reads used for the initial assembly were mapped to the binned contigs using BBmap of the BBmap package 35.43 (http://sourceforge.net/projects/bbmap/) using stringent settings (approximate minimum identity=0.98) or CLC (sand filter Crenothrix). Mapped reads were reassembled (only for the lacustrine Crenothrix) using SPAdes 3.5.0 (Bankevich et al., 2012) with mismatch corrector enabled and default parameters. Quality of the reassembled bins was assessed using CheckM 1.05 running the lineage-specific workflow (Parks et al., 2015). Annotation of the Crenothrix D3 draft genome was performed using RAST (Aziz et al., 2008). CDS prediction and automated pre-annotation of the two Wolfenbüttel sand filter Crenothrix genome sequence bins were performed using the PROKKA pipeline (Seemann, 2014) with an in-house extended protein reference database. The annotation of key metabolic pathways was manually refined.
The Whole Genome Shotgun project of lacustrine Crenothrix sp. D3 has been deposited at DDBJ/ENA/GenBank under the accession MBQZ00000000. The version described in this paper is version MBQZ01000000. Reads (Lake Zug and Lake Rotsee) have been deposited at the Sequence Read Archive under BioProject PRJNA325574. The two sand filter Crenothrix metagenomic assemblies are available in the European Nucleotide Archive (ENA) under the study accession number PRJEB19189.
Phylogenetic analyses
Full-length amino-acid sequences of bacterial PmoA and AmoA protein sequences were retrieved from the Integrated Microbial Genomes database (IMG-ER; Markowitz et al., 2009) using Pfam family PF02461. Previously published protein sequences of ‘unusual’ PmoA of C. polyspora (accession ABC59822–ABC59827; Stoecker et al., 2006), partial PmoA of C. fusca (accession ABL64049; Vigliotta et al., 2007) AmoA sequences belonging to Candidatus Nitrospira nitrosa, (accession CUS31358; van Kessel et al., 2015) as well as Candidatus Nitrospira inopinata (accession CUQ66826; Daims et al., 2015) were added to the reference set. After removing duplicate sequences, protein sequences were aligned using Clustal Omega 1.2.0 (Sievers et al., 2011) and default parameters. A phylogenetic tree (135 taxa) was calculated using RAxML 8.2.6 (Stamatakis, 2014) and parameters: -f a -k -x 48020621 -p 6809427 -N 100 -T 8 -m PROTGAMMAWAG.
Partial Crenothrix 16S rRNA gene sequences were retrieved from the Crenothrix draft genomes using RNAmmer 1.2 (Lagesen et al., 2007), aligned using the SILVA incremental aligner (SINA) 1.2.11 (Pruesse et al., 2012) and imported to the SILVA SSU NR99_123 database (Quast et al., 2013) using ARB 6.1 (Ludwig et al., 2004). Phylogenetic trees of the 16S rRNA gene sequences were calculated using RAxML 7.7.2 integrated in ARB with the GAMMA model of rate heterogeneity and the GTR substitution model with 100 bootstraps.
Results and discussion
Crenothrix in Lake Rotsee and Lake Zug
To investigate the potential occurrence of filamentous Crenothrix bacteria in two stratified lakes and their involvement in the lacustrine methane cycle, we first recorded geochemical evidence for methane oxidation in situ. Concentration profiles recorded in Lake Rotsee and Lake Zug over the course of 3 years suggested a zone of methane consumption that persistently coincided with the oxycline (profiles from Lake Rotsee 2013 are shown in Oswald et al. (2015), from 2014 in Supplementary Figure 1; profiles from Lake Zug 2012, 2013 and 2014 are shown in Oswald et al., 2016). Concurrently, incubations with 13CH4 confirmed high rates of methane oxidation at the oxycline (Oswald et al., 2015, 2016; Supplementary Figure 1). These incubations were set up under both oxic and anoxic conditions. In Lake Rotsee, oxic incubation conditions were obtained either by addition of air or solely by incubation of anoxic water in the light. In the latter case, aerobic methane oxidation was presumably sustained by oxygenic photosynthesis (Milucka et al., 2015; Oswald et al., 2015). In Lake Zug, oxic incubations were solely supplemented with air and incubated in the dark. These different incubation set ups reflected the different nature of the two lakes, Lake Rotsee has a shallow, sun-lit oxycline, whereas the oxycline of Lake Zug is very deep and dark. Additionally, anoxic Lake Zug incubations supplemented with nitrate were also set up as Lake Zug had the appropriate environment to test for methane-dependent denitrification (Supplementary Table 4).
We then analyzed the microbial community at the Lake Rotsee oxycline by 16S rRNA gene amplicon sequencing in 2 consecutive years (2013 and 2014; Supplementary Figure 2). Along with gamma-proteobacterial Methylococcaceae (Methylobacter, Methylocaldum, Methylomonas and Methyloglobulus species), CABC2E06 (an uncultured Methylococcales clone; Wang et al., 2012; Quaiser et al., 2014), and the marine methylotrophic group, also sequences belonging to Crenothrix were retrieved. On the basis of the number of recovered sequences, Crenothrix-related organisms were 2–5-fold less abundant than Methylococcaceae and comprised 0.06–0.1% of the total bacterial sequences in situ. However, it is possible that the true abundance of Crenothrix in situ was higher than what the 16S rRNA gene abundances suggest, as, for example, DNA extraction biases might strongly select against these thickly sheathed microorganisms.
We could additionally confirm the presence of Crenothrix in both lakes by CARD-FISH with two oligonucleotide probes reported to target Crenothrix, Mgamma669 and Creno445 (Eller et al., 2001; Stoecker et al., 2006). The more specific oligonucleotide probe Creno445 bound only sporadically, when the hybridization stringency was strongly reduced (Supplementary Figure 3). On the other hand, the Mgamma669 probe hybridized most of the conspicuous filaments in all analyzed samples from both lakes (in situ water as well as incubations, Figure 1; Supplementary Figure 3) even though some filaments did not hybridize even with this more general probe (for example, Supplementary Figures 3a, b). With both probes, we observed two hybridized cell morphotypes—filaments and single round cells (Figure 1; Supplementary Figure 3). Both morphotypes have been observed for Crenothrix spp. previously and it has been proposed that the smaller round cells represent reproductive cells that bud from the ends of vegetative cell filaments (Cohn, 1870; Völker et al., 1977). However, given the compromised specificity of the Creno445 probe at low stringency and the broad specificity of the Mgamma669 probe, it is also possible that the hybridized single cells represented other gamma-MOB, reportedly targeted by the Mgamma669 probe (for example, Methylobacter or Methylomonas; Eller et al., 2001). Therefore, the here-reported Crenothrix cell counts and biovolumes are solely based on counts of Creno445- or Mgamma669-hybridized filaments and thus represent conservative estimates. Overall, in all analyzed incubations from both lakes total Crenothix biovolumes increased over time (Supplementary Figure 4b). This confirms that Crenothrix was growing under both oxic and anoxic conditions.
Whereas unicellular gamma-MOB had consistently cell sizes of ca. 2 μm, the individual cells in Crenothrix-like filaments reached an average length of ca. 5 μm (Figure 1; Supplementary Figures 3a and 5a). The average length and width of Lake Rotsee Crenothrix filaments was ca. 45 and ca. 1.5 μm, respectively, with individual filaments reaching >100 μm length (Supplementary Figure 3). Filaments were often intertwined and bunched together, as observed previously (Cohn, 1870; Völker et al., 1977). In Lake Rotsee, the biovolume of Crenothrix was about eight-fold higher than that of unicellular gamma-MOB at depths corresponding to the highest observed methane oxidation rates (in 2012 and 2013; Supplementary Figure 4a). Only in 2014 unicellular gamma-MOB biomass contribution was higher than that of Crenothrix (Supplementary Figure 4). We speculate that these differences might be connected to the complex life cycle of Crenothrix (Supplementary Discussion). In Lake Zug, the filaments were shorter but more consistent in terms of length, reaching an average length and width of ca. 28 and 1.4 μm (in 2013) and ca. 20 and ca. 1.4 μm (in 2014), respectively.
Methanotrophic growth of Crenothrix
To confirm that the observed cell growth (that is, increase in cell numbers and biovolume over time; Supplementary Figure 4b) was methane-derived, samples from the 13CH4-supplemented incubations were further analyzed by nanoSIMS. Filamentous bacteria hybridized with the Mgamma669 probe consistently constituted the highest 13C-enriched population in all three investigated incubations (Lake Rotsee oxic, Lake Zug oxic and Lake Zug anoxic; Figure 1; Supplementary Figure 5). The 13C enrichment confirmed that 13CH4 was assimilated into cell biomass, such as is common for gamma-proteobacterial methanotrophs (Trotsenko and Murrell, 2008). In some of the images, fragmentation of filaments into single vegetative cells was apparent, even though the uptake of 13C appeared homogenously spread throughout the whole filament. In both lakes, Crenothrix filaments appeared to be colonized by other non-identified bacteria, which did not show comparably strong enrichment in 13C and might thus represent heterotrophic epibionts (Figure 1). In contrast, the single round cells (hybridized with Mgamma669 probe) were similarly enriched in 13C as the Crenothrix filaments (Figures 1a–c), supporting the speculation that these cells belong to methanotrophic bacteria and might potentially represent reproductive Crenothrix cells.
In the Lake Rotsee oxic incubation, the uptake of methane-derived carbon by Crenothrix filaments was comparable to that of ‘classical’ unicellular gamma-MOB (13C enrichment of 22±4.8 at % and 29±4.1 at %, respectively; Table 1; Figure 1; Supplementary Figure 2). However, due to its larger biovolume Crenothrix assimilated ca. 4–6-fold more methane than the ‘classical’ gamma-MOB in the same incubation (1.73 or 1.18 μmol methane l−1 d−1 and 0.27 μmol methane l−1 d−1, respectively; Table 1). These numbers are based on average filament biovolumes and cell counts determined by CARD-FISH at the beginning of the incubation and do not take into account any increase in cell numbers over time, as the incubation conditions might have differently affected the growth of the different MOB. However, even if we take into account the increase of cell numbers over time, overall contribution of Crenothrix to methane uptake in Lake Rotsee was still higher than that of the unicellular gamma-MOB, even though the difference was not so pronounced (ca. 1.4 higher based on Tend cell counts).
Crenothrix filaments in Lake Zug oxic incubations were also active and assimilated methane at rates of ca. 0.04 μmol methane l−1 d−1 (Table 1; Figures 1d–f). This is much lower than the overall contribution of Crenothrix in Lake Rotsee, which is largely due to their lower abundance (1.1E+03 cells per ml) and smaller average biovolume (ca. 30 μm3).
Additionally, Crenothrix was also active in our anoxic denitrifying incubations where not enough oxygen was present to account for measured methane oxidation rates (2.7 μmol l−1 d−1 13CO2 produced in 15NO3-supplemented incubation, a ca. 10-fold increase compared to control incubation without any added electron acceptor (0.234 μmol l−1 d−1 13CO2 produced)). The methane-dependent growth under oxygen-deficient conditions was evidenced as cell biomass enrichment in both 13C (from 13C-CH4; Figures 1g–i) and 15N (from 15N-nitrate; Supplementary Figure 6), even though the methane uptake rates were somewhat lower (0.03 μmol methane l−1 d−1) than those in incubations supplemented with oxygen (Table 1).
Metagenomic analyses of Lake Rotsee and Lake Zug
Due to the strong dominance of eukaryotic sequences in Lake Rotsee, we were not able to assemble a genomic bin of Crenothrix from any of the sequenced samples (Supplementary Table 1a).
On the other hand, in the Lake Zug metagenomes eukaryotic sequences were almost completely absent and the relative abundance of Crenothrix-related sequences was considerably higher (Supplementary Table 1a). Therefore, a metagenome from a Lake Zug anoxic incubation (sample Z3, Supplementary Table 4) was used for the assembly of a Crenothrix genome.
The Crenothrix D3 draft genome was binned by exploiting the differential coverage of contigs in metagenomes obtained from the in situ metagenome of Lake Zug and two different incubations (an oxygen-supplemented and an anoxic, nitrate-supplemented; Supplementary Figure 7a; see also Materials and Methods section and Supplementary Table 4 for sample details). We retrieved several bins representing gamma-MOB from the Lake Zug assembly (data not shown). The metagenomic sequences within these two bins were also present in our Lake Rotsee metagenomes, as indicated by their respective coverage (Supplementary Figure 7b). 16S rRNA gene retrieved from one of these bins putatively belonged to a Methylobacter (Figure 2a). The other bin contained a partial 16S rRNA gene (909 bp) that clustered closely with C. polyspora (Figure 2a), even though the level of similarity (95% identity) suggests that the Lake Zug Crenothrix is a different species. Most closely related environmental sequences were retrieved from groundwater and habitats highlighted primarily for iron richness (Bruun et al., 2010), yet apparently containing methane (Kojima et al., 2009; Kato et al., 2013).
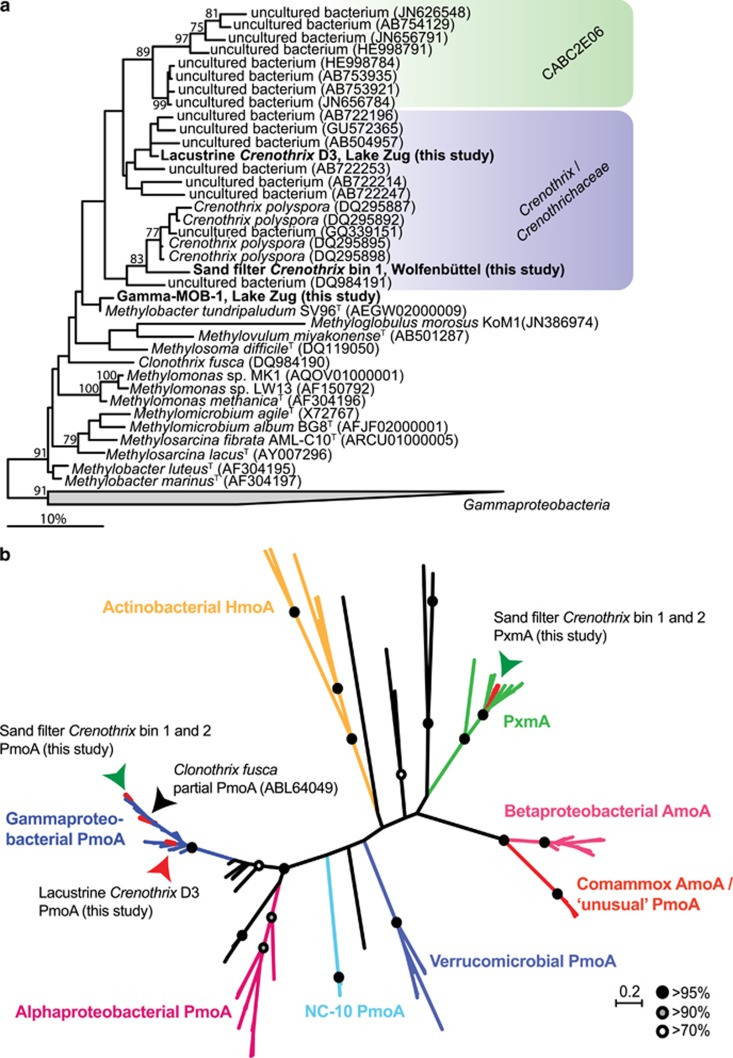
Figure 2.
Phylogenetic tree of Crenothrix 16S rRNA gene and PmoA amino-acid sequences retrieved from Lake Zug and sand filters of the Wolfenbüttel waterworks. (a) Phylogenetic tree of partial 16S rRNA gene sequence retrieved from the lacustrine Crenothrix (909 bp) and from one sand filter Crenothrix (817 bp, bin 1) draft genomes. Note that the 16S rRNA gene sequence of Lake Zug ‘lacustrine’ Crenothrix (but not of the sand filter Crenothrix) is monophyletic with clade CABC2E06. The tree was calculated with the RAxML maximum likelihood program implemented in the ARB package without constraining the alignment by a filter or weighting mask. Bootstrap values >70 (out of 100 resamplings) are shown in front of each node. The taxonomic affiliations indicated by the colored boxes are based on the SILVA SSU reference database (release 123; (Pruesse et al., 2007)). Fourteen type strains spread among gamma-proteobacteria were used as an outgroup. Nucleotide accession numbers are listed in brackets. The bar shows an estimated nucleotide sequence divergence of 10%. (b) Maximum likelihood phylogenetic tree of bacterial PmoA/AmoA amino-acid sequences (135 taxa) showing affiliation of PmoA sequences recovered from the Lake Zug Crenothrix bin (red arrow) as well as of the two sand filter Crenothrix genome bins (green arrows). All three Crenothrix PmoA sequences clustered within the ‘classical’ gamma-proteobacterial PmoA branch. Bootstrap support of total 100 bootstraps are shown in black (>95%), gray (>90%) and white (>70%) circles. Scale bar indicates substitutions per site.
Retrieval of the Crenothrix D3 16S rRNA gene sequence from the Lake Zug metagenome allowed us to also investigate the reasons behind the poor performance of the Creno445 probe. The comparison of the probe binding region on the 16S rRNA gene sequence revealed that the Creno445 FISH probe (length: 18 nt) had five mismatches with the partial 16S rRNA gene from our metagenomic Crenothrix D3 bin (Supplementary Table 2). Interestingly, out of 47 16S rRNA gene sequences in the SILVA database (NR99, release 123) that were assigned to Crenothrix/Crenothrichaceae, only seven sequences (including four C. polyspora sequences published by Stoecker et al. (2006)) contained less than five mismatches. Thus it seems that while the Creno445 probe is very specific to C. polyspora, it might not be suitable for environmental detection of other Crenothrix strains and species. In comparison, the lacustrine Crenothrix 16S rRNA gene had only a single mismatch with the Mgamma669 probe, which explains the comparably better performance of this (not Crenothrix-specific) probe on our samples.
Interestingly, the clade CABC2E06, which forms an apparent sister group to Crenothrix based on the 16S rRNA tree (Figure 2a), had an identical number of mismatches to both probes. As the 16S rRNA gene sequences assigned to this group were retrieved from both Lake Rotsee (Supplementary Figure 2) and Lake Zug (data not shown), it is feasible that the CABC2E06 bacteria in these samples were also hybridized by the Mgamma669 probe. Additionally, if the CABC2E06 bacteria were filamentous, they may have been included in the here-reported cell and biovolume counts.
Genome-inferred C1 metabolism of lacustrine Crenothrix D3
In the Crenothrix D3 draft genome from Lake Zug (Supplementary Table 1b), we searched for pMMO genes. We found all genes encoding for pMMO, which were organized in the arrangement pmoCAB, such as is common for gamma-proteobacterial type I MOB (Trotsenko and Murrell, 2008). The phylogenetic analysis of the PmoA amino-acid sequence showed that the sequence fell within the PmoA group of other known gamma-MOB, including the PmoA sequence of the other described filamentous methane oxidizer, C. fusca (Figure 2b). However, the presence of conventional gamma-proteobacterial pmoA in the lacustrine Crenothrix strain was inconsistent with the findings of ‘unusual’ pmoA previously reported for C. polyspora based on PCR and quantitative PCR (Stoecker et al., 2006). Our Crenothrix D3 draft genome did not contain any ‘unusual’ pmoA; in fact, no ‘unusual’ pmoA or amoA has been retrieved in any of the other gamma-MOB-assigned bins either.
We thus decided to address this discrepancy by obtaining metagenomic data from the original samples used in the Stoecker et al. (2006) study. Two samples obtained in 2004 from the rapid sand filters of the Wolfenbüttel waterworks (Germany) were analyzed and, after differential coverage binning, genomic information of two Crenothrix strains was obtained (Supplementary Figure 8). A partial 16S rRNA sequence retrieved from one sand filter Crenothrix bin (bin 1; 817 bp) was 98% identical to the C. polyspora 16S rRNA sequence. As the sample reportedly contained high proportions of C. polyspora, it is feasible that (at least one of) the sand filter Crenothrix was in fact C. polyspora. However, throughout this manuscript we refer to these organisms as sand filter Crenothrix, without a species name. The sand filter and the lacustrine Crenothrix likely represented different species as indicated by the average sequence identities of their shared genes (Supplementary Discussion).
Both genomes of the sand filter Crenothrix species contained a pmoCAB operon (gene similarities between both bins 96–99%) and a pxmABC operon (gene similarities between both bins 93–99%). PmoA encoded by the genes from the pmoCAB operon clustered together with other gamma-proteobacterial PmoA sequences (Figure 2b) and the affiliation of the pxmABC operon with the sequence-divergent pxm cluster was confirmed by a phylogenetic analysis of pxmA (Tavormina et al., 2011; Figure 2b). PxmA has been suggested to play a role in methane oxidation under hypoxic and denitrifying conditions by Methylomonas denitrificans and Methylomicrobium album (Kits et al., 2015a, 2015b). It thus appears that Crenothrix might be another denitrifying methanotroph containing both pmoCAB and pxmABC operons. Importantly, no ‘unusual’ pmoA could be detected in the sand filter Crenothrix bins. However, the ‘unusual’ pmoA sequence previously assigned to C. polyspora was detected in a different bin, clearly belonging to the completely nitrifying Nitrospira, apparently co-occurring with C. polyspora in the sample (Daims et al., 2015; van Kessel et al., 2015; Pinto et al., 2016). This finding is discussed in more detail in the Supplementary Discussion. It is interesting to note that whereas all three Crenothrix PmoA sequences fell within the ‘classical’ gamma-proteobacterial PmoA branch, the lacustrine Crenothrix PmoA clustered separate from the sand filter Crenothrix bins 1 and 2 and C. fusca (Figure 2b). Comparison of the 16S rRNA gene and PmoA amino-acid trees suggested that the PmoA of the lacustrine Crenothrix might have been obtained laterally from another gamma-proteobacterial methanotroph. This is supported by the fact that transposase genes were located immediately up- and downstream of the lacustrine Crenothrix pmoCAB operon on the respective contig (data not shown).
In addition to the gene cluster encoding for pMMO, we also retrieved a full gene cluster for soluble methane monooxygenase (sMMO; smmoXYBZDC) in the lacustrine Crenothrix and in one sand filter Crenothrix bin. This enzyme is relatively rare in gamma-proteobacterial methanotrophs (Murrell, 2010) and was not found in C. polyspora previously (Stoecker et al., 2006), presumably due to the mismatches between the applied PCR primers and the respective target regions in the mmoX gene. We cannot conclusively prove involvement of sMMO in methane oxidation by Crenothrix; however, as the substrate range of sMMO seems much broader than that of pMMO (Dalton, 2005; Semrau et al., 2011), it is feasible that Crenothrix might have the capacity to utilize other C-compounds as suggested previously (Stoecker et al., 2006). This could explain the reported occurrence of Crenothrix in, for example, natural bitumen deposits (Saidi-Mehrabad et al., 2013). All three retrieved genomes (two sand filter Crenothrix genomes as well as the lacustrine Crenothrix D3 genome) further contained all necessary genes for complete oxidation of methane to CO2 (Supplementary Discussion; Figure 3).
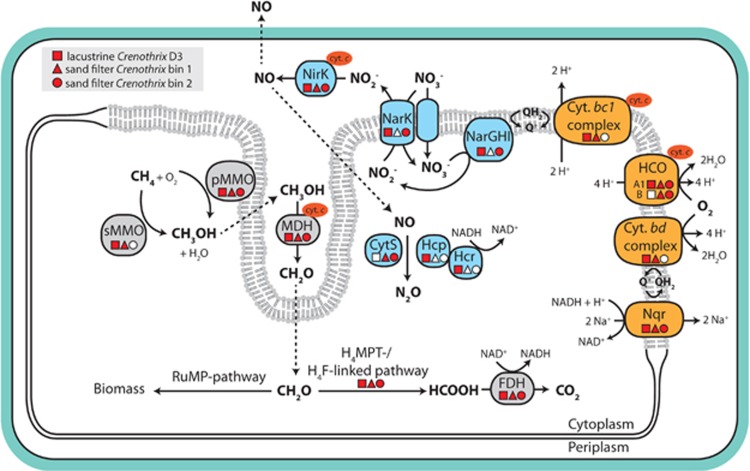
Figure 3.
Genome-inferred metabolic potential of Crenothrix for respiration and methane oxidation. Predicted metabolic potential of the lacustrine Crenothrix as well as of the two sand filter Crenothrix species with respect to its CH4 and N metabolism inferred from the three draft genomes. Indicated are the methane oxidation pathway (gray boxes), the aerobic respiratory chain (orange boxes) and the pathway for nitrate respiration (blue boxes). Genes that were found in the respective Crenothrix genomes (square: lacustrine Crenothrix D3; triangle: sand filter Crenothrix bin 1; circle: sand filter Crenothrix bin 2) are depicted in red, not found in white. Cyt. bc1 complex, cytochrome bc1 complex; Cyt. bd complex, cytochrome bd complex (cydABCD); cyt c., cytochrome c; CytS, cytochrome c’-beta; FDH, formate dehydrogenase; H4F, tetrahydrofolate; H4MPT, tetrahydromethanopterin; HCO, heme copper oxygen reductase (COXI-III); Hcp, hybrid cluster protein; Hcr, NADH-dependent Hcp reductase; MDH, methanol dehydrogenase (xoxF); Nar, nitrate reductase (narGHI); NarK, nitrate/nitrite antiporter (narK); NirS, copper-containing nitrite reductase (nirS); Nqr, sodium-translocating NADH:quinone oxidoreductase; pMMO, particulate methane monooxygenase (pmoCAB); Q, ubiquinone; RuMP, ribulose monophosphate; sMMO, soluble methane monooxygenase (smmoXYBZDC).
Like many other type I methanotrophs (Chistoserdova and Lidstrom, 2013), Crenothrix might use the RuMP pathway for C1 assimilation from formaldehyde, as genes for all necessary enzymes were found in all three draft genomes (Figure 3). On the other hand, the serine cycle apparently missed genes encoding for hydroxypyruvate reductase and malate thiokinase. Crenothrix had the genomic potential for mixed acid fermentation to succinate and potentially acetate (gene encoding for phosphate acetyltransferase was missing in lacustrine Crenothrix D3 genome and one sand filter Crenothrix, but was putatively present in the other sand filter Crenothrix bin) and hydrogen production (via NAD-reducing hydrogenase, hoxFUYH; only present in the lacustrine Crenothrix). Pyruvate, which serves as the starting point for fermentation, could be generated from formaldehyde via enzymes of the RuMP and pyrophosphate-mediated glycolytic pathway that was encoded in all three Crenothrix genomes. Mixed acid fermentation and H2 production via these pathways has been shown to be a major route of methane-derived carbon respiration in methanotrophs growing under oxygen limitation (Kalyuzhnaya et al., 2013).
Aerobic and anaerobic respiration by Crenothrix
In agreement with the demonstrated cell growth and activity in our oxic incubations, all three Crenothrix genomes encoded a multitude of aerobic respiratory chain complexes, such as a sodium-pumping NADH:ubiquinone oxidoreductase (Na+-NQR), the M and L subunits of the NADH:quinone oxidoreductase, the bc1 complex, an A1-type heme copper cytochrome c oxidase, a type B heme copper cytochrome c oxidase (only the sand filter Crenothrix) and a cytochrome bd oxidase that might potentially act as a high-affinity terminal oxidase (Figure 3).
Additionally, the draft genome of the lacustrine Crenothrix D3 as well as one of the sand filter Crenothrix strains also contained a partial pathway for the respiration of nitrate. We retrieved genes encoding for a membrane-bound respiratory nitrate reductase (narGHI), a nitrite/nitrate antiporter (narK) as well as a periplasmic multi-copper nitrite reductase (nirK). Genes encoding for nitric oxide (NO) and nitrous oxide (N2O) reductases (norBC and q-type nor, and nosZ, respectively) were not found in any of the three bins. Yet, interestingly, all three Crenothrix genomes encoded proteins for alternative pathways of NO detoxification to N2O. In the genome of Crenothrix D3, a gene cluster containing hcp and hcr genes was found. The hcp gene encodes for a unique hybrid cluster protein (Hcp), which has recently been shown to act as a high-affinity NO reductase in Escherichia coli, producing N2O as the end product (Wang et al., 2016). The Hcp sequence retrieved from the Crenothrix D3 genome contained the six highly conserved residues involved in 4Fe-2S-2O cluster coordination (Aragao et al., 2008) as well as a glutamic acid residue (E492 of E. coli Hcp) essential for NO reductase activity (Wang et al., 2016). Overall, the Crenothrix D3 Hcp shared 49% amino-acid identity with the NO-reducing Hcp of E. coli. The hcr gene, located immediately downstream from hcp, encodes for the Hcr protein and acts as a NADH-dependent Hcp reductase (van den Berg et al., 2000), while simultaneously protecting Hcp from nitrosylation by its substrate, NO (Wang et al., 2016). The hcp/hcr genes in Crenothrix D3 genome were preceded by norR, a transcriptional regulator of three different enzymes (NO reductase, flavorubredoxin and flavohaemoglobin) that all utilize NO as a substrate (Rodionov et al., 2005). We thus speculate that, despite being routinely annotated as a hydroxylamine reductase, the Hcp/Hcr system in Crenothrix could in fact act as a NO reductase and substitute Nor-type NO reductases under denitrifying conditions. In the two sand filter Crenothrix genome bins no homologs of Hcp were found. However, both bins (but not the lacustrine Crenothrix genome bin) contained a homolog of cytochrome c'-beta, a member of the cytochrome P460 family found in, for example, gamma-proteobacterial methane oxidizers (Zahn et al., 1996; Campbell et al., 2011) and gamma- and beta-proteobacterial ammonia oxidizers (Bergmann and Hooper, 2003; Klotz et al., 2006). Cytochrome c'-beta can reduce NO to N2O (Elmore et al., 2007). Interestingly, in one of the bins this gene (cytS) was located directly downstream of the haoA and haoB genes encoding for hydroxylamine dehydrogenase. As both the Hcp and the cytochrome c’-beta are predicted to be cytoplasmic proteins and NO is produced in the periplasm (by NirK), it is feasible that their activities are not coupled and (some) NO might escape out of the cell (Figure 3).
The experimentally demonstrated and genome analysis-supported metabolic potential for methane-dependent growth under nitrate-reducing conditions cannot serve as a final proof of nitrate reduction by Crenothrix in Lake Zug. However, it is interesting to speculate that such metabolic versatility might expand the habitat of these facultative anaerobic bacteria, potentially enabling them to survive periods of oxygen starvation by switching to using nitrate as an electron acceptor for methane oxidation. Denitrification is an emerging feature of gamma-MOB, which has been supported by genomics and was also experimentally demonstrated (Hoefman et al., 2014; Kalyuzhnaya et al., 2015; Skennerton et al., 2015; Kits et al., 2015a, 2015b). It has been proposed that respiration of nitrate might enable aerobic gamma-MOB to colonize anoxic waters (Chistoserdova, 2015; Knief, 2015). In Lake Rotsee and Lake Zug, Crenothrix was indeed found in the anoxic waters below the oxycline in at least 2 consecutive years. Its abundance in anoxic lake waters suggests that it might successfully compete with more obligate anaerobic methane oxidizers, such as archaeal methanotrophs (Haroon et al., 2013) or ‘Candidatus Methylomirabilis oxyfera’ (Ettwig et al., 2010).
Conclusions
Members of the genus Crenothrix are rare methane oxidizers, which are not available in pure or enrichment cultures and will not be readily picked up in environmental samples by the currently available specific FISH probe (Creno445). The ambiguity surrounding their pmoA has further complicated the in situ detection using molecular methods. In the past, this has hampered our understanding of these peculiar organisms and possibly led us to underestimate their role in the biogeochemical nutrient and element cycles.
In our study, we could unambiguously demonstrate a key role for these organisms in the mitigation of methane emissions from two stratified lakes. In Lake Rotsee, Crenothrix even contributed more to methane uptake than the ‘classical’ unicellular gamma-MOB. In up to 3 consecutive years Crenothrix was recurrently found throughout the stratification period of Lake Rotsee and Lake Zug, and thus appears to be a stable part of the indigenous microbial community. Our data are also the first to demonstrate that Crenothrix is capable of growing as a planktonic species in the lake water column. Given the capacity of Crenothrix to rapidly grow up into large biomass, its participation in methane cycling also in other relevant habitats should be considered.
Acknowledgments
We would like to thank Christian Dinkel, Carole Guggenheim and Jasmine Berg for support during field campaigns and Bernd Bendinger for providing the sand filter Crenothrix samples. We thank Philipp Hach, Hannah Marchant, Bernhard Fuchs, Gaute Lavik, Michaela Steinberger and Petra Pjevac for technical support, Craig Herbold for ANI and AAI analyses, Laura Bristow for critical reading of the manuscript, and Boran Kartal for his input regarding the Hcp. This study was supported by the Max-Planck Society, the ETH, the Eawag and the Swiss National Science Foundation (SNF grant no. 135299, 153091 & 128707). MW was supported by an ERC Advanced Grant (NITRICARE, 294343).
Author contributions
JM, MMMK and CJS designed research, KO and AB performed field measurements, incubation experiments and sampling, JSG performed metagenomic analyses on lake water samples and phylogenetic analyses; HD, MW and MA performed metagenomic analysis on the sand filter Crenothrix sample; SL and DT performed FISH and nanoSIMS measurements, BW, MMMK and CJS contributed material and analysis tools; KO, JSG and JM wrote the paper with input from all co-authors.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Aragao D, Mitchell EP, Frazao CF, Carrondo MA, Lindley PF. (2008). Structural and functional relationships in the hybrid cluster protein family: structure of the anaerobically purified hybrid cluster protein from Desulfovibrio vulgaris at 1.35 A resolution. Acta Crystallogr D 64: 665–674. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastviken D, Cole J, Pace M, Tranvik L. (2004). Methane emissions from lakes: dependence of lake characteristics, two regional assessments, and a global estimate. Global Biogeochem Cyc 18: 1–12. [Google Scholar]
- Bergmann DJ, Hooper AB. (2003). Cytochrome P460 of Nitrosomonas europaea. Eur J Biochem 270: 1935–1941. [DOI] [PubMed] [Google Scholar]
- Bodelier PLE, Meima-Franke M, Hordijk CA, Steenbergh AK, Hefting MM, Bodrossy L et al. (2013). Microbial minorities modulate methane consumption through niche partitioning. ISME J 7: 2214–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, Pel R et al. (1998). Direct linking of microbial populations to specific biogeochemical processes by C-13-labelling of biomarkers. Nature 392: 801–805. [Google Scholar]
- Bowman JP. (2005). Order VII. Methylococcales ord. nov. In: Garrity G, Brenner DJ, Krieg NR, Staley JR (eds). Bergey's Manual of Systematic Bacteriology, 2nd edn. Springer: New York, USA, pp 248–270..
- Bruun AM, Finster K, Gunnlaugsson HP, Nornberg P, Friedrich MW. (2010). A comprehensive investigation on iron cycling in a freshwater seep including microscopy, cultivation and molecular community analysis. Geomicrobiol J 27: 15–34. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Nyerges G, Kozlowski JA, Poret-Peterson AT, Stein LY, Klotz MG. (2011). Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. Fems Microbiol Lett 322: 82. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Lidstrom ME. (2013) Aerobic methylotrophic prokaryotes. In: Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E (eds), The Prokaryotes 4th edn Springer: Berlin, Heidelberg, pp 267–285.
- Chistoserdova L. (2015). Methylotrophs in natural habitats: current insights through metagenomics. Appl Microbiol Biot 99: 5763–5779. [DOI] [PubMed] [Google Scholar]
- Cohn F. (1870). Über den Brunnenfaden (Crenothrix polyspora mit Bemerkungen über die mikroskopische Analyse des Brunnenwassers. Beitr Biol Pflanzen 1: 108–131. [Google Scholar]
- Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528: 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton H. (2005). The Leeuwenhoek Lecture 2000 - the natural and unnatural history of methane-oxidizing bacteria. Philos Trans R Soc Lond B Biol Sci 360: 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr N, Glaser B, Kolb S. (2010). Methanotrophic communities in Brazilian ferralsols from naturally forested, afforested, and agricultural sites. Appl Environ Microb 76: 1307–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewniak L, Maryan N, Lewandowski W, Kaczanowski S, Sklodowska A. (2012). The contribution of microbial mats to the arsenic geochemistry of an ancient gold mine. Environ Pollut 162: 190–201. [DOI] [PubMed] [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR, Wheeler TJ HMMER development team. (2013). HMMER 3.1b1 (May 2013). Available at: http://hmmer.org.
- Eller G, Stubner S, Frenzel P. (2001). Group-specific 16S rRNA targeted probes for the detection of type I and type II methanotrophs by fluorescence in situ hybridisation. Fems Microbiol Lett 198: 91–97. [DOI] [PubMed] [Google Scholar]
- Elmore BO, Bergmann DJ, Klotz MG, Hooper AB. (2007). Cytochromes P460 and c′-beta: a new family of high-spin cytochromes c. FEBS Lett 581: 911–916. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548. [DOI] [PubMed] [Google Scholar]
- Haroon MF, Hu SH, Shi Y, Imelfort M, Keller J, Hugenholtz P et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500: 567–570. [DOI] [PubMed] [Google Scholar]
- Hoefman S, van der Ha D, Boon N, Vandamme P, De Vos P, Heylen K. (2014). Niche differentiation in nitrogen metabolism among methanotrophs within an operational taxonomic unit. BMC Microbiol 14: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DD. (1902). A new species of CrenothrixC. manganifera. Trans Am Microsc Soc 23: 31–39. [Google Scholar]
- Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A et al. (2013). Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4: 2785. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lamb AE, McTaggart TL, Oshkin IY, Shapiro N, Woyke T et al. (2015). Draft genome sequences of gammaproteobacterial methanotrophs isolated from Lake Washington sediment. Genome Announc 3: e00103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Kirkegaard RH, Albertsen M. (2016). mmgenome: a toolbox for reproducible genome extraction from metagenomes. bioRxiv: 059121.
- Kato S, Chan C, Itoh T, Ohkuma M. (2013). Functional gene analysis of freshwater iron-rich flocs at circumneutral pH and isolation of a stalk-forming microaerophilic iron-oxidizing bacterium. Appl Environ Microb 79: 5283–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirf MK, Dinkel C, Schubert CJ, Wehrli B. (2014). Submicromolar oxygen profiles at the oxic–anoxic boundary of temperate lakes. Aquat Geochem 20: 39–57. [Google Scholar]
- Kits KD, Campbell DJ, Rosana AR, Stein LY. (2015. a). Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Front Microbiol 6: 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kits KD, Klotz MG, Stein LY. (2015. b). Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp nov type strain FJG1. Environ Microbiol 17: 3219–3232. [DOI] [PubMed] [Google Scholar]
- Klotz MG, Arp DJ, Chain PSG, El-Sheikh AF, Hauser LJ, Hommes NG et al. (2006). Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl Environ Microb 72: 6299–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Fukuhara H, Fukui M. (2009). Community structure of microorganisms associated with reddish-brown iron-rich snow. Syst Appl Microbiol 32: 429–437. [DOI] [PubMed] [Google Scholar]
- Kolk LA. (1938). A comparison of the filamentous iron organisms, Clonothrix fusca Roze and Crenothrix polyspora Cohn. Am J Bot 25: 11–17. [Google Scholar]
- Lagesen K, Hallin P, Rødland E, Stærfeldt H, Rognes T, Ussery D. (2007). RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. (2009). IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25: 2271–2278. [DOI] [PubMed] [Google Scholar]
- Milucka J, Kirf M, Lu L, Krupke A, Lam P, Littmann S et al. (2015). Methane oxidation coupled to oxygenic photosynthesis in anoxic waters. ISME J 9: 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molisch H. (1910). Die Eisenbakterien: Gustav Fischer, Jena. Available at: http://library.wur.nl/WebQuery/clc/speccol/181818.
- Murrell JC. (2010). The aerobic methane oxidizing bacteria (methanotrophs). In: Timmis KN (ed). Handbook of Hydrocarbon and Lipid Microbiology. Springer Berlin Heidelberg: Berlin, Heidelberg, Germany, pp 1953–1966..
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105: 17861–17866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshkin IY, Beck DAC, Lamb AE, Tchesnokova V, Benuska G, McTaggart TL et al. (2015). Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J 9: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald K, Milucka J, Brand A, Littmann S, Wehrli B, Kuypers MMM et al. (2015). Light-dependent aerobic methane oxidation reduces methane emissions from seasonally stratified lakes. PLoS ONE 10: e0132574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald K, Milucka J, Brand A, Hach P, Littmann S, Wehrli B et al. (2016). Aerobic gammaproteobacterial methanotrophs mitigate methane emissions from oxic and anoxic lake waters. Limnol Oceanogr 61: S101–S118. [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R. (2002). Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microb 68: 3094–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AJ, Marcus DN, Ijaz UZ, Bautista-de lose Santos QM, Dick GJ, Raskin L. (2016). Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere 1: pii: e00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM. (2012). Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol 14: 1009–1023. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Bodi X, Dufresne A, Naquin D, Francez AJ, Dheilly A et al. (2014). Unraveling the stratification of an iron-oxidizing microbial mat by metatranscriptomics. PLoS ONE 9: e102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. (2005). Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol 1: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze E. (1896). Le Clonothrix, un nouveau type generique de Cyanophycees. J Bot 10: 325–330. [Google Scholar]
- Saidi-Mehrabad A, He Z, Tamas I, Sharp CE, Brady AL, Rochman FF et al. (2013). Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J 7: 908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Semrau JD, DiSpirito AA, Vuilleumier S. (2011). Facultative methanotrophy: false leads, true results, and suggestions for future research. Fems Microbiol Lett 323: 1–12. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skennerton CT, Ward LM, Michel A, Metcalfe K, Valiente C, Mullin S et al. (2015). Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (family Methylothermaceae) indicates multiple adaptations to oxygen limitation. Front Microbiol 6: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecker K, Bendinger B, Schoning B, Nielsen PH, Nielsen JL, Baranyi C et al. (2006). Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci USA 103: 2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MSM, Klotz MG. (2011). A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep 3: 91–100. [DOI] [PubMed] [Google Scholar]
- Trotsenko YA, Murrell JC. (2008). Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol 63: 183–229. [DOI] [PubMed] [Google Scholar]
- van den Berg WAM, Hagen WR, van Dongen WMAM. (2000). The hybrid-cluster protein (‘prismane protein’) from Escherichia coli. Eur J Biochem 267: 666–676. [DOI] [PubMed] [Google Scholar]
- van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B et al. (2015). Complete nitrification by a single microorganism. Nature 528: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigliotta G, Nutricati E, Carata E, Tredici SM, De Stefano M, Pontieri P et al. (2007). Clonothrix fusca Roze 1896, a filamentous, sheathed, methanotrophic gamma-proteobacterium. Appl Environ Microbiol 73: 3556–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker H, Schweisfurth R, Hirsch P. (1977). Morphology and ultrastructure of Crenothrix polyspora Cohn. J Bacteriol 131: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vine CE, Balasiny BK, Rizk J, Bradley CL, Tinajero-Trejo M et al. (2016). The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Mol Microbiol 100: 877–892. [DOI] [PubMed] [Google Scholar]
- Wang JJ, Krause S, Muyzer G, Meima-Franke M, Laanbroek HJ, Bodelier PLE. (2012). Spatial patterns of iron- and methane-oxidizing bacterial communities in an irregularly flooded, riparian wetland. Front Microbiol 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenburg DA, Guinasso NL Jr. (1979). Equilibrium solubilities of methane, carbon monoxide, and hydrogen in water and sea water. Journal of Chemical and Engineering Data 24: 356–360. [Google Scholar]
- Wolfe RS. (1960). Observations and studies of Crenothrix polyspora. J Am Water Works Assoc 52: 915–918. [Google Scholar]
- Zahn JA, Arciero DM, Hooper AB, Dispirito AA. (1996). Cytochrome c′ of Methylococcus capsulatus bath. Eur J Biochem 240: 684–691. [DOI] [PubMed] [Google Scholar]
- Zhou JZ, Bruns MA, Tiedje JM. (1996). DNA recovery from soils of diverse composition. Appl Environ Microb 62: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.