Abstract
Introduction
Evolutionarily selected over billions of years for their interactions with biomolecules, natural products have been and continue to be a major source of pharmaceuticals. In the 1990s, pharmaceutical companies scaled down their natural product discovery programs in favor of synthetic chemical libraries due to major challenges such as high rediscovery rates, challenging isolation, and low production titers. Propelled by advances in DNA sequencing and synthetic biology technologies, insights into microbial secondary metabolism provided have inspired a number of strategies to address these challenges.
Areas covered
This review highlights the importance of genomics and metagenomics in natural product discovery, and provides an overview of the technical and conceptual advances that offer unprecedented access to molecules encoded by biosynthetic gene clusters.
Expert opinion
Genomics and metagenomics revealed nature’s remarkable biosynthetic potential and her vast chemical inventory that we can now prioritize and systematically mine for novel chemical scaffolds with desirable bioactivities. Coupled with synthetic biology and genome engineering technologies, significant progress has been made in identifying and predicting the chemical output of biosynthetic gene clusters, as well as in optimizing cluster expression in native and heterologous host systems for the production of pharmaceutically relevant metabolites and their derivatives.
Keywords: Genome mining, biosynthetic gene clusters, secondary metabolites, metagenomics, bioinformatics
1. Introduction
Humans have long recognized the rich repertoire of molecules produced by microorganisms as a fertile source of therapeutics. Evolutionarily selected for interactions with biomolecules, the remarkably diverse and complex core chemical scaffolds of natural products have a higher chance to be biologically active than compounds from combinatorial synthesis approaches. In fact, more than half of clinical drugs from 1981 to 2014 are derived from or inspired by natural products [1]. In the pre-genomic era, most natural product discovery efforts employed a ‘top-down’ approach driven by the screening biological samples for desirable bioactivities, followed by compound isolation and characterization [2]. Yet by the 1990s, such strategies had largely failed to uncover new natural products as pharmaceutical companies struggled with high rediscovery rates and de-emphasized their natural product discovery efforts. Driven by the rapidly decreasing cost and increasing throughput of DNA sequencing technologies [3], significant progress in genomics has renewed interest in natural product discovery [4]. Rapidly expanding microbial genomic and metagenomic datasets reveal a vast number of biosynthetic gene clusters in nature, which are predicted to encode far more natural products than what we have characterized to date [5]. Uncovering novel natural products is not only fascinating but also highly pertinent in face of today’s burgeoning drug resistance and health problems.
The conventional ‘top-down’ discovery approach can only provide us access to a small fraction of microbial biosynthetic potential given that majority of microorganisms cannot be isolated or cultured [6, 7]. Furthermore, even in culturable organisms, the encoded secondary metabolites of many biosynthetic gene clusters (BGCs) are unknown [8, 9]. This may be due to strong down-regulation of product biosynthesis at the transcriptional, translational and/or post-translational levels in the absence of the right activating cues in the laboratory. Alternatively, secondary metabolites that are produced at very low yields may escape detection and characterization. The ability to efficiently and strategically access these vast unexplored chemical resources will be invaluable to drug discovery. Besides relying on serendipitous discoveries of bioactive compounds, natural product discovery is now increasingly driven by genomics and focused on BGCs that are predicted to encode novel biomedically relevant molecules [4]. A better understanding of genotype-chemotype relationships informs researchers about the chemical logic of natural product biosynthesis and guides ‘bottom-up’ approaches that begin with genetic manipulation for the “detectable” production of target metabolites. Natural product discovery, which used to be predominantly an analytical chemistry problem, has become a challenge in genomics and molecular biology on how to manipulate relevant genes and sequences to produce the desired encoded metabolites.
In this review, we highlight key technical and conceptual advances in genomics-driven natural product discovery. These include bioinformatics-guided identification of BGCs in genomes and metagenomes, pathway and host engineering strategies for the activation of silent gene clusters in native and heterologous systems, as well as combinatorial biosynthesis for generating natural product analogs. This is not meant to be an exhaustive review and we apologize if key studies were inadvertently left out.
2. Genome-mining for natural product BGCs
The development of next-generation sequencing has significantly accelerated the sequencing of microbial genomes at much reduced costs, leading to an exponential growth of genomic sequencing data with over 30,000 sequenced bacterial genomes currently deposited in public archives [10]. However, an immediate bottleneck is our current ability to readily analyze and process such large volumes of data. For natural product drug discovery, this means to identify potential secondary metabolites BGCs that encode for novel bioactive metabolites from microbial genomes. Toward this end, a variety of genome-mining tools have been developed.
AntiSMASH (antibiotics and secondary metabolite analysis shell) allows in silico identification of BGCs in bacterial and fungal genomes [11, 12]. Studies using antiSMASH have revealed numerous BGCs in bacteria and fungi [13, 14, 15, 16], many of which encode novel bioactive metabolites. Notable features of antiSMASH version 3.0 include more comprehensive analysis of gene clusters, annotation of key residues in biosynthetic enzymes, and the inclusion of a lantipeptide-focused module [12, 17]. In addition, there are genome-mining tools specific for particular biosynthetic pathways or species. SMURF (secondary metabolite unknown region finder) focuses on genome-mining in fungi [18], while BAGEL3 is developed for the analysis of bacteriocin encoding-genes and relatively unexplored classes of posttranslationally modified peptides [19]. Multiple tools including NP.searcher and ASMPKS are available for analyzing PKS (polyketide synthase)/NRPS (non-ribosomal peptide synthetase) pathways and predicting their substrates [20, 21, 22]. Recognizing knowledge gap between isolated PKs/NRPs and their gene clusters (i.e. only 10% of PKs/NRPs are associated with gene clusters), Dejong et al. introduced a retro-biosynthetic in silico analysis platform to link known PKS and NRPS metabolites to emergent gene clusters [23]. The platform includes a retro-biosynthetic analysis component (GRAPE) and an alignment algorithm for cheminformatics (GARLIC), and is useful for identifying clusters encoding for known and novel molecules. Furthermore, a consortium effort to standardize BGC annotation yielded the Minimal Information about a Biosynthetic Gene cluster (MIBiG) format, which encourages more systematic and consistent characterization of BGCs by the community [12, 24]. Readers are referred to recent reviews for detailed discussion and comparison of genome-mining strategies focused on natural product discovery [25, 26, 27, 28].
Plant-derived natural products are also important drug sources [1], but compared to bacterial and fungal systems, genomics-driven natural product discovery in plants is in the early stages. Genome and transcriptome analyses led to the recent recognition of BGCs as common genomic features in plants and supported the regulated production of many bioactive natural products, including alkaloids and terpenoids [29, 30]. With an expanding list of plant genomes, natural product biosynthesis pathway discovery is increasingly genomics-driven compared to the pregenomic approach of starting with the biochemical characterization of a biosynthetic gene. There are already efforts to standardize the analyses and curation of plant BGCs with their microbial counterparts to support future genome mining endeavors [24, 31].
As predictive software programs become increasingly robust and streamlined for genome mining (Figure 1), researchers will not need expertise in bioinformatics to perform a variety of analyses, yet one should be mindful of the limitations. Since most tools employ predictive rules based on characterized pathways, they are geared towards identification of major families of biosynthetic gene clusters such as PKSs/NRPSs, they are less reliable with regard other pathways. Moreover, most software programs have been unable to accurately predict metabolite structures [25]. Nevertheless, BGCs identified in silico are excellent starting points for downstream experimental investigations.
Figure 1. Workflow of genome-mining for natural product drug discovery.
Genome-mining software such as antiSMASH is able to analyze the sequenced genome in silico, identify potential biosynthetic gene clusters and predict core structures of encoded metabolites. The predicted BGCs are starting points for downstream experimental activation and validation.
3. Accessing silent natural product gene clusters
Secondary metabolism is tightly regulated [32] and the fact that many BGCs remain “silent” under laboratory conditions presents a major challenge to “activate” these BGCs and assess the therapeutic potential of their encoded natural products. As metabolic profiling using advanced analytical methods continue to uncover new compounds that have eluded detection due to low production yields (Section 4.2), the activation of BGCs that are not or inadequately expressed under laboratory conditions have also resulted in the discovery of many diverse natural products exhibiting a range of bioactivities [5]. Here we highlight the major strategies to activate BGC expression with a focus on natural product discovery. Readers are referred to recent reviews for details on the metabolic, pathway and genome engineering approaches in native and heterologous host systems for secondary metabolite production [33, 34, 35].
3.1. Activation of silent BGCs in native hosts
Native hosts are endogenous producers of the natural products of interest. Logically, they possess sufficient and necessary cellular factors for metabolite biosynthesis, including relevant precursors, pathway regulators, and transporters. In principle, under the ‘right’ conditions with the appropriate biological and/or environmental cues, metabolite production can be elicited in native hosts. Some native hosts can successfully express silent gene clusters when grown under alternative conditions, while others require targeted genetic manipulations such as the introduction of heterologous promoters [36, 37] or perturbation of transcriptional and post-transcriptional regulatory mechanisms (vide infra). The development of genome editing and genome engineering technologies in natural product-relevant organisms will greatly enhance the scalability of these activation strategies in native hosts [34]. Notably, genetic manipulation can be challenging for non-model native producing hosts. While developing genetic tools for uncharacterized wild type host organisms may be tedious and time consuming, the chances of obtaining BGC-encoded secondary metabolites from native producers, which likely possess all the metabolic and biosynthetic requirements, are high.
3.1.1. Variation of growth conditions and small molecule inducers
Systematic variation of growth conditions, also known as the OSMAC (one-strain, many compounds) strategy, has traditionally been employed to explore the biosynthetic capabilities of isolated strains. For Aspergillus nidulans, simple variation of fermentation conditions such as media and culturing period can dramatically affect its metabolite profile, leading to the identification of several potential anti-cancer compounds [38, 39]. But more often than not, specific conditions are required to elicit the expression of target gene clusters in microbes. In the discovery of lugdunin, a novel peptide antibiotic produced by the human commensal Staphylococcus lugdunensis, Zipperer et al. noted that metabolite production was only elicited when S. lugdunensis was cultivated under iron-limiting conditions on solid agar [40]. Other physical triggers of metabolite production such as rare earth elements have been reported in Streptomyces spp. [41, 42]. Nonetheless, screening of different conditions in multiple strainsoften suffers from limited reproducibility and can quickly become impractical when considering a range of variables. In addition, obscure conditions cannot be predicted and require empirical studies. Notably, innovative culture strategies have been developed for natural product discovery. Designed to isolate single microbial cells in diffusion chambers that mimic the natural environment, iChip allows the recovery of up to 50% of the microorganisms in environmental samples [43]. Use of iChip led to the identification of lassomycin, a ribosomally synthesized cyclic peptide antibiotic with activity against Mycobacterium tuberculosis [44], and teixobactin, a new class of antibiotics made by a newly identified species Eleftheria terrae that shows broad-spectrum antimicrobial activity without detectable resistance [45]. In addition, a high-throughput assay to identify small molecule elicitors for targeted BGCs in Burkholderia thailandensis was recently reported [46]. Since microorganisms naturally reside in a complex environment and constantly interact with other species, co-cultivation of different microorganisms can also induce expression of silent BGCs [47, 48, 49, 50].
3.1.2. Manipulation of regulators
Genome mining studies have uncovered global and pathway-specific transcriptional regulators that can be manipulated for BGC activation. Overexpression of transcriptional activators leads to production of “hidden” metabolites [51, 52]., including the production of an unprecedented 51-membered glycosylated macrolide [53]. On the other hand, deletion of pathway-specific repressors has been demonstrated to activate a number of silent BGCs and trigger metabolite production [54, 55, 56]. Likewise, manipulation of transcription factors in plants has yielded the overproduction of desired metabolites [57]. For biosynthetic pathways without obvious regulators, global alteration of gene expression may be employed. In streptomycetes, specific mutations in RNA polymerase or ribosomal proteins can affect gene expression and production of new antibacterial compounds [58]. In addition, a reporter strain-based mutant selection strategy has been developed, enabling unbiased screening of activation conditions. Using this selection, two novel aminoglycosides were discovered in Streptomyces sp. PGA64 [59].
3.1.3. Perturbation of epigenetic control
The importance of epigenetic modifications such as DNA/histone methylation and acetylation in regulation of eukaryotic gene expression has been recognized in recent years [60]. Manipulation of fungal epigenome potentially allows BGC activation. In A. nidulans, deletion of epigenetic modification enzymes such as histone deacetylase (HDAC) or histone methyltransferase has been shown to activate several silent BGCs and dramatically change its metabolite profile [61]. In addition, chemical perturbation of the fungal epigenome and led to the discovery of new metabolites [62, 63]. However, current strategies for epigenetic modulation result in global changes, which cannot be predicted. With advances in DNA sequencing and editing technologies, the precise location and function of an epigenetic marker in a fungal genome are likely to be recognized and manipulated to enable targeted BGC activation.
3.2. Activation of silent BGCs in heterologous hosts
To access silent BGCs in genetically recalcitrant or uncultured microorganisms, heterologous expression of BGCs in a genetically tractable organism represents an attractive approach. For functional expression in heterologous hosts, target BGCs often require additional refactoring, such as replacement of the native promoters with well-characterized promoters, insertion of appropriate ribosomal binding sites or terminators. Since de novo synthesis and genetic manipulation of large gene clusters (>40 kb) can be challenging, a number of synthetic biology tools have been developed to facilitate the reconstruction of biosynthetic pathways in heterologous hosts.
3.2.1. Direct capturing and refactoring of gene clusters
Advances in recombinant DNA and synthetic biology technologies allow BGCs to be directly captured into compatible vectors and refactored for expression in heterologous hosts [33]. For example, RecET-mediated linear-linear recombination in E. coli is employed to capture large (>50 kb) megasynthases into expression vectors [64]. Similarly, yeast-based transformation-associated recombination (TAR) allowed the capture and activation of a 67 kb NRPS cluster from Saccharomonospora in Streptomyces coelicolor to produce a new antimicrobial lipopeptide, taromycin A [65]. Use of the RNA-guided Cas9 nuclease circumvents the requirement for unique restriction sites flanking the target BGCs and facilitates direct cloning of large clusters (up to 100 kb) by Gibson assembly or TAR [66, 67]. Recently, the combined use of TAR with CRISPR/Cas9 in a yeast-based promoter-engineering platform mCRISTAR enabled the efficient multiplex replacement of eight promoters to activate the tetarimycin A cluster in S. albus [68].
Besides direct capture, various methods have been developed for scarless assembly of BGCs from de novo synthesized DNA fragments and PCR products [69]. These assembly techniques should also facilitate the engineering of biosynthetic gene clusters for combinatorial biosynthesis (Section 6) as well as cluster refactoring for heterologous expression. For example, Luo et al. utilized yeast DNA assembler to simultaneously assemble and refactor a PKS-NRPS cluster from Streptomyces griseus by inserting a constitutive promoter in front of each of the six biosynthetic genes. Heterologous expression of the refactored pathway in Streptomyces lividans yielded new polycyclic tetramate macrolactams [70].
3.2.2. Optimization of heterologous hosts
When choosing a heterologous host for expression, it is important to consider whether the production host possesses the necessary metabolic precursors, enzymatic machinery and appropriate regulatory systems for target BGCs. Uncoordinated expression of biosynthetic genes may result in imbalanced metabolic flux and production of toxic intermediates. For instance, Escherichia coli, which is one of the widely used Gram-negative bacteria for heterologous expression, has been engineered to express phosphopantetheine transferases (PPTases) for PKS/NRPS activation and production of PK/NRP natural products [71]. In order to improve secondary metabolite production, it is often necessary to optimize the heterologous production hosts. Multiplexed automated genome engineering (MAGE), which enables facile introduction of genetic diversity at targeted loci during DNA replication, has been used to effectively increase lycopene production titer in E. coli [72]. The CRISPR/Cas9 system was recently reconstituted and used to delete genes and entire BGCs in multiple Streptomyces [73, 74, 75, 76], paving the way towards genome-minimized hosts for heterologous expression of biosynthetic pathways from actinomycetes. Conceivably, extension of these valuable technologies to optimize heterologous host systems will facilitate BGC characterization and natural product drug discovery.
4. Integrated strategies to access genome-encoded small molecules
4.1. Bioinformatics-guided synthetic approach
Given the fact that genetic manipulation of native and heterologous hosts can be time-consuming, a new paradigm in natural product drug discovery which bypasses the need for strain cultivation, gene cluster expression and product isolation from fermentation broths was reported recently. Based on the primary sequence of NRPS clusters in the human microbiome, Chu et al. predicted and chemically synthesized a small library of peptides for the encoded NRP and identified humimycin, a potent anti-MRSA (methicillin-resistant Staphylococcus aureus) peptide with a new mechanism of action [77]. This work represents the first example of the synthetic-bioinformatic natural product (syn-BNP) approach for drug discovery and may be especially helpful in metagenomics studies (Section 5). However, as mentioned earlier, in contrast to well-annotated information on PKS/NRPS clusters, prediction algorithms for other types of BGCs are relatively limited; in those cases, structures of the synthetic molecules can significantly deviate from the encoded natural products and hence may fail to elicit the desired bioactivity. Whether the syn-BNP approach can be generalized to most BGCs depends on future development for more accurate bioinformatics predictions.
4.2. Mass spectrometry-guided genome mining
Driven by advanced analytical methods, mass spectrometry-guided genome mining involves the iterative matching de novo MSn sequence tags to genomic features, using retro-biosynthetic logic in order to connect secondary metabolites to their BGCs [78, 79]. Matched BGC sequence information may be harnessed to further elucidate compound structures and/or to identify additional molecular features for searching. Mass spectrometry-guided genome mining has been extended to connect groups of structurally related molecules with entire BGC families [80]. Notably, the genetic tools and pathway engineering strategies described in Section 3 will be invaluable to metabolomics-guided genome mining efforts by the functional validation and characterization of BGCs and their metabolites. Furthermore, when coupled with principal-component analysis of metabolite profiles, genetic mutants can be used to identify key molecular features that correspond to a BGC of interest [37]. While metabolomics-guided genome mining has been largely employed for the discovery of peptide (ribosomal and non-ribosomal) natural products due to relatively well-characterized biosynthetic logic, the approach can be extended to other groups of secondary metabolites such as glycosylated natural products [81] and possibly polyketides.
5. Metagenomics-driven drug discovery
More than 99% of microorganisms in the environment have resisted laboratory cultivation and this uncultured microbial majority represents a vast chemical treasure trove [6, 7]. Metagenomics, which involves the direct capture and analysis of environmental DNA (eDNA), allows culture-independent and unbiased access to microbial biosynthetic potential otherwise missed by traditional methods requiring the isolation and cultivation of pure microbial cultures [82, 83]. In this section, we highlight the technical and conceptual advances in metagenomics that led to the discovery of new natural products from diverse environmental niches including soil, marine environments and the human body. Readers are referred to recent reviews for more details [28, 84].
Given that BGCs constitute only a small fraction of microbial genomes [85, 86], the ability to efficiently search metagenomic libraries for rare clones containing relevant biosynthetic genes involved in secondary metabolism and encoding for novel natural products is imperative. Screening is especially challenging for large complex metagenomes such as soil, which can contain up to 105 unique species and require multimillion-membered mega-libraries for sufficient coverage [87, 88]. Conventional function- and sequence-based methods involve screening libraries for easily observable phenotypes or the presence of target DNA sequences, respectively (Figure 2a).
Figure 2. Metagenome-driven drug discovery with an alternative targeted sequence-based pipeline.
(a) Environmental DNA (eDNA) is extracted, cloned and ligated into a shuttle vector, and then transformed into a host cell to create a metagenomic library. Conventional function- and sequence-based methods involve screening metagenomic libraries for easily observable phenotypes or the presence of target DNA sequences, respectively. Target biosynthetic pathways are identified, assembled and functionally expressed through a variety of methods to obtain the encoded natural product for characterization. (b) Alternatively, PCR is used to profile the biosynthetic pathways present in crude eDNA samples. The PCR amplicons are phylogenetically organized to predict the chemical output of the biosynthetic pathways. Samples predicted to harbor novel or target biosynthetic gene clusters are prioritized for library construction, clone recovery, and heterologous production. Reproduced and adapted from [84] with permissions from Springer.
Function-based metagenomics does not require prior knowledge of genetic information and has been successfully applied to uncover novel chemical cores and interesting biotransformations associated with their biosynthesis. Facile visual screens for pigment production or antibiosis (zone of inhibition) led to the discovery of structurally distinct antibiotics such as turbomycin [89], N-acyl amino acids derivatives [90, 91] and isocyanide-containing compounds [92, 93], which represent some of the first natural products identified from metagenomics libraries. Depending on the target compounds and library size, other functional screens include direct metabolite analysis by LC-MS methods [94, 95], reporter/biosensor-based screens for metabolite-responsive gene expression [96, 97] and enzymatic assays [98, 99]. Enzyme activities unique to secondary metabolism can be harnessed to greatly improve the efficiency of identifying eDNA clones with functional biosynthetic gene pathways for downstream phenotypic screens. Required for the posttranslational activation of NRPSs and PKSs, PPTases were harnessed in a phage display study to recover NRPS and PKS sequences from environmental samples [100]. By coupling pigment production or cellular growth to PPTase function, complementation of PPTase activity can be used to screen or select for eDNA clones that contain PKS and/or NRPS gene clusters [87, 101]. Notably, the scope of function-based screening is limited by the insert sizes of cosmid/fosmid eDNA libraries as well as the choice of library hosts, which may not support functional expression of heterologous BGCs for various reasons including codon bias, lack of biosynthetic precursors or enzymatic activity, and incompatible regulatory systems.
In contrast, sequence-based metagenomics does not require functional reconstitution of biosynthetic pathways in library hosts and allows for the discovery of products of large BGCs (e.g. multimodular PKS and/or PKS clusters) that exceed the insert size for cosmid/fosmid or bacterial artificial chromosome libraries. Conventional sequence-based screens involve PCR with degenerate primers targeting relatively uncommon biosynthetic genes to identify eDNA clones of interest (Figure 2a). In an alternative approach, barcoded PCR amplicons of common conserved biosynthetic features (e.g. adenylation, condensation, ketosynthase domains) within environmental samples are sequenced to generate sequence tags that reflect microbial genomic diversity, which can be used to prioritize metagenome mining efforts for the discovery of novel and/or biomedically relevant BGCs and metabolites. Akin to the reconstruction of bacterial phylogeny from 16S rRNA sequences, phylogenetic analysis software such as eSNaPD (environmental Surveyor of Natural Product Diversity) and NaPDos (Natural Product Domain Seeker) classify eDNA-derived gene clusters by comparing the sequence similarity of environmental sequence tags to those from characterized BGCs [102, 103]. By recovering eDNA sequences related to known BGCs, this strategy has been successfully employed to discover new bioactive glycopeptide, epoxyketone and anthracycline congeners [87, 104, 105]. New chemical functionalities can also be discovered using this method. A new subclass of natural tryptophan dimers possessing a pyrrolinium indolocarbazole core was uncovered by focusing on eDNA-derived gene clusters with sequence tags that are not closely associated with characterized tryptophan dimer BGCs [88]. In general, while targeted metagenome mining may not be suitable for the discovery of fundamentally distinct chemical entities, it can be used to survey the biosynthetic potential of environmental samples as well as identify and recover novel variants of pharmaceutically relevant BGCs for downstream analyses (Figure 2b).
With advances in next-generation sequencing, bulk eDNA can be directly sequenced and assembled. This approach has been successfully employed to uncover BGCs of interest and genomes of bacterial symbionts that produce patellazoles [106] and chemotherapeutic ET-473 [107]. Yet due to challenges in assembling short reads, which may be partially addressed by pair-end sequencing or increasing sequence coverage, shotgun metagenomics is largely limited to relatively simple metagenomes or pre-enriched (e.g. filtration, differential centrifugation) samples. Rapid progress in third-generation long-read sequencing technologies circumvent the difficulties in assembling highly repetitive genomic sequences and in the near future should enable the assembly of entire BGCs, and potentially genomes, that are represented in metagenomes [108]. Until then, short-read sequencing may be applied to survey the biosynthetic genes present in the microbiome and guide the design of degenerate PCR primers to complement and expand the scope of sequence-based metagenomics. Single-cell genome sequencing allows access to genomes that are underrepresented in metagenomic samples as well as the assembly of genomes from completely uncharacterized microorganisms [109, 110, 111]. Using a combination of deep sequencing, single cell sorting and whole genome amplification, Wilson et al. uncovered a new bacterial taxon Entotheonella that accounts for the production of almost all bioactive polyketides and polypeptides isolated from its host T. swinhoei [14]. The existence of multiple additional BGCs in Entotheonella genomes suggests that the metabolic repertoire of these widely distributed symbionts may rival that of Actinobacteria.
Efficient translation of genetic sequences into actual chemical compounds is critical to the metagenomics approach for natural product discovery. For synthetically accessible natural products whose structures can be reliably predicted from nucleic acid sequences (e.g. NRPs), chemical synthesis circumvents the need for heterologous production systems and accelerates translation of genomes and metagenomes into chemical compounds that can be screened for desired bioactivities [77]. In most cases, accessing the biosynthetic potential of uncultured microorganisms will require heterologous hosts to facilitate expression of eDNA-derived BGCs and production of the encoded metabolite. Enabled by the availability of broad host range shuttle plasmids, the use of distantly related hosts including diverse Proteobacteria [112, 113] and Streptomyces [94, 114] for functional metagenomics provides access to a wider range of bioactive compounds by increasing the likelihood of matching eDNA-derived BGCs with genetically and biochemically compatible hosts for functional expression. Sequence-based metagenomics also benefits from the use of gifted heterologous hosts with the innate ability to functionally express a variety of target biosynthetic pathways. In addition, multiplex pathway and genome engineering technologies to improve heterologous expression strains and optimize cluster expression (Section 3) promise to further expand the scope of uncultured biosynthetic potential that can be accessed and harnessed.
6. Combinatorial biosynthesis
The explosion of BGC sequence information offers critical insights into how remarkably complex natural products covering vast biologically relevant chemical structure space are assembled from a limited set of simple building blocks [115]. BGCs generally encode two groups of biosynthetic enzymes – one group generates key biosynthetic precursors and assembles the core scaffold while the other group derivatizes the scaffolds [116]. Understanding nature’s logic of encoding chemical diversity will enable rational engineering of biosynthetic pathways to obtain analogs of privileged natural product scaffolds or novel natural product-like scaffolds that may be challenging to synthesize chemically for drug discovery [116, 117].
Guided by comparative genomic studies focused on biosynthetic genes, rational engineering or directed evolution of biosynthetic enzymes responsible for generating biosynthetic precursors, assembling core chemical structures and tailoring of scaffolds to expand or alter their substrate selectivity is one of the main strategies for diversification of natural product scaffolds. For example, scaffold diversification of terpenes to create non-native terpenes can be achieved by manipulating isoprenoid precursor supplies and genetic engineering of terpene synthases [118, 119]. The use of promiscuous tailoring enzymes for the derivatization (e.g. alkylation, acylation, oxidation, glycosylation) of natural product scaffolds allows exploration of a defined chemical structure-function space. Naturally occurring and engineered P450s and glycosyltransferases exhibiting broad substrate specificities have been successfully employed for late-stage derivatization of terpenes, PKs and NRPs [115]. New sulfated glycopeptide congeners were obtained in vitro and in vivo by exploiting eDNA-derived sulfotransferases frequently associated with glycopeptide BGCs [120].
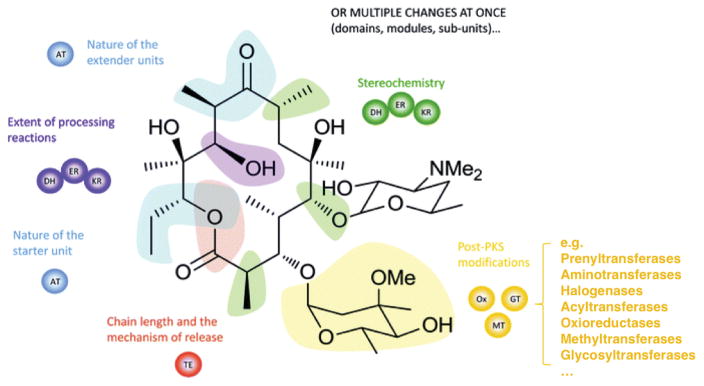
The “assembly line” biosynthetic logic of modular PKS/NRPS megaenzymes makes them attractive targets for rational engineering of complex PK/NRP scaffolds [121, 122, 123]. Engineering strategies include mutagenesis, insertion/swapping of domains, modules or subunits to alter 1) type of starter and extender units, 2) extent and stereochemistry of scaffold processing, 3) chain length and release mechanism, and 4) post-PKS/NRPS modifications (Figure 3). By deleting and swapping domains, Sugimoto and colleagues reprogrammed an aureothin BGC into one that produces luteoreticulin, a related compound with distinct biological activities, and other novel derivatives [124]. Advances in synthetic biology strategies such as DNA assembler and other approaches described in section 3.2.1 greatly accelerate genetic manipulation of biosynthetic pathways [125]. Despite early promise [126, 127], the generation of large libraries of PK/NRP analogs by combinatorial biosynthesis is generally hindered by low yielding or non-functional constructs and the paucity of high throughput screens/selections for active pathways. Recently, leveraging on efficient yeast homologous recombination and short homology stretches between pikromycin and erythromycin PKS genes, Chemler et al. created a hybrid PKS library and identified active chimeric enzymes that were capable of producing novel macrolactone analogs [128]. Future application of this approach should shed light on the structure-activity relationship for these megaenzymes and inform genetic engineering efforts. With increasing structural information [121, 122] as well as the availability of algorithms to guide combinatorial library design [129] and capture beneficial variants [130], directed evolution strategies will likely be useful in generating libraries of active PKS/NRPS hybrids for PK/NRP diversification [131, 132].
Figure 3. Diversification of polyketide scaffolds by genetic engineering of PKS clusters.
Possible modifications to polyketide structures are colored according to the responsible PKS domains or stand-alone (post-PKS) enzymes. The changes are not mutually exclusive and can be made in combination. Reproduced and adapted from [121] with permissions from The Royal Society of Chemistry.
While this section focuses on the concept of derivatization and diversification of natural product scaffolds by genetic engineering strategies guided by comparative genomics, genetic engineering of BGCs can also be used in combination with other established strategies to diversify natural product scaffolds. In mutasynthesis, strains are engineered to uptake and incorporate unusual biosynthetic precursors to generate natural product analogs with potentially improved phamacoproperties [133, 134]. Semi-synthesis melds biological and chemical synthetic routes to produce complex natural products such biomedically relevant terpenoids and FDA approved anti-cancer drug ET-743 [135, 136, 137, 138]. Notably, PKSs and NRPSs have been engineered to incorporate alternative starter/extension units harboring fluorine atoms known to improve drug pharmacoproperties [139], or orthogonally reactive handles such as terminal alkynes for combinatorial late-stage derivatization [140, 141].
7. Conclusion
Natural products are still a major source of inspiration for clinical drugs despite the scaling down of natural product discovery efforts by pharmaceutical companies in the mid-1990s due to high rediscovery rates and challenging product synthesis. Rejuvenation of natural product research in recent years can be largely attributed to advances in DNA sequencing, genomics/metagenomics, synthetic biology and genome editing technologies. Analyses of microbial genomes and metagenomes reveal that we have barely explored the chemical and functional diversity of microorganisms. Even in actinobacteria that have been the research focus for decades, majority of the BGCs has not been linked to their secondary metabolites [8], underscoring the potential for new discoveries even in the most extensively screened species. Bioinformatic tools have been developed to prioritize and identify relevant BGCs from genomes/metagenomes, predict the structures of their chemical products, integrate genomic with metabolomic and biological information to identify genotype-chemotype relationships, as well as prioritize downstream characterization efforts. New DNA capture and assembly methods as well as multiplexed genome engineering technologies facilitate the optimization of heterologous expression systems and biosynthetic pathways for the production of secondary metabolites. These developments pave a way towards more systematic and targeted discovery of novel natural products starting from genomic information.
8. Expert opinion
With the rapidly increasing amount of DNA sequence information, the promise of better quality genomes/metagenomes using third-generation and single-cell genome sequencing platforms, as well as the significant headway made in classification of BGCs and their chemical outputs, it is clear that the major bottleneck in natural product discovery lies in our ability to link BGCs with their cognate secondary metabolites [34]. Activation of silent BGCs in native producing hosts will require the development of separate sets of genetic tools for non-model organisms. Advances have been made in the development of alternative heterologous expression hosts for BGCs from unknown or inaccessible sources but success hinges upon host-BGC compatibility and often involves time-consuming capture or cloning, assembly and refactoring and/or optimization of large and unwieldy clusters. Until we better understand the determinants governing host-BGC compatibility, a diverse collection of robust, orthogonal heterologous hosts maximizes the probability of finding a host-BGC combination for successful expression. The development of new host systems will require identification and engineering of gifted strains that are genetically and metabolically predisposed to express and produce the encoded molecules from a diverse set of BGCs [112, 113, 114]. With many bacteria phyla lacking cultured representatives [14, 110], advanced culture methods and separate sets of genetic tools may be needed to obtain compatible expression hosts to more readily access the biosynthetic potential of these underexplored groups of microorganisms [45]. Significant progress has been made in the development of molecular parts and tools for the genetic manipulation of Streptomyces spp., a prolific group of bacteria known to produce bioactive metabolites, but there is still much room for improvement before “plug-and-play” natural product discovery becomes a reality, and one can expect the same for other emerging host systems. Combinatorial genome-scale engineering tools, while currently limited to model organisms like E. coli, will be invaluable towards the engineering of more robust production systems for different BGC families when transferred to natural product-relevant hosts [34]. Last but not least, entire biosynthetic gene clusters can be codon-optimized/-randomized, refactored, synthesized de novo and assembled for optimal expression in designated heterologous hosts [69, 142], although current gene synthesis costs precludes routine adoption of this approach.
One of the main underlying assumptions in genomics- and metagenomics-driven natural product discovery is that BGC sequence diversity reflects biosynthetic and chemical diversity, and by extension the pharmacological activity profiles of the metabolites. Homology to characterized clusters has been used as a proxy for “chemical dereplication” and to prioritize BGCs of interest for characterization [143, 144]. Based on phylogenetic similarity between amplicon sequences of key biosynthetic features within BGCs, the sequence-tag strategy for targeted metagenomics has been successful in identifying new congeners and subclasses of biomedically relevant natural products (Section 5). Nonetheless, there are indications that cluster sequence or architecture divergence does not always translate into chemical structure variation. For example, biosynthetic genes may not be physically clustered [145, 146]. From an evolution perspective, enzymes may already be engaged in a biosynthetic pathway before the respective genes are physically recruited into the BGC through a mechanism that is yet to be determined [116]. Acyl carrier protein (ACP)-less PKS clusters suggest an alternative biosynthetic route or the possible recruitment of ACPs from outside the cluster [117]. Additionally, highly divergent BGCs in terms of both cluster architecture and gene sequence can have similar chemical outputs. Global analysis of prokaryotic BGCs revealed that two distinct clades of BGCs with limited homology actually produce highly similar and conserved aryl polyenes [143]. On the contrary, PKS clusters containing phylogenetically related sets of AT and KS protein domains can give rise to polyketides with distinct core structures [116]. With individual BGC families evolving differently [116], the choice of biosynthetic features for phylogenetic profiling and inference of chemical output, especially for sequence-tag metagenomics, is crucial but may not be obvious without sufficient empirical information. Comparing larger regions or entire BGCs, though not always possible, will facilitate the classification of BGCs that can be linked to secondary metabolites groups and their associated pharmacological activities [143].
While this review focuses on harnessing genomic and metagenomic data to prioritize and facilitate the discovery of new chemical entities, chemical (e.g. compound isolation, structure identification) and biological characterization of natural products are just as critical to the drug discovery process. In fact, structure elucidation and determination of compound mode-of-action represent some of major bottlenecks in the natural product discovery process. Liquid chromatography-, mass spectrometry-, nuclear magnetic resonance-based technologies have progressed significantly for general chemical profiling of complex natural product samples as well as structure determination of large complex molecules [147, 148, 149, 150]. Given the inherent low-throughput nature of structure elucidation, especially of large complex molecules with unknown chemical scaffolds, it is critical that such efforts are channeled towards compounds with desired bioactivity and modes-of-action. Conceptually distinct from the conventional high throughput bioassays, multiparametric screening cell-, image- and sequencing-based strategies generate information-rich bioactivity fingerprints have been successfully used to prioritize and classify natural products based on their modes of action [150]. For effective classification, however, many of these multiparametric screening platforms require a sizable reference set of bioactivity profiles that may not be readily accessible to everyone. There are recent community-wide efforts to integrate multiple sequence databases and relevant web services, as well as standardize BGC annotation and their chemical outputs to facilitate comparative analyses of large datasets [12, 24, 151]. These repositories are publicly shared and curated to improve chemical characterization as well as foster collaboration, emphasizing the need to tackle existing challenges (e.g. high rediscovery rates, genes-to-chemical bottleneck, low production titers) in natural product discovery as a community. Similar repositories for standardized biological characterization data, integrated with genomic and chemical information data, will be invaluable towards uncovering chemical feature(s) with desired bioactivities from nature. It is possible that we will be able to reliably deduce compound pharmacological profiles from analysis of BGCs as the list and diversity of experimentally validated BGCs linked to their secondary metabolites with known bioactivity expand.
Overall, this is an exciting time for natural product discovery, which was mostly a random “hit-or-miss” endeavor in the pregenomics era. Genomics and metagenomics have given us a glimpse of the remarkable biosynthetic potential and vast chemical inventory in nature that we can prioritize and systematically mine for target natural products. As various technologies mature to clone and functionally express target BGCs in heterologous systems or activate silent BGCs in their native hosts, we will be able to more rapidly and cost effectively translate genome information into chemical compounds for drug screens. Public repositories with integrated web services establish the framework for genome-driven natural product discovery efforts and our predictive ability for the chemical output of BGCs will continue to improve as the list and diversity of experimentally characterized clusters and their encoded metabolites expand. These advances open up opportunities to understand the intricacies of natural product biosynthesis and reengineering of BGCs for the diversification and derivatization of natural products during drug discovery.
Article highlights.
A substantial proportion of pharmaceuticals are derived from or inspired by natural products.
Advances in DNA sequencing reveal silent biosynthetic gene clusters and uncultured microorganisms as vast untapped resources for novel bioactive chemical scaffolds.
A collection of in silico tools supports mining of genomes and metagenomes for biosynthetic pathways that potentially encode novel pharmaceutically relevant molecules.
Driven by advances in synthetic biology and genome engineering tools, pathway and host engineering strategies facilitate the functional expression and characterization of biosynthetic gene clusters.
Understanding the chemical logic of biosynthetic pathways enables rational biosynthetic pathway engineering for the diversification and derivatization of privileged natural product scaffolds.
Genomics- and metagenomics-driven natural product discovery continues to uncover new bioactive chemical entities and promises to revitalize waning drug pipelines in the near future.
Acknowledgments
Funding:
The authors gratefully acknowledge financial support from the National Institutes of Health (GM077596) (to H Zhao), the A*STAR Visiting Investigator Program (to H Zhao), and the National Research Foundation, Singapore (NRF2013-THE001-094) to (MM Zhang).
Footnotes
Declaration of Interest:
The authors are all employees of the Agency for Science, Technology and Research (A*STAR), Singapore. They have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Bibliography
Papers of special note have been highlighted either of interest (•) or of considerable interest (••) to readers.
- 1••.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–61. doi: 10.1021/acs.jnatprod.5b01055. This comprehensive review documents the significant impact of natural products on drug discovery and development from 1981 to 2014. [DOI] [PubMed] [Google Scholar]
- 2.Katz L, Baltz RH. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–76. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 3.Mardis ER. DNA sequencing technologies: 2006–2016. Nature Protocols. 2017;12:213–8. doi: 10.1038/nprot.2016.182. [DOI] [PubMed] [Google Scholar]
- 4.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–29. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 5•.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–23. doi: 10.1038/nrmicro3496. This review discusses the different strategies developed to activate and characterize silent biosynthetic gene clusters from bacteria and fungi. [DOI] [PubMed] [Google Scholar]
- 6.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–40. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 7.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 8.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–84. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wenzel SC, Müller R. Myxobacteria—‘microbial factories’ for the production of bioactive secondary metabolites. Mol Biosystems. 2009;5:567–74. doi: 10.1039/b901287g. [DOI] [PubMed] [Google Scholar]
- 10.Land M, Hauser L, Jun S-R, Nookaew I, Leuze MR, Ahn T-H, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics. 2015;15:141–61. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nuc Acid Res. 2011;39:W339–46. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nuc Acid Res. 2015;43:W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Shen X, Peng H, Hu H, Wang W, Zhang X. Comparative genomic analysis and phenazine production of Pseudomonas chlororaphis, a plant growth-promoting rhizobacterium. Genom Data. 2015;4:33–42. doi: 10.1016/j.gdata.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UA, Heycke N, Schmitt S, Rinke C, Helfrich EJ, Brachmann AO, Gurgui C, Wakimoto T, Kracht M, Crusemann M, Hentschel U, Abe I, Matsunaga S, Kalinowski J, Takeyama H, Piel J. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 15.Umemura M, Koike H, Machida M. Motif-independent de novo detection of secondary metabolite gene clusters-toward identification from filamentous fungi. Front Microbiol. 2015;6:371. doi: 10.3389/fmicb.2015.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letzel AC, Pidot SJ, Hertweck C. A genomic approach to the cryptic secondary metabolome of the anaerobic world. Nat Prod Rep. 2013;30:392–428. doi: 10.1039/c2np20103h. [DOI] [PubMed] [Google Scholar]
- 17.Blin K, Kazempour D, Wohlleben W, Weber T. Improved lanthipeptide detection and prediction for antiSMASH. PLoS One. 2014;9:e89420. doi: 10.1371/journal.pone.0089420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Gen Biol. 2010;47:736–41. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nuc Acid Res. 2013;41:W448–53. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li MHT, Ung PMU, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tae H, Kong EB, Park K. ASMPKS: an analysis system for modular polyketide synthases. BMC Bioinformatics. 2007;8:327. doi: 10.1186/1471-2105-8-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tae H, Sohng JK, Park K. Development of an analysis program of type I polyketide synthase gene clusters using homology search and profile hidden Markov model. J Microbiol Biotechnol. 2009;19:140–6. doi: 10.4014/jmb.0809.554. [DOI] [PubMed] [Google Scholar]
- 23••.Dejong CA, Chen GM, Li H, Johnston CW, Edwards MR, Rees PN, Skinnider MA, Webster AL, Magarvey NA. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat Chem Biol. 2016;12:1007–14. doi: 10.1038/nchembio.2188. This report presents a retrobiosynthetic in silico analysis platform that identifies biosynthetic gene clusters encoding for known polyketides and non-ribosomal peptides. [DOI] [PubMed] [Google Scholar]
- 24••.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, De Bruijn I, Chooi YH, Claesen J, Coates RC. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–31. doi: 10.1038/nchembio.1890. This paper proposes standardized annotation and deposition of data on biosynthetic gene clusters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 2014;304:230–5. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 26••.Medema MH, Fischbach MA. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–48. doi: 10.1038/nchembio.1884. This perspective compares the various algorithms developed to identify biosynthetic gene clusters and predict their chemical output, as well as networking strategies to integrate genomic, chemical and biological data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Lee TAJ, Medema MH. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Gen Biol. 2016;89:29–36. doi: 10.1016/j.fgb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes–a review. Nat Prod Rep. 2016;33:988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 29.Kellner F, Kim J, Clavijo BJ, Hamilton JP, Childs KL, Vaillancourt B, Cepela J, Habermann M, Steuernagel B, Clissold L. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015;82:680–92. doi: 10.1111/tpj.12827. [DOI] [PubMed] [Google Scholar]
- 30.Nützmann HW, Huang A, Osbourn A. Plant metabolic clusters–from genetics to genomics. New Phytologist. 2016;211:771–89. doi: 10.1111/nph.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kautsar SA, Duran HGS, Blin K, Osbourn A, Medema MH. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. 2016:083535. doi: 10.1093/nar/gkx305. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–15. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Zhang MM, Wang Y, Ang EL, Zhao H. Engineering microbial hosts for production of bacterial natural products. Nat Prod Rep. 2016;33:963–87. doi: 10.1039/c6np00017g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA. Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol. 2016;14:135–49. doi: 10.1038/nrmicro.2015.24. This review presents key advances in synthetic biology and how they can be applied to build and optimize natural product biosynthesis pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connor SE. Engineering of secondary metabolism. Ann Rev Genet. 2015;49:71–94. doi: 10.1146/annurev-genet-120213-092053. [DOI] [PubMed] [Google Scholar]
- 36.Olano C, García I, González A, Rodriguez M, Rozas D, Rubio J, Sánchez-Hidalgo M, Braña AF, Méndez C, Salas JA. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microbial Biotechnol. 2014;7:242–56. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortina NS, Krug D, Plaza A, Revermann O, Müller R. Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew Chem Int Ed Engl. 2012;51:811–6. doi: 10.1002/anie.201106305. [DOI] [PubMed] [Google Scholar]
- 38.Scherlach K, Schuemann J, Dahse H-M, Hertweck C. Aspernidine A and B, prenylated isoindolinone alkaloids from the model fungus Aspergillus nidulans. J Antibiot. 2010;63:375–7. doi: 10.1038/ja.2010.46. [DOI] [PubMed] [Google Scholar]
- 39.Scherlach K, Hertweck C. Discovery of aspoquinolones A–D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Org Biomol Chem. 2006;4:3517–20. doi: 10.1039/b607011f. [DOI] [PubMed] [Google Scholar]
- 40.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brötz-Oesterhelt H, Grond S, Peschel A, Krismer B. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–6. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 41.Kawai K, Wang G, Okamoto S, Ochi K. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol Lett. 2007;274:311–5. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka Y, Hosaka T, Ochi K. Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2) J Antibiot. 2010;63:477–81. doi: 10.1038/ja.2010.53. [DOI] [PubMed] [Google Scholar]
- 43.Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, Epstein SS. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol. 2010;76:2445–50. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gavrish E, Sit CS, Cao S, Kandror O, Spoering A, Peoples A, Ling L, Fetterman A, Hughes D, Bissell A, Torrey H, Akopian T, Mueller A, Epstein S, Goldberg A, Clardy J, Lewis K. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem Biol. 2014;21:509–18. doi: 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45•.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–9. doi: 10.1038/nature14098. This study reports a new class of antibiotics that is produced by a soil microorganism isolated using the iChip microfluidics device. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. PNAS. 2014;111:7266–71. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol. 2011;77:400–6. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Konig CC, Scherlach K, Schroeckh V, Horn F, Nietzsche S, Brakhage AA, Hertweck C. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. Chembiochem. 2013;14:938–42. doi: 10.1002/cbic.201300070. [DOI] [PubMed] [Google Scholar]
- 49.Adnani N, Vazquez-Rivera E, Adibhatla SN, Ellis GA, Braun DR, Bugni TS. Investigation of interspecies interactions within marine Micromonosporaceae using an improved co-culture approach. Mar Drugs. 2015;13:6082–98. doi: 10.3390/md13106082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4:e00459–13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–7. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 52.Rachid S, Sasse F, Beyer S, Müller R. Identification of StiR, the first regulator of secondary metabolite formation in the myxobacterium Cystobacter fuscus Cb f17. 1. J Biotechnol. 2006;121:429–41. doi: 10.1016/j.jbiotec.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 53.Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. PNAS. 2011;108:6258–63. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sidda JD, Song L, Poon V, Al-Bassam M, Lazos O, Buttner MJ, Challis GL, Corre C. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem Sci. 2013;5:86–9. [Google Scholar]
- 55.O’Rourke S, Wietzorrek A, Fowler K, Corre C, Challis GL, Chater KF. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol Microbiol. 2009;71:763–78. doi: 10.1111/j.1365-2958.2008.06560.x. [DOI] [PubMed] [Google Scholar]
- 56.Rachid S, Gerth K, Müller R. NtcA—A negative regulator of secondary metabolite biosynthesis in Sorangium cellulosum. J Biotechnol. 2009;140:135–42. doi: 10.1016/j.jbiotec.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 57.Jirschitzka J, Mattern DJ, Gershenzon J, D’Auria JC. Learning from nature: new approaches to the metabolic engineering of plant defense pathways. Curr Opin Biotechnol. 2013;24:320–8. doi: 10.1016/j.copbio.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 58.Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol. 2009;27:462–4. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 59.Guo F, Xiang S, Li L, Wang B, Rajasärkkä J, Gröndahl-Yli-Hannuksela K, Ai G, Metsä-Ketelä M, Yang K. Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Met Eng. 2015;28:134–42. doi: 10.1016/j.ymben.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Cichewicz RH. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat Prod Rep. 2009;27:11–22. doi: 10.1039/b920860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bok JW, Chiang Y-M, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–4. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–7. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 63.Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org Biomol Chem. 2009;7:435–8. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]
- 64.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Müller R, Stewart AF. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–6. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 65.Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. PNAS. 2014;111:1957–62. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee NC, Larionov V, Kouprina N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nuc Acid Res. 2015;43:e55. doi: 10.1093/nar/gkv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang W, Zhao X, Gabrieli T, Lou C, Ebenstein Y, Zhu TF. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun. 2015:6. doi: 10.1038/ncomms9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Kang HS, Charlop-Powers Z, Brady SF. Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Syn Biol. 2016;5:1002–10. doi: 10.1021/acssynbio.6b00080. This study describes a multiplex promoter engineering strategy to refactor natural product biosynthetic gene clusters for heterolgous expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Luo Y, Enghiad B, Zhao H. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat Prod Rep. 2016;33:174–82. doi: 10.1039/c5np00085h. This review compares between the different direct DNA cloning and assembly methods available for the reconstruction and refactoring of biosynthetic gene clustes for heterologous expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Neubauer P. Escherichia coli as a cell factory for heterologous production of nonribosomal peptides and polyketides. New Biotechnol. 2014;31:579–85. doi: 10.1016/j.nbt.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 72.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–8. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cobb RE, Wang Y, Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Syn Biol. 2015;4:723–8. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Syn Biol. 2015;4:1020–9. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 75.Huang H, Zheng G, Jiang W, Hu H, Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochimica et Biophysica Sinica. 2015;47:231–43. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 76.Zeng H, Wen S, Xu W, He Z, Zhai G, Liu Y, Deng Z, Sun Y. Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA (sm) combined system. App Microbiol Biotechnol. 2015;99:10575–85. doi: 10.1007/s00253-015-6931-4. [DOI] [PubMed] [Google Scholar]
- 77.Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, Reddy BV, Charlop-Powers Z, Zebroski HA, Gallardo-Macias R, Jaskowski M, Satish S, Park S, Perlin DS, Freundlich JS, Brady SF. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat Chem Biol. 2016;12:1004–6. doi: 10.1038/nchembio.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78••.Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. This work describes the iterative matching of de novo MSn molecular features of peptide natural products to the genomic features within biosynthetic clusters to establish chemotype-genotype relationships. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuchs SW, Sachs CC, Kegler C, Nollmann FI, Karas M, Bode HB. Neutral loss fragmentation pattern based screening for arginine-rich natural products in Xenorhabdus and Photorhabdus. Anal Chem. 2012;84:6948–55. doi: 10.1021/ac300372p. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen DD, Wu C-H, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C. MS/MS networking guided analysis of molecule and gene cluster families. PNAS. 2013;110:E2611–E20. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kersten RD, Ziemert N, Gonzalez DJ, Duggan BM, Nizet V, Dorrestein PC, Moore BS. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. PNAS. 2013;110:E4407–E16. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–9. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 83.Seow K, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson CR, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–8. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katz M, Hover BM, Brady SF. Culture-independent discovery of natural products from soil metagenomes. J Ind Microbiol Biotechnol. 2016;43:129–41. doi: 10.1007/s10295-015-1706-6. [DOI] [PubMed] [Google Scholar]
- 85.Garcia JA, Fernández-Guerra A, Casamayor EO. A close relationship between primary nucleotides sequence structure and the composition of functional genes in the genome of prokaryotes. Mol Phylogen Evol. 2011;61:650–8. doi: 10.1016/j.ympev.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 86.Bentley SD, Chater KF, Cerdeno-Tarraga A-M, Challis GL, Thomson N, James KD, Harris DE, Quail MA, Kieser H, Harper D. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2) Nature. 2002;417:141–7. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 87•.Charlop-Powers Z, Banik JJ, Owen JG, Craig JW, Brady SF. Selective enrichment of environmental DNA libraries for genes encoding nonribosomal peptides and polyketides by phosphopantetheine transferase-dependent complementation of siderophore biosynthesis. ACS Chem Biol. 2013;8:138–43. doi: 10.1021/cb3004918. This research harnesses functional complementation of phosphopantetheine transferase activity to selectively enrich for eDNA clones containing PKS/NRPS-containing biosynthetic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang FY, Ternei MA, Calle PY, Brady SF. Targeted metagenomics: finding rare tryptophan dimer natural products in the environment. JACS. 2015;137:6044–52. doi: 10.1021/jacs.5b01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol. 2002;68:4301–6. doi: 10.1128/AEM.68.9.4301-4306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brady SF, Chao CJ, Clardy J. New natural product families from an environmental DNA (eDNA) gene cluster. JACS. 2002;124:9968–9. doi: 10.1021/ja0268985. [DOI] [PubMed] [Google Scholar]
- 91.Brady SF, Chao CJ, Clardy J. Long-chain N-acyltyrosine synthases from environmental DNA. Appl Environ Microbiol. 2004;70:6865–70. doi: 10.1128/AEM.70.11.6865-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brady SF, Bauer JD, Clarke-Pearson MF, Daniels R. Natural products from isnA-containing biosynthetic gene clusters recovered from the genomes of cultured and uncultured bacteria. JACS. 2007;129:12102–3. doi: 10.1021/ja075492v. [DOI] [PubMed] [Google Scholar]
- 93.Brady SF, Clardy J. Cloning and heterologous expression of isocyanide biosynthetic genes from environmental DNA. Angew Chem Int Ed Engl. 2005;44:7063–5. doi: 10.1002/anie.200501941. [DOI] [PubMed] [Google Scholar]
- 94.Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J. Novel natural products from soil DNA libraries in a streptomycete host. Org Lett. 2000;2:2401–4. doi: 10.1021/ol005860z. [DOI] [PubMed] [Google Scholar]
- 95.Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem. 2005;6:1760–5. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 96.Williamson LL, Borlee BR, Schloss PD, Guan C, Allen HK, Handelsman J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl Environ Microbiol. 2005;71:6335–44. doi: 10.1128/AEM.71.10.6335-6344.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Uchiyama T, Abe T, Ikemura T, Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- 98.Uchiyama T, Miyazaki K. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol. 2009;20:616–22. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 99.Colin P-Y, Kintses B, Gielen F, Miton CM, Fischer G, Mohamed MF, Hyvönen M, Morgavi DP, Janssen DB, Hollfelder F. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat Commun. 2015;6:10008. doi: 10.1038/ncomms10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang K, He J, Yang M, Yen M, Yin J. Identifying natural product biosynthetic genes from a soil metagenome by using T7 phage selection. Chembiochem. 2009;10:2599–606. doi: 10.1002/cbic.200900297. [DOI] [PubMed] [Google Scholar]
- 101.Owen JG, Robins KJ, Parachin NS, Ackerley DF. A functional screen for recovery of 4′-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ Microbiol. 2012;14:1198–209. doi: 10.1111/j.1462-2920.2012.02699.x. [DOI] [PubMed] [Google Scholar]
- 102••.Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7:e34064. doi: 10.1371/journal.pone.0034064. This report presents a phylogenomic tool to classify biosynthetic diversity based on sequence similarities of select biosynthetic domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103••.Owen JG, Reddy BVB, Ternei MA, Charlop-Powers Z, Calle PY, Kim JH, Brady SF. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. PNAS. 2013;110:11797–802. doi: 10.1073/pnas.1222159110. Phylogentic relationships of natural product sequence tags are used to predict gene content and chemical output of biosynthetic gene clusters to prioritize clusters of interest for characterization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Owen JG, Charlop-Powers Z, Smith AG, Ternei MA, Calle PY, Reddy BVB, Montiel D, Brady SF. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. PNAS. 2015;112:4221–6. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang HS, Brady SF. Arimetamycin A: improving clinically relevant families of natural products through sequence-guided screening of soil metagenomes. Angew Chem Int Ed Engl. 2013;52:11063–7. doi: 10.1002/anie.201305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Genome streamlining and chemical defense in a coral reef symbiosis. PNAS. 2012;109:20655–60. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schofield MM, Jain S, Porat D, Dick GJ, Sherman DH. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ Microbiol. 2015;17:3964–75. doi: 10.1111/1462-2920.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Koren S, Phillippy AM. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol. 2015;23:110–20. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 109.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–88. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 110.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 111.Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. PNAS. 2007;104:11889–94. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112•.Craig JW, Chang F-Y, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol. 2010;76:1633–41. doi: 10.1128/AEM.02169-09. This functional metagenomics study demonstrates that different expression host strains enable access molecules encoded by distinct sets of biosynthetic pathways in environmental DNA libraries. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Craig JW, Chang FY, Brady SF. Natural products from environmental DNA hosted in Ralstonia metallidurans. ACS Chem Biol. 2009;4:23–8. doi: 10.1021/cb8002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iqbal HA, Low-Beinart L, Obiajulu JU, Brady SF. Natural product discovery through improved functional metagenomics in Streptomyces. JACS. 2016;138:9341–4. doi: 10.1021/jacs.6b02921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.King JR, Edgar S, Qiao K, Stephanopoulos G. Accessing Nature’s diversity through metabolic engineering and synthetic biology. F1000Res. 2016;5:397. doi: 10.12688/f1000research.7311.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116••.Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol. 2014;10:e1004016. doi: 10.1371/journal.pcbi.1004016. This study offers insights into the evolution of different biosynthetic gene cluster families and proposes strategies to rationally engineer biosynthetic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hillenmeyer ME, Vandova GA, Berlew EE, Charkoudian LK. Evolution of chemical diversity by coordinated gene swaps in type II polyketide gene clusters. PNAS. 2015;112:13952–7. doi: 10.1073/pnas.1511688112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zerbe P, Bohlmann J. Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 2015;33:419–28. doi: 10.1016/j.tibtech.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 119.Brück T, Kourist R, Loll B. Production of macrocyclic sesqui- and diterpenes in heterologous microbial hosts: a systems approach to harness Nature’s molecular diversity. ChemCatChem. 2014;6:1142–65. [Google Scholar]
- 120.Banik JJ, Craig JW, Calle PY, Brady SF. Tailoring enzyme-rich environmental DNA clones: a source of enzymes for generating libraries of unnatural natural products. JACS. 2010;132:15661–70. doi: 10.1021/ja105825a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weissman KJ. Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology. Nat Prod Rep. 2016;33:203–30. doi: 10.1039/c5np00109a. [DOI] [PubMed] [Google Scholar]
- 122.Winn M, Fyans J, Zhuo Y, Micklefield J. Recent advances in engineering nonribosomal peptide assembly lines. Nat Prod Rep. 2016;33:317–47. doi: 10.1039/c5np00099h. [DOI] [PubMed] [Google Scholar]
- 123.Olano C, Méndez C, Salas JA. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat Prod Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- 124•.Sugimoto Y, Ding L, Ishida K, Hertweck C. Rational design of modular polyketide synthases: morphing the aureothin pathway into a luteoreticulin assembly line. Angew Chem Int Ed Engl. 2014;53:1560–4. doi: 10.1002/anie.201308176. The first example where a biosynthetic pathway is engineered to produce a different natural product from another organism. [DOI] [PubMed] [Google Scholar]
- 125.Shao Z, Luo Y, Zhao H. Rapid characterization and engineering of natural product biosynthetic pathways via DNA assembler. Mol BioSystems. 2011;7:1056–9. doi: 10.1039/c0mb00338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol. 2005;23:1171–6. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 127.Sherman DH. The Lego-ization of polyketide biosynthesis. Nat Biotechnol. 2005;23:1083–4. doi: 10.1038/nbt0905-1083. [DOI] [PubMed] [Google Scholar]
- 128••.Chemler JA, Tripathi A, Hansen DA, O’Neil-Johnson M, Williams RB, Starks C, Park SR, Sherman DH. Evolution of efficient modular polyketide synthases by homologous recombination. JACS. 2015;137:10603–9. doi: 10.1021/jacs.5b04842. Using homologous recombination in yeast, this study generated active chimeric polyketide synthases capable of producing macrolactone cogeners, including a novel macrolide analog. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meyer MM, Hochrein L, Arnold FH. Structure-guided SCHEMA recombination of distantly related β-lactamases. Protein Eng Des Sel. 2006;19:563–70. doi: 10.1093/protein/gzl045. [DOI] [PubMed] [Google Scholar]
- 130.Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol. 2007;25:338–44. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 131.Fischbach MA, Lai JR, Roche ED, Walsh CT, Liu DR. Directed evolution can rapidly improve the activity of chimeric assembly-line enzymes. PNAS. 2007;104:11951–6. doi: 10.1073/pnas.0705348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Evans BS, Chen Y, Metcalf WW, Zhao H, Kelleher NL. Directed evolution of the nonribosomal peptide synthetase AdmK generates new andrimid derivatives in vivo. Chem Biol. 2011;18:601–7. doi: 10.1016/j.chembiol.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yan Y, Chen J, Zhang L, Zheng Q, Han Y, Zhang H, Zhang D, Awakawa T, Abe I, Liu W. Multiplexing of combinatorial chemistry in antimycin biosynthesis: expansion of molecular diversity and utility. Angew Chem Int Ed Engl. 2013;52:12308–12. doi: 10.1002/anie.201305569. [DOI] [PubMed] [Google Scholar]
- 134.Bode HB, Meiser P, Klefisch T, Cortina NSdJ, Krug D, Göhring A, Schwär G, Mahmud T, Elnakady YA, Müller R. Mutasynthesis-Derived myxalamids and origin of the isobutyryl-CoA starter unit of Myxalamid B. Chembiochem. 2007;8:2139–44. doi: 10.1002/cbic.200700401. [DOI] [PubMed] [Google Scholar]
- 135.Paddon CJ, Westfall PJ, Pitera D, Benjamin K, Fisher K, McPhee D, Leavell M, Tai A, Main A, Eng D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–32. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 136.Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat Chem Biol. 2014;10:837–44. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ajikumar PK, Xiao W-H, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–4. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cuevas C, Pérez M, Martín MJ, Chicharro JL, Fernández-Rivas C, Flores M, Francesch A, Gallego P, Zarzuelo M, de la Calle F. Synthesis of ecteinascidin ET-743 and phthalascidin Pt-650 from cyanosafracin B. Org Lett. 2000;2:2545–8. doi: 10.1021/ol0062502. [DOI] [PubMed] [Google Scholar]
- 139.Walker MC, Thuronyi BW, Charkoudian LK, Lowry B, Khosla C, Chang MC. Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways. Science. 2013;341:1089–94. doi: 10.1126/science.1242345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhu X, Liu J, Zhang W. De novo biosynthesis of terminal alkyne-labeled natural products. Nat Chem Biol. 2015;11:115–20. doi: 10.1038/nchembio.1718. [DOI] [PubMed] [Google Scholar]
- 141.Kries H, Wachtel R, Pabst A, Wanner B, Niquille D, Hilvert D. Reprogramming nonribosomal peptide synthetases for “clickable” amino acids. Angew Chem Int Ed Engl. 2014;53:10105–8. doi: 10.1002/anie.201405281. [DOI] [PubMed] [Google Scholar]
- 142.Smanski MJ, Bhatia S, Zhao D, Park Y, Woodruff LB, Giannoukos G, Ciulla D, Busby M, Calderon J, Nicol R. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol. 2014;32:1241–9. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 143.Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–21. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Donia Mohamed S, Cimermancic P, Schulze Christopher J, Wieland Brown Laura C, Martin J, Mitreva M, Clardy J, Linington Roger G, Fischbach Michael A. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–14. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. PNAS. 2015;112:13150–4. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lo H-C, Entwistle R, Guo C-J, Ahuja M, Szewczyk E, Hung J-H, Chiang Y-M, Oakley BR, Wang CC. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. JACS. 2012;134:4709–20. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Molinski TF. NMR of natural products at the ‘nanomole-scale’. Nat Prod Rep. 2010;27:321–9. doi: 10.1039/b920545b. [DOI] [PubMed] [Google Scholar]
- 148.Carter GT. NP/MS since 1970: from the basement to the bench top. Nat Prod Rep. 2014;31:711–7. doi: 10.1039/c3np70085b. [DOI] [PubMed] [Google Scholar]
- 149.Jarmusch AK, Cooks RG. Emerging capabilities of mass spectrometry for natural products. Nat Prod Rep. 2014;31:730–8. doi: 10.1039/c3np70121b. [DOI] [PubMed] [Google Scholar]
- 150.Kurita KL, Linington RG. Connecting phenotype and chemotype: high-content discovery strategies for natural products research. J Nat Prod. 2015;78:587–96. doi: 10.1021/acs.jnatprod.5b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151••.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–37. doi: 10.1038/nbt.3597. This article describes a community-wide effort to put together a publicly accessible and curated repository for mass spectrometry data to facilitate natural product discovery and chemical dereplication. [DOI] [PMC free article] [PubMed] [Google Scholar]