Aging is a complicate process that is far away from being understood but increasingly being investigated. The term is majorly used for multicellular organisms because singe cells, especially cell lines used in laboratories, are often immortal. Nevertheless, also multicellular organisms are based on single cells. Therefore, the understanding of how organisms age requires the understanding of how single cells age. In the recent years and with the progress in single-cell molecular biology, a certain genetic determination of aging transcriptional signatures and aging patterns were recognized, for example.1 However, from studies of genetically identical organisms it became evident that other factors than genetic regulation have a significant influence on cellular aging.2 Therefore, more physiological measures as read-outs for cell aging have been proposed.3 This is a very sensible concept but even if cells are in the Go-phase of the cell cycle and can be regarded as senescent, protein translation and other modes of biosynthesis mask the intrinsic aging process of particular subcellular components. To really understand the post mature aging requires a cell model with vastly quiescent translation and biosynthesis. Even in this case, one must distinguish single-cell aging after cell replacement into an (artificial) environment like a primary cell culture on the one hand and a natural behavior with systemic interactions on the other hand.
We like to discuss the classical example of biosynthetically quiescent cells, the mammalian red blood cells (RBCs). In the last century, RBCs were the preferred model cells to investigate membrane transport.4 With the start of the genomic era and progress in genetic manipulation, RBCs lost their popularity as model cells because with the enucleation of the late erythroblast in mammals, which is regarded as the birth of the RBC, these cells lose their transcriptional apparatus.5 This putative disadvantage can be regarded as an advantage in investigating particular aspects of post mature aging since it defines a starting point of an aging process of an average 120 days in human RBCs until these cells get cleared.6,7 Even if there is a minor translational activity left in reticulocytes, the translation activity decreases within a few days.8 The situation is obviously different in non-mammalian RBCs containing a nucleus and hence transcription and translation activity.
RBCs have in fact been, and still are, a favorite subject of investigation of cellular senescence.9, 10 Although RBCs lose their subcellular organelles, they maintain a plethora of cellular functions like anaerobic glycolysis, the pentose phosphate shunt,9 cellular signaling11 and possibly even a variant of programmed cell death called eryptosis.12
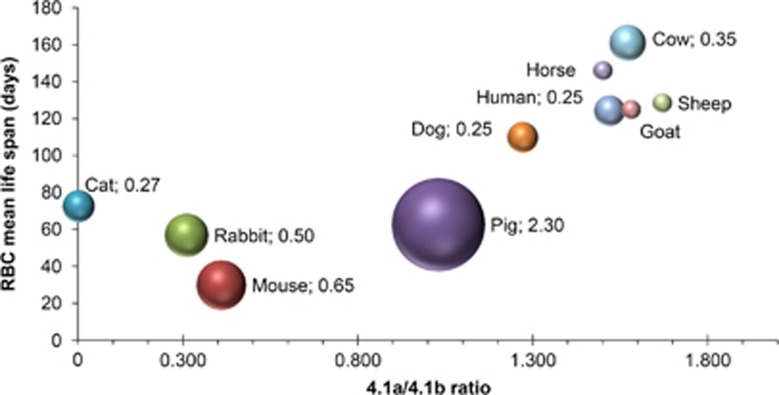
RBCs contain a number of biomarkers for age and senescence, like activity of the plasma membrane redox system, cysteine influx or enzymatic activity.10 However, it is not our intention to review senescence markers in RBCs. In contrast, we like to highlight a protein that can be used to determine exact age independent from other physico-chemical parameters such as cell density or invasive labeling techniques. This is the protein 4.1a/4.1b ratio (see Figure 1), which can be regarded as a 'molecular clock'. Although this ‘time stamp’ is a useful marker, it does not allow any conclusions concerning the mechanistic view of aging.
Figure 1.
Bubble graph showing the correlation between protein Band 4.1a/4.1b ratio and RBC life span in a number of mammals. Band 4.1 protein, whose modification from the naturally synthesized ‘4.1b’ form to the ‘4.1a’ form trough deamidation of a single asparaginyl residue, can be regarded as a molecular clock.6 The mean 4.1a/4.1b ratio of the total population of circulating RBCs from a given animal was plotted against the mean erythrocyte survival time of that animal. The 4.1a/4.1b values were originally measured by the authors of the article from which these data were derived;13 the S.D. was omitted for the sake of clarity, but it can be found in the original article. In the case of the cat, the expressed protein 4.1b does not undergo conversion to 4.1a because Ser, not Asn is present at position 502 (the deamidating Asn in protein 4.1 in humans and other mammals). Therefore, 4.1a/4.1b ratio cannot be measured in cat RBCs. The survival time is given as mean of two or more values obtained in the literature, as reported in the above-cited article. It must be reminded that the mean life span of RBCs results from two different kinetics of clearance: a first-order random destruction and a time-dependent destruction. In various mammals, the two components are present to a variable extent with a prevalent time-dependent clearance in human RBCs and an almost complete random clearance in mice. Although information on this aspect is sparse in the literature, the data from the last most accurate comparative account of RBC life span have been used to give an estimate of this aspect. Thus, the entity of the random component of RBC clearance, in % of RBCs removed per day, is given, where available, as the value to the right of the species’ name, which is also reflected in the size of the bubble (data of random destruction taken from Vacha14)
For RBCs, a number of candidate hallmarks of senescence have been described, measured and considered necessary and sufficient for the clearance of old RBCs;9 however, a unifying view as to the identity of the real molecular determinants of RBC senescence and death is still lacking. Previous evidence pointed to an involvement of intracellular Ca2+ in the aging process.15 With the advent of the free-radical theory of aging,16 the concept that RBCs undergo oxidative damage and the immunological evidence of the appearance of neo-antigens on the surface of old RBCs merged into an unified view of an oxidation-based degenerative senescence followed by an auto-immune mechanism for RBC clearance.7, 9 However, there are still many not-understood peculiarities in RBC aging and clearance: the exact identity of the neo-antigen(s) is not yet clear, alternatives to the immunoglobulin-based mechanism have been proposed,7, 17 an explanation for the different components of random destruction and time-dependent clearance in the kinetics of removal of old RBCs in different mammals is still lacking (see also Figure 1). Another most prominent phenomenon whose existence is still awaiting confirmation is neocytolysis.18 By looking at the aging process from a systemic point of view, though, some insight can be gained. By examining how the erythropoietic system is constructed, from the erythroblastic island in the bone marrow, to the spleen and liver specialized endothelial system, to the vascular bed, and the reticulo-endothelial system, the RBCs is continuously interacting with multiple 'systems'. One possible view is that RBCs are actually cleared from the circulation before they have accumulated molecular damage and have become dysfunctional, that is, before eryptosis is induced. A great deal of circumstantial evidence point to this direction. It is starting from early studies of asplenic subjects, where the whole maturation process from reticulocyte to erythrocyte (and also the normal modification of a mature RBC) is completely deranged. Furthermore, it continues with the fact that normal old RBCs appear to have normal ATP and GSH contents,6 a normal biconcave shape, which ensures good deformability properties, a hemoglobin content that is quantitatively and possibly qualitatively the same as in the newborn cell, and are 'only' smaller in size and dehydrated. Looking at the cardiovascular system, the RBC is one of the most stressed cell types and most risky, should it lose its proper functionality, as for sickle RBCs, spherocytes, paroxysmal nocturnal hemoglobinuria RBCs, G6PD deficient RBCs in favism, to name only a few pathological conditions.
The selective pressure for having a healthy cardiovascular system in an organism in its fertile age has evolved a number of gimmicks to prevent damage to RBCs (anti-oxidant buffers and enzymes) and to locally repair it (redox enzymes: glutathione peroxidase, glutathione reductase, peroxiredoxins, methemolgobin reductase). In the absence of a biosynthetic machinery that turns over the single molecules, it has evolved the ability of removing damaged components with the 'pitting' function of the spleen and the reticulo-endothelial macrophages, which continuously keep the RBC in check. It is by this mechanism that RBCs are maintained in a functional state until the very end of their life and go back to the bone marrow to die.6 It is because of this that the in vitro aged RBCs, suffering from the storage lesion for being kept under blood bank conditions for a fraction of their potential life span in vivo, differ so much from the old circulating RBCs.
Therefore, RBCs in vivo may well serve as a unique cellular aging model to investigate molecular properties altered in the process of aging as outlined below with their intrinsic advantages, i.a. their limited lifetime and the easy accessibility of these primary cells. However, experimentally this may require either singe-cell proteomics for the protein 4.1a/b ratio or to mark reticulocytes at a certain time point and transfuse them back into the circulation to be able to know their age.19 Another interesting aspect of RBCs as model cells in cell aging research is comparative physiology and the variability of the average lifetime of RBCs as plotted against the protein 4.1a/b ratio in Figure 1 also considering random clearance of RBCs.
Although every cell type belonging to a multicellular or multi-organ living being may have its own peculiar individual features in terms of propensity to accumulate and display senescence properties, the proteins in the RBCs are among the longest lived intracellular molecules and are not subjected to turnover, they contribute to the multifactorial aging process of the whole organism. Is the lack of turnover of these proteins due to selective pressure not playing a role on proteins that deteriorate relatively late with respect to the reproductive age of the subject? Recent studies on aging have found peculiarly long-lived proteins, which appear to escape turnover such as myelin and histone proteins.20 But these are found in post mitotic cells of essential organs and because they are subtracted, for reasons that are still mysterious, to the general rule of change, they may be among the molecular culprits on which to focus the attention.
Acknowledgments
This work was supported by the European Seventh Framework Program under grant agreement number 602121 (CoMMiTMenT) and the European Framework ‘Horizon 2020’ under grant agreement number 675115 (RELEVANCE).
Footnotes
The authors declare no conflict of interest.
References
- Enge M et al. bioRxiv 2017. (e-pub ahead of print; https://doi.org/10.1101/108043).
- Johnson TE. Science 1990; 249: 908–912. [DOI] [PubMed]
- Mendenhall A, Driscoll M, Brent R. Aging Cell 2016; 15: 4–13. [DOI] [PMC free article] [PubMed]
- Bernhardt I, Ellory JC (eds). Red Cell Membrane Transport in Health and Disease. Springer: Heidelberg, Germany, 2003. [Google Scholar]
- Minetti G et al. Blood Rev 2013; 27: 91–101. [DOI] [PubMed]
- Brovelli A, Minetti G. In: Bernhardt I, Ellory JC (eds). Red Cell Membrane Transport in Health and Disease. Springer: Heidelberg, Germany, 2003, pp 673–690.
- Lutz HU, Bogdanova A. Front Physiol 2013; 4: 387. [DOI] [PMC free article] [PubMed]
- Rapoport SM. The Reticulocyte. RC Press: Boca Raton, FL, USA, 1986. [Google Scholar]
- Clark MR. Physiol Rev 1988; 68: 503–554. [DOI] [PubMed]
- Kumar D, Rizvi SI. Rejuvenation Res 172014: 446–452. [DOI] [PMC free article] [PubMed]
- Minetti G, Low PS. Curr Opin Hematol 1997; 4: 116–121. [DOI] [PubMed]
- Lang KS et al. Cell Physiol Biochem 2005; 15: 195–202. [DOI] [PubMed]
- Inaba M, Maede Y. Biochim Biophys Acta 1988; 944: 256–264. [DOI] [PubMed]
- Vacha JRed Cell life-span. In: Agar NS, Board PG (eds). Red Blood Cells of Domestic Mammals. Elsevier: Amsterdam, The Netherlands, 1983. pp 67–132. [Google Scholar]
- Bogdanova A et al. Int J Mol Sci 2013; 14: 9848–9872. [DOI] [PMC free article] [PubMed]
- Beckman KB, Ames BN. Physiol Rev 1998; 78: 547–581. [DOI] [PubMed]
- Bratosin D et al. Biochimie 1998; 80: 173–195. [DOI] [PubMed]
- Risso A et al. Front Physiol 2014; 5: 54. [DOI] [PMC free article] [PubMed]
- Wang J et al. PLoS ONE 2013; 8: e67697. [DOI] [PMC free article] [PubMed]
- Toyama BH, Hetzer MW. Nat Rev 2013; 14: 55–61. [DOI] [PMC free article] [PubMed]