Abstract
During the brain development, the process of neural stem cells (NSCs) proliferation and differentiation is precisely regulated. The deficiency in the embryonic brain development will cause serious developmental disorders. Epigenetic modifications play critical roles in controlling proliferation and differentiation in different types of stem cells. Histone variants, as one of epigenetic regulators, have been reported to be associated with many bioprocesses. Among different variants, H3.3 is one of the important epigenetic regulators, but its role in embryonic NSCs remains unclear. Here we demonstrate that H3.3 is intrinsically required for NSCs proliferation and differentiation. Suppression of the H3.3 mediated by shRNAs causes the reduction of the PAX6-positive NSCs proliferation, and promotes the premature terminal mitosis and neuronal differentiation. Particularly, the level of the H4K16ac is selectively reduced in the H3.3 knockdown NSCs. We further confirm that H3.3 is directly interacted with the MOF, a specific H4K16 acetyltransferase. Interestingly, H3.3/MOF increases the level of H4K16ac by a mutual cooperation manner. However, the H3.3K36R mutant could not increase the level of H4K16ac. RNA-seq data show the GLI1, a transcriptional regulator, is downregulated in H3.3 knockdown NSCs. Furthermore, the neurogenesis phenotype of the GLI1 knockdown is consistent with the H3.3 knockdown. Overexpression of the H3.3, MOF, and GLI1 could rescue the abnormal phenotype caused by H3.3 knockdown in the embryonic brain, but H3.1 or H3.3K36R overexpression can not rescue it. Taken together, these results suggest that H3.3 cooperates with MOF to increase the level of the H4K16ac and the GLI1, and then regulates the NSCs proliferation and differentiation.
The formation of normal functional mammalian brain requires precise regulation of proliferation and differentiation of neural stem cells (NSCs).1, 2, 3, 4 In the neocortex, radial glial (RG) cells give birth to self-renewing cells and intermediate progenitor (IP) cells. IP cells subsequently produce neurons.5, 6 During the development, the NSCs acquire the specific neuronal traits7 to produce different types of neurons, which are segregated into specific cell layers to form the basic framework of the cerebral cortex.8
Epigenetic modifications on histones or histone variants play an important role in the DNA replication. Most mammals contain two similar H3 family members named canonical H3 and replacement H3.3. The histone variant H3.3 is highly conserved evolutionarily.9 In most animals, H3.3 differs from H3 by just four amino acid substitutions,10 but it has been confirmed to play specific and crucial roles in the regulation of chromatin dynamics and transcription.11 H3.3 is deposited into transcribed genes and gene regulatory elements and considered as a symbol of transcriptional activated genes.12, 13 However, previous study has also shown that the H3.3 is enriched in the repressed genes.14 These findings indicate that the function of the H3.3 in the different system needs to be further investigated. During the development, the histone H4 contains some different post-transcriptional modification. Among them, Lys16 of histone H4 (H4K16) is different from other acetylated residues: its distribution on each chromosome is average;15 it forms a block of the transmission of histone hypoacetylation and the silencing of the gene expression. Previous study has shown that H4K16ac is tightly associated with self-renewal and differentiation of the embryonic stem cell.16 Furthermore, whether there is a crosstalk between H3.3 and H4K16ac in regulating the development of the embryonic brain remains largely unexplored.
There are three family members of the GLI transcription factors, but the function of different Gli protein is distinct.17 Previous study about the GLI1 mainly focuses on its role in the cancer.18 It is also reported that GLI1 plays an important role in the remyelination.19 GLI1 is an active protein during the different bioprocess, and it could be activated by a variety of environment factors such as the DNA damage,20 and cytokines.21 However, it remains largely unknown whether GLI1 affects stem cells fate and differentiation in the vertebrate brain during the early development.
To investigate the function of histone H3.3 in early brain development, we downregulate its expression in embryonic NSCs via in utero electroporation (IUE) of H3.3 shRNA, and the data show that H3.3 knockdown decreases the proliferation of RG cells, and increases the ratio of the neurons during the cortical development via reducing the acetylation on the H4K16. The results demonstrate that H3.3 could interact with MOF, the acetyltransferase of the H4K16, and H3.3/MOF corporately increases the level of H4K16ac by a mutual cooperation way. Also, the reduction of the H3.3 decreases the recruitment of the MOF, and then decreases the level of the H4K16ac. The decreasing level of the H3.3 and H4K16ac leads to the reduction of GLI1 expression, which ultimately controls the proliferation and neuronal differentiation during cortical development. Together, we demonstrate an important signaling link between the histone variant H3.3, H4 acetylation, and GLI1 on the regulation of brain development.
Results
Histone H3.3 Is expressed in the NSCs
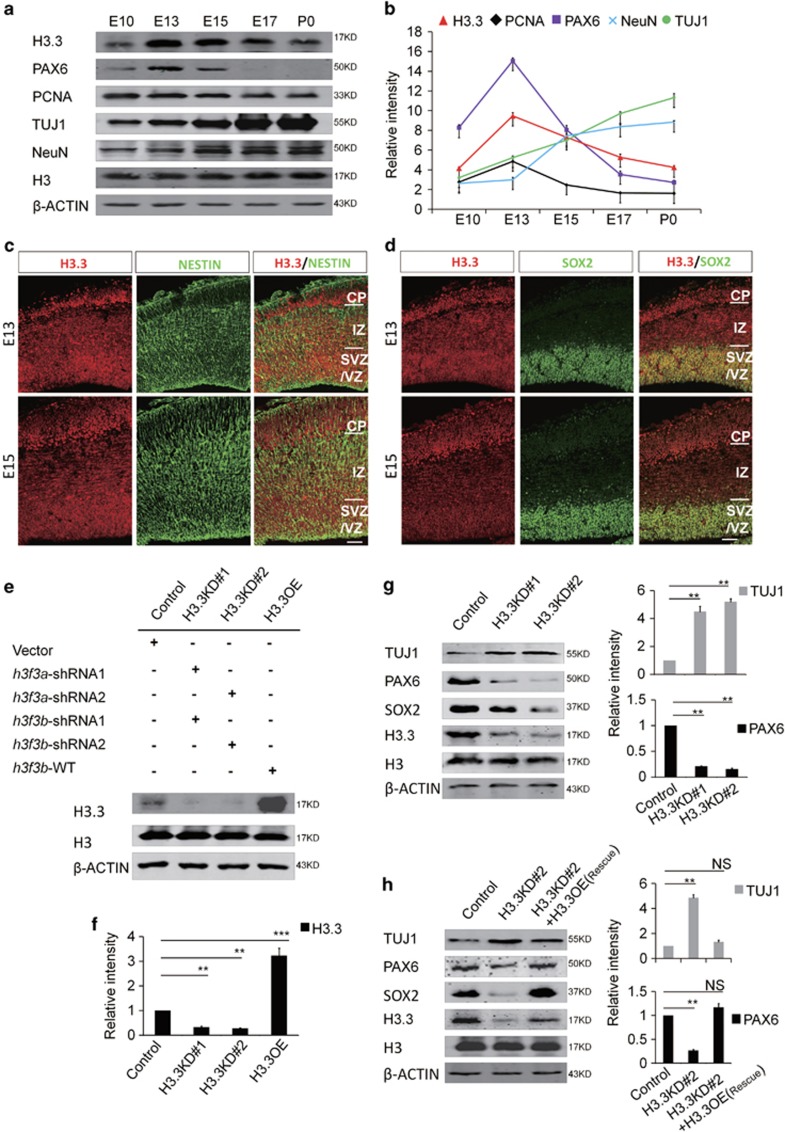
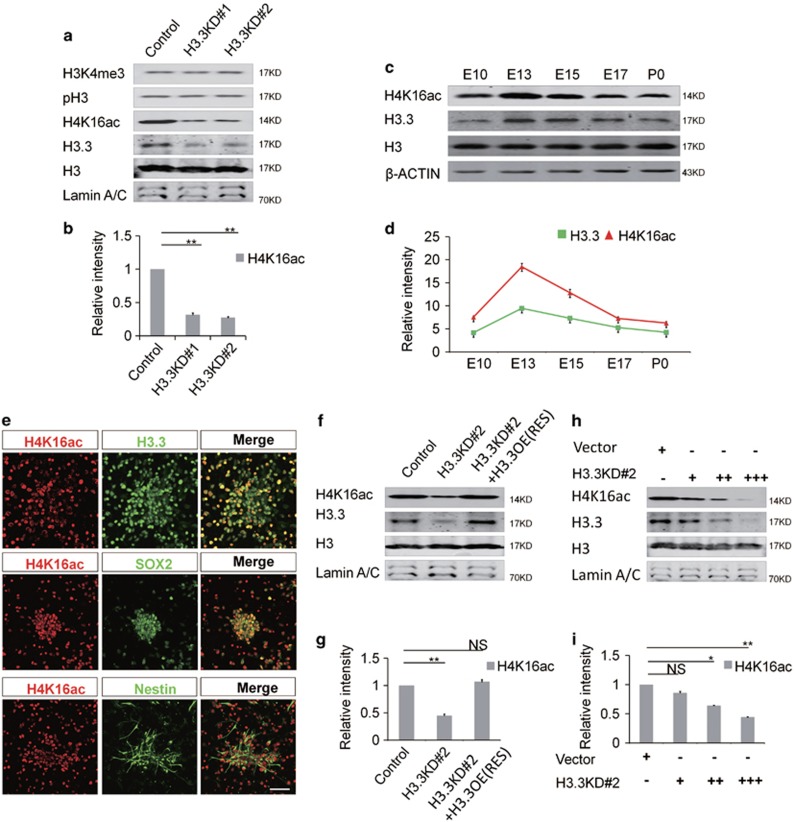
Previous study has shown that the level of histone H3.3 is generally accumulated with age. However, the expression of H3.3 during the embryonic development remains unexplored. We performed western blot to check the H3.3 expression pattern in brain tissue during different developmental periods. Our results showed that the expression of H3.3 is dynamic during early brain development. The results showed that histone H3.3 was detectable at E10, and its expression was gradually increased until E13.5. After that, the expression was reduced between E15.5 and P0 (Figure 1a). The expression pattern of H3.3 was similar with PAX6, which is a progenitor cell marker. This result suggests that Histone H3.3 may play a key role in early embryonic brain development (Figure 1b).
Figure 1.
Histone H3.3 Is expressed in embryonic neural progenitor cells. (a) H3.3 and different neurogenesis markers are present in cortical lysates collected at various time points (E10, E13, E15, E17, and P0). (b) A scatter diagram shows the change trend of H3.3 and different neurogenesis markers. (n=3; bar represents mean±S.E.M). (c) E13 and E15 embryonic brain sections were co-stained with anti-H3.3 and anti-NESTIN antibodies. The scale bar represents 25 μm. (d) E13 and E15 embryonic brain sections were co-stained with anti-H3.3 and anti-SOX2 antibodies. The scale bar represents 25 μm. (e,f) In the NSCs, the level of H3.3 protein is reduced by shRNA-mediated interference (H3.3KD#1: h3f3a-shRNA-1 combine with h3f3b-shRNA-1; H3.3KD#2: h3f3a-shRNA-2 combine with h3f3b-shRNA-2). H3.3 protein is increased by h3f3b-WT overexpression (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01, ***P<0.001; β-ACTIN served as loading control). (g) Protein level of neural stem cell markers, including PAX6 and SOX2 are reduced in the H3.3 knockdown NSCs versus the control. However, neuronal differentiation marker TUJ1 is increased (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; β-ACTIN served as loading control). (h) Protein level changes of PAX6, SOX2, and TUJ1 caused by H3.3KD#2 (h3f3a-shRNA-2 targeting to UTR and h3f3b-shRNA-2) knockdown can be rescue by H3.3 (h3f3a) overexpression in NSCs (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; β-ACTIN served as loading control)
In the embryonic cortex, the immunostaining result showed that the histone H3.3 protein co-localized with NESTIN- and SOX2-positive neuronal progenitor cells residing in the VZ/SVZ at E13.5 and E15.5 (Figures 1c and d). Moreover, we found that H3.3 co-localized with NESTIN- and SOX2-positive cultured NSCs (Supplementary Figure 1a). Since H3.3 protein was encoded by h3f3a and h3f3b genes simultaneously, we generated two h3f3a-specific short hairpin RNA (shRNA) (h3f3a shRNA-1 and h3f3a shRNA-2) and two h3f3b -specific short hairpin RNA (shRNA) (h3f3b shRNA-1 and h3f3b shRNA-2) plasmids. The knockdown efficiency was tested and the combination of h3f3a shRNA-1 with h3f3b shRNA-1 (H3.3KD#1) and h3f3a shRNA-2 with h3f3b shRNA-2 (H3.3KD#2) could efficiently silence H3.3 expression in vivo and in vitro (Figure 1e,Supplementary Figures 1b and 1c). We also constructed H3.3-overexpression plasmid to overexpress H3.3 expression in embryonic NSCs (Figures 1e and f). We found that H3.3 knockdown decreased the expression of neural stem cell markers PAX6 and SOX2, and increased the expression of neurogenesis marker TUJ1 (Figure 1g). Also, H3.3 (h3f3a) overexpression could rescue the abnormal phenotype caused by H3.3 knockdown (h3f3a shRNA-2 targeting to UTR and h3f3b shRNA-2) (Figure 1h). These results suggest that H3.3 participates in the process of the early neurogenesis.
H3.3 regulates neural progenitor cell proliferation
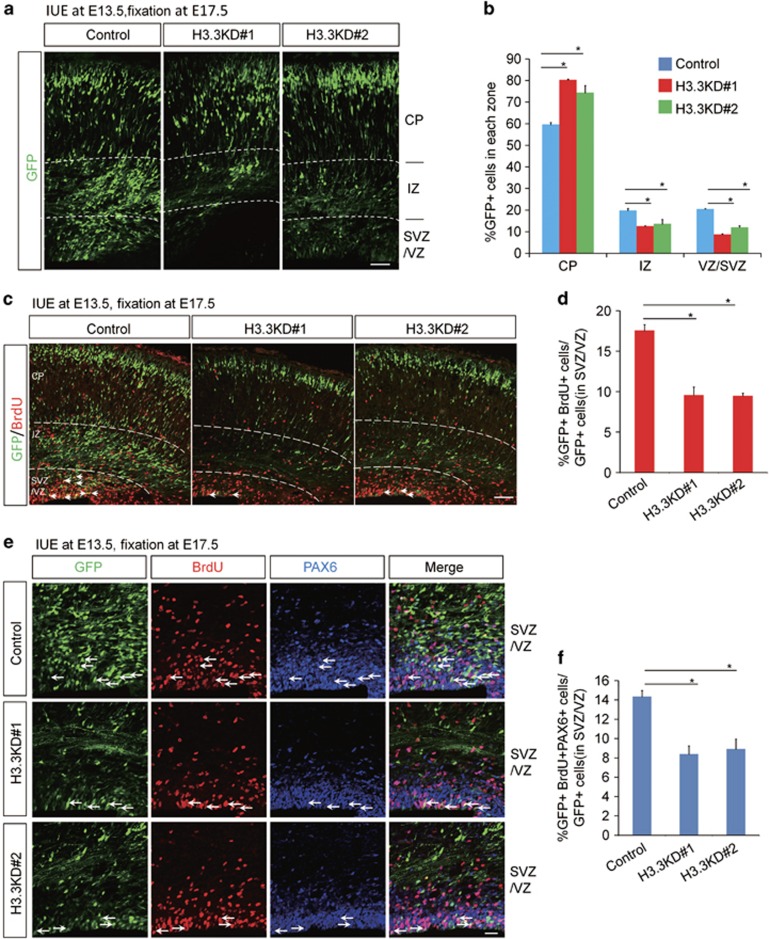
We employed in utero electroporation (IUE) to introduce H3.3 shRNA cocktails with a Venus-GFP into neural progenitor cells of the developing cortex in E13.5 mouse embryos, and brains were collected at E17.5 for phenotypic analysis. We found that knockdown of H3.3 resulted in an obvious change in cell distribution compared to controls. There was a significant loss of GFP-positive cells in the proliferation zone ventricular/subventricular zone (VZ/SVZ), and the GFP-positive cells in the intermediate zone (IZ) were also reduced. H3.3 knockdown caused an augmentation of GFP-positive cells in the cortical plate (CP) (Figures 2a and b). To test whether the reduction of GFP-positive cells in VZ/SVZ was due to decreased neural progenitor cells proliferation, pregnant dams were injected with BrdU 2 h prior to brains collection at E17.5. Knockdown of H3.3 resulted in a 40% decrease of BrdU and GFP double-positive cells (Figures 2c and d), and similar result was obtained in vitro (Supplementary Figures 3a and 3b). However, there is no difference between control and canonical histone H3.1-overexpression, at the same time, the phenotype caused by H3.3 knockdown could not be rescued by H3.1-overexpression (Supplementary Figure 2). Our results showed that BrdU and PAX6 double-positive dividing NSCs in H3.3 knockdown group was decreased 42% compared to the control (Figures 2e and f). These results suggest that H3.3 is required for the proliferation of NSCs.
Figure 2.
H3.3 regulates the proliferation of the NSCs. (a,b) H3.3 knockdown causes GFP-positive cell positioning changes in embryonic brain. The empty shRNA control or H3.3KD plasmid was electroporated into E13.5 embryonic mouse brains, and the phenotype was analyzed at E17.5. The percentage of GFP-positive cells in each region is shown (n=3 independent experiments; error bars represent mean±S.E.M; *P<0.05). The scale bar represents 50 μm. (c,d) BrdU incorporation is reduced in H3.3 knockdown plasmids-electroporated sections. The mouse brain was electroporated at E13.5 and BrdU (50 mg/kg) was injected 2 h prior to killing at E17.5. The white arrows indicate GFP and BrdU double-positive cells. The bar graph shows the percentage of BrdU and GFP double-positive cells relative to the total GFP-positive cells in the VZ/SVZ (n=3 independent experiments; *P<0.05, bars represent mean±S.E.M). The scale bar represents 50 μm. (e,f) The percentage of GFP+BrdU+PAX6+ cells is decreased in H3.3 knockdown brains. The mouse brain was electroporated at E13.5 and BrdU (50 mg/kg) was injected 2 h prior to killing at E17.5. The white arrows indicate GFP+BrdU+PAX6+ cells. The bar graph shows the percentage of GFP+BrdU+PAX6+ cells relative to the total GFP-positive cells in the VZ/SVZ (n=3 independent experiments; *P<0.05; bars represent mean±S.E.M). The scale bar represents 20 μm
H3.3 regulates neural progenitor cell differentiation and neuronal development
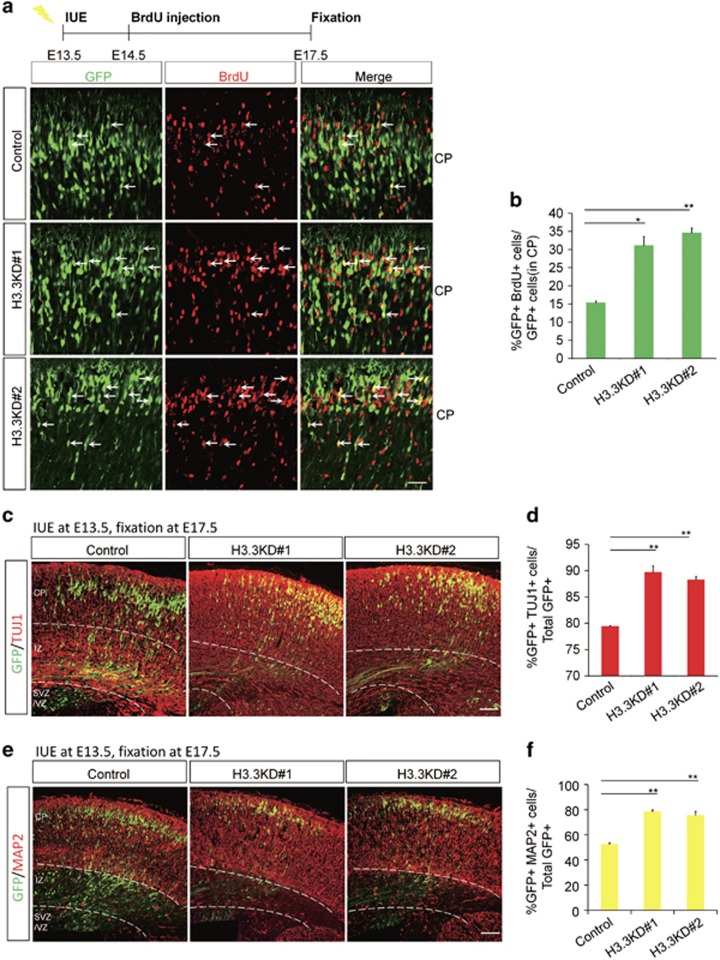
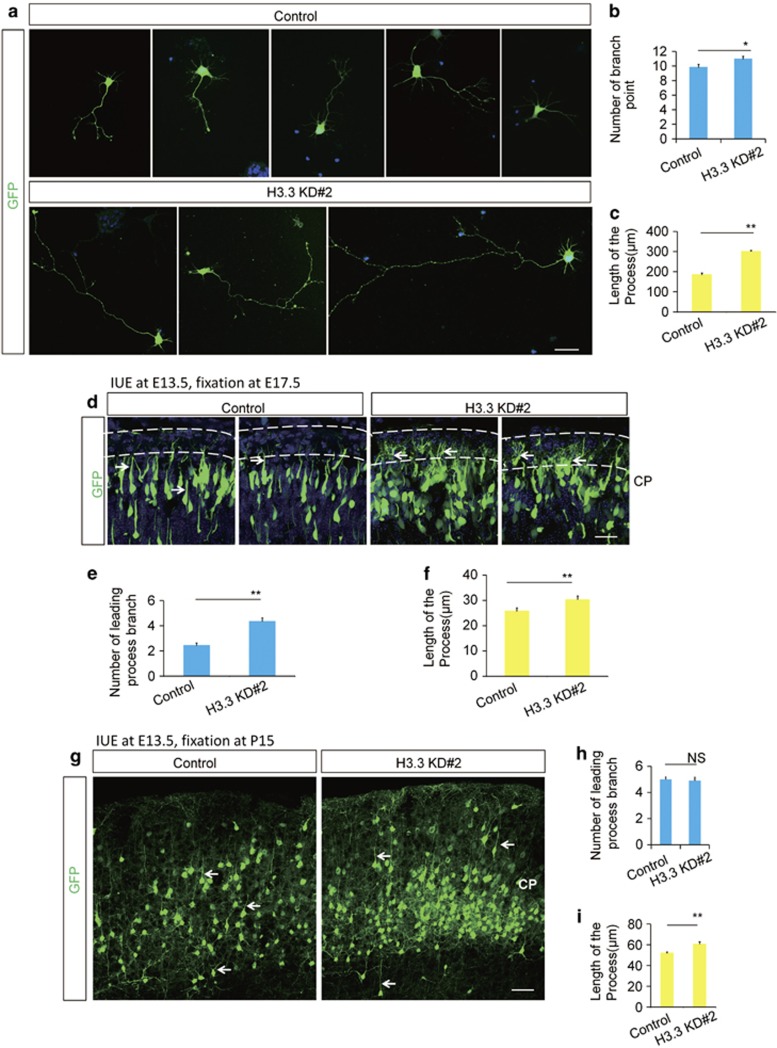
To determine whether H3.3 regulates premature NSCs terminal mitosis to influence the neurogenesis process during embryonic cortical development, BrdU birth-dating experiment was performed. As the timeline, a single BrdU injection (50 mg/kg) was administered to pregnant dams 24 h after IUE, and the brains were collected at E17.5 for phenotypic analysis. Because only those NSCs at S phase of mitosis could absorb the BrdU, NSCs in their final mitotic division at the time of BrdU injection were labeled with BrdU permanently, and they could carry the BrdU-label during the process of the neurogenesis and migrate to the CP. Double-labeled (BrdU+GFP+) cells of newly born neurons in the CP showed a two-fold increase in H3.3 knockdown brains compared with the control (Figures 3a and b). To test whether H3.3 knockdown directly leads to increased neuronal differentiation, we immunostained electroporated brain sections with neuronal markers of TUJ1 and MAP2. The ratio of GFP-TUJ1 double-positive cells revealed that the knockdown of H3.3 led to a significant increase in the percentage of GFP-labeled cells that were also positive for TUJ1 (Figures 3c and d), and in vitro experiment showed the similar result (Supplementary Figures 3c and 3d). And the quantification of GFP-MAP2 double-positive cells revealed that H3.3 knockdown lead to an obvious increase in the percentage of MAP2+ neurons (Figures 3e and f). These results demonstrated that H3.3 knockdown resulted in an increase in neuronal differentiation. Further study revealed that the abnormal phenotype caused by H3.3 knockdown was not induced by apoptosis and neuronal migration (Supplementary Figure 4). We also found that the neuronal development was abnormal in H3.3 knockdown neurons. The length of the neuronal process in cultured neurons increased about 1.5 times in the H3.3 knockdown group versus the control (Figures 4a–c), and the structure of the leading process was more complex in H3.3 knockdown brain at E17 (Figures 4d–f) and P15 (Figures 4g–i). All the data indicate that H3.3 is involved in neural stem cell differentiation and neuronal development.
Figure 3.
H3.3 Knockdown promotes neural progenitor differentiation. (a,b) Timeline of the BrdU birth-dating experiment. Images show brain sections from control or H3.3 knockdown mice were immunostained with an anti-BrdU antibody. The white arrows indicate GFP and BrdU double-positive cells. The scale bar represents 25 μm. The bar graph shows the percentage of BrdU and GFP double-positive cells relative to total GFP-positive cells in the CP (n=3 independent experiments; *P<0.05; bars represent mean±S.E.M). (c–f) H3.3 knockdown increases neuronal differentiation. E17.5 brain sections were stained for TUJ1 (c) or MAP2 (e) after the electroporation of the control or H3.3 knockdown plasmid into the brain at E13.5. The quantification of the percentage of GFP and TUJ1 double-positive cells (d) or GFP and MAP2 double-positive cells (f) relative to the total GFP-positive cells is shown as a bar graph (n=3 independent experiments; *P<0.05; bars represent mean±S.E.M). The scale bar represents 50 μm
Figure 4.
H3.3 Knockdown causes the abnormal neuronal development. (a–c) H3.3 regulates the morphological maturation of neurons in vitro. GFP-positive cells isolated from 2 days post-electroporation brains by H3.3 knockdown (H3.3KD#2) and control were cultured for 72 h. The bar graph shows the number of branch point (b) and the length of the process(c) in vitro. (*P<0.05, bars represent mean±S.E.M). The scale bar represents 5 μm. (d–f) H3.3 regulates the morphological maturation of neurons in vivo at E17.5. The bar graph shows the number of branch point (e) and the length of the process (f) in vivo. (n=11 cells from 3 brains for each condition; *P<0.05, **P<0.01, bars represent mean±S.E.M). The scale bar represents 50 μm. (g–i) H3.3 regulates the neuronal development in vivo at P15. The bar graph shows the number of branch point (h) and the length of the process (I) in vivo. (n=3 independent experiments; **P<0.01, bars represent mean±S.E.M). The scale bar represents 50 μm
H3.3 knockdown decreases the H4K16 acetylation
Loss of H3.3 has been shown to be associated with the abnormal nuclear morphology,22 which suggests that H3.3 may play an important role in histone modification. Because H3.3 is associated with the activation of transcription, we detected some markers of transcriptional activation chromatin (H3K4me3, H3S10p and H4K16ac) by western blot after H3.3 knockdown to determine whether there is crosstalk between them. Interestingly, the protein level of H4K16ac, a hallmark of hyperactive chromatin, was significant downregulated after H3.3 knockdown in NSCs. But other histone modifications levels were not changed (Figures 5a and b). And the immunostaining showed that the H4K16ac was significance reduced in NSCs when H3.3 was repressed (Supplementary Figures 5a and 5b). For further study, we tested whether there is relationship between H3.3 and H4K16ac by western blot and immunostaining. Our results showed that, during the process of the neurogenesis, the protein expression of the H4K16ac was consistent with the H3.3. Like the expression pattern of H3.3, H4K16ac was detectable at E10, and its expression was gradually increased until E13.5. After that, the expression was decreased between E15.5 and P0 (Figures 5c and d). The immunostaining showed that H4K16ac was co-localized with H3.3, NESTIN, and SOX2 (Figure 5e). And we found that H3.3(h3f3a) overexpression could rescue the reduction of the H4K16ac caused by H3.3 knockdown (Figures 5f and g). Further study showed that there was a dose–response relationship between H3.3 and H4K16ac. When H3.3 was gradually decreased by different dose of H3.3 shRNA cocktail, the level of the H4K16ac was reduced as well (Figures 5h and i). Thus, H3.3 regulates the level of H4K16ac during brain development.
Figure 5.
H3.3 Knockdown decreases the acetylation of H4K16. (a,b) Protein levels of different transcriptional activation markers are detected in the H3.3 knockdown NSCs versus the control. Among them, H4K16 is selectively downregulated (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control). (c,d) H4K16ac, H3.3, are detected at various time points (E10, E13, E15, E17, and P0) in cortical lysates. Samples for western blot analysis are shown. A scatter diagram shows the change trend of H4K16ac, H3.3, and different neurogenesis markers (n=3 independent experiments; bar represents mean±S.E.M; β-ACTIN served as loading control). (e) NSCs were isolated from the E12.5 mice brains and cultured in the proliferative medium for 24 h. The cells were co-stained with anti-H4K16ac and anti-H3.3 antibodies; anti-H4K16ac anti-SOX2 antibodies; anti-H4K16ac and anti-NESTIN antibodies. Scale bar represents 25μm. (f,g) Protein level change of H4K16ac can be rescued by H3.3 overexpression in NSCs (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control). (h,i) The level of H4K16ac was gradually reduced by gradually increased dose of H3.3KD#2 shRNA-mediated interference in NSC cells (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control)
H3.3 and MOF promote the acetylation in the collaborative role
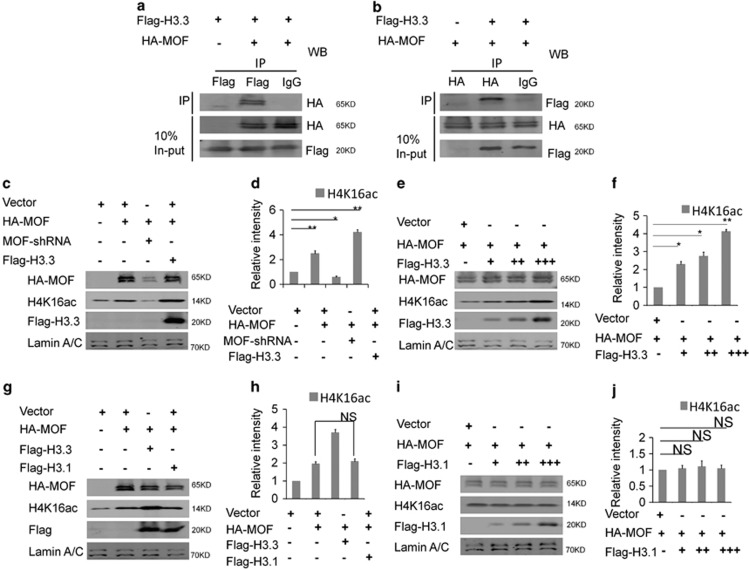
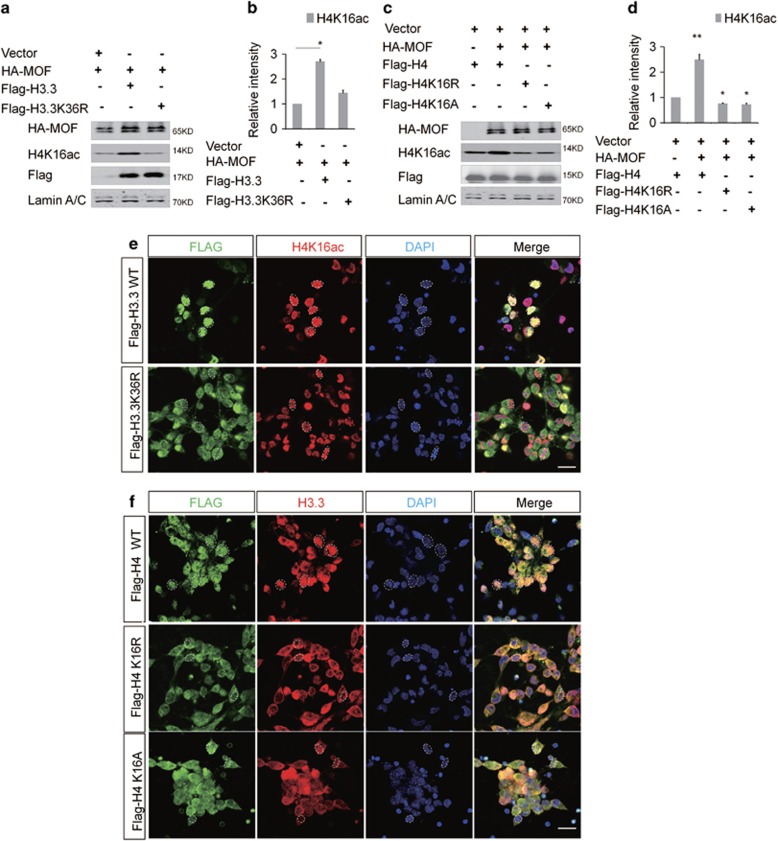
To test whether the reduction of the H4K16ac was due to mof expression or not, the Mof mRNA was detected and there was no significant change between H3.3 knockdown and control (Supplementary Figure 5c). The co-immunoprecipitation results showed that H3.3 interacted with the MOF directly (Figures 6a and b). And the western blot result showed H3.3 and MOF promoted the acetylation in the collaborative role. When H3.3 and MOF were co-expressed in NSC, the level of H4K16ac was increased versus MOF alone (Figures 6c and d). We also found that there was a dose–response relationship between H3.3 and MOF in promoting the acetylation of the H4K16. The level of H4K16ac was gradually augmented with the gradual increase dose of H3.3 (Figures 6e and f). However, the level of the H4K16ac did not change when H3.1 was overexpressed (Figures 6g–j). Previous study has demonstrated that H3K36me3, a transcriptional activation marker, is enriched at the H3.3 versus H3.23 Our results showed the overexpression of H3.3 could increase the level of H3K36me3 (Supplementary Figure 5d and 5e). In turn, the augmentation of the H4K16ac promoted the level of the H3K36me3 (Supplementary Figure 5f). The H3K36me3 was co-localized with the H3.3 in NSCs (Supplementary Figure 5g). To further investigate the relationship between H3.3 and MOF, H3.3 mutant (H3.3K36R) was constructed by changing amino acid of Lys36 to Arg36 of the H3.3. The western result showed that H3.3K36R failed to promote the acetylation like H3.3-WT (Figures 7a and b). We also constructed H4 mutant of H4K16R and H4K16A by changing Lys16 to Arg16 or Ala16, the level of H4K16ac was decreased when H4K16R or H4K16A was overexpressed (Figures 7c and d). NSCs were infected with H3.3 or H3.3K36R, and the immunostaining results showed that there was significantly different distribution between H3.3K36R, and H3.3. H3.3 was mostly distributed in the nucleus area, but H3.3K36R was distributed in the whole cell area. Consistent with the western bolt, the fluorescence intensity of the H4K16ac in H3.3-infected NSCs is stronger than the group of H3.3K36R. And we also found more abnormal nuclear morphology in Flag-H3.3K36R infected NSCs versus the Flag-H3.3-infected NSCs (Figure 7e). We also observed that the distribution of Flag-H4K16R and H4K16A was different with H4 infected NSCs, respectively. H4 was distributed in the nucleus area, and H4K16R and H4K16A were mostly distributed in the whole cell area. Moreover, the distribution of H3.3 was also abnormal in H4K16R- and H4K16A-infected NSCs versus the H4 (Figure 7f). These results suggest the modifications of Lys36 of H3.3 and Lys16 of H4 are the important epigenetic regulation sites in NSCs.
Figure 6.
H3.3 interacts with MOF directly, and H3.3/MOF promotes the acetylation of H4K16 in the collaborative role. (a) HA-MOF was immunoprecipitated by Flag-H3.3. The indicated plasmids were transfected into 293T cells. Protein lysates were divided into two parts: one for the in-put and the other for the indicated Flag-labeled magnetic beads IP. The immunoprecipitated proteins were probed with anti-HA antibodies to detect HA-MOF. (b) Flag-H3.3 was immunoprecipitated by HA-MOF. The indicated plasmids were transfected into 293 T cells. The immunoprecipitated proteins were probed with anti-Flag antibodies to detect Flag-H3.3. (c,d) H3.3 and MOF promote the acetylation of the H4K16 in the collaborative role. The indicated viruses were delivered into NSCs, and the protein level of H4K16ac was detected (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control). (e,f) H3.3 and MOF promote the acetylation of the H4K16 in the collaborative dose-response role. The indicated viruses were delivered into NSC, and the protein level of H4K16ac was detected (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control). (g–j) H3.1 could not promote the acetylation of the H4K16. The indicated viruses were delivered into NSCs, and the protein level of H4K16ac was detected (n=3 independent experiments; bar represents mean±S.E.M; Lamin A/C served as loading control)
Figure 7.
H3.3K36 and H4K16 Mutation Influence the H4K16 Acetylation. (a, b) H3.3K36R could not promote the acetylation of the H4K16 as H3.3-WT. The indicated viruses were delivered into NSC, and the protein level of H4K16ac was detected (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control). (c,d) Overexpression of H4K16R or H4K16A could not promote the acetylation of H4K16 in NSCs. The indicated viruses were delivered into NSCs, and the protein level of H4K16ac was detected (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; Lamin A/C served as loading control). (e) Abnormal distribution of Flag-H3.3K36R decreases the fluorescence intensity of the H4K16ac in NSCs versus the Flag-H3.3WT. (f) Abnormal distribution of Flag-H4K16R (or H4K16A) versus the Flag-H4WT
GLI family members are highly suppressed in H3.3 knockdown NSCs
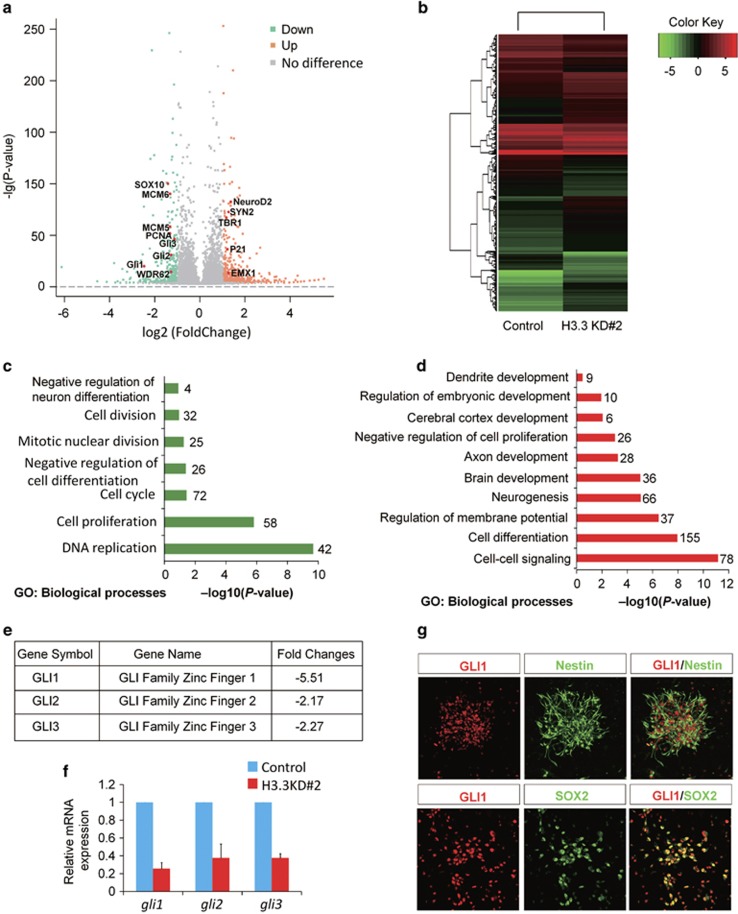
Next, to investigate the gene expression changes in H3.3 knockdown cells, we performed RNA-sequencing (RNA-seq) to analyze the genome-wide changes by H3.3 dysfunction. The NSCs were obtained from the E12.5 embryonic cortex, and the H3.3 knockdown virus cocktail or control virus was delivered into the cells. The cells were cultured for 4 days, and then the RNA was extracted for RNA-seq. The volcano map (Figure 8a) and the heat map (Figure 8b) showed that the gene profiling expression was changed between the H3.3 knockdown and control. The genes analysis revealed that transcript levels of 1081 genes were upregulated or downregulated by more than two-fold between the H3.3 knockdown and the control. GO term analysis of downregulated genes in samples showed a significant enrichment of biological processes related to NSCs proliferation (for example: cell division, cell proliferation, negative regulation of neuron differentiation, and mitotic nuclear division) (Figure 8c). GO term analysis of upregulated genes showed a significant enrichment of biological processes related to neuronal differentiation (for example: neurogenesis, cerebral cortex development, brain development, cell differentiation, and dendrite development) (Figure 8d). After further analyzing the data of the RNA-seq, we found that all members in the GLI Zinc finger family were changed more than 2-fold (Figure 8e). To confirm whether the GLI family members’ expression was downregulated, we performed realtime-PCR to detect the expression of Gli1, Gli2, and Gli3 in control and H3.3 knockdown NSCs. The data showed that there was a strong reduction of Gli1, Gli2, and Gli3 expression. Consistent with the RNA-sequencing results, Gli1 was the most decreased gene among them (Figure 8f). Moreover, the immunostaining results showed that GLI1 co-localized with NESTIN- and SOX2 in cultured neuronal progenitor cells, which were isolated from E12.5 mouse brains and cultured in proliferative medium for one day (Figure 8g). These results indicate that GLI1 may be the downstream target of H3.3 involved in early neurogenesis.
Figure 8.
GLI zinc finger family is highly suppressed in H3.3 knockdown NSCs. (a,b) The volcano map (a) and the heat map (b) show that the gene profiling expression. Genes analysis reveals that transcript levels of 1081 genes were upregulated or downregulated by more than two-fold between the H3.3 knockdown and the control. (c,d) Enrichment analysis for biological process. Gene ontology (GO) terms associated with the genes downregulated (c) or upregulated (d) in H3.3 knockdown NSCs versus the control. (e) RNA-seq analysis shows the mRNAs expression level of GLI family members are dramatic reduced in H3.3 knockdown NSCs versus the control. (f) Realtime-PCR analysis of GLI family members gene expression in H3.3 knockdown NSCs versus the control (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01). (g) GLI1 is co-localized with the NSCs markers of NESTIN and SOX2. NSCs were isolated from the E12.5 mice brains and cultured in the proliferative medium for 24 h. The cells were co-stained with anti-GLI1 and anti-NESTIN; anti-GLI1 and anti-SOX2 antibodies. Scale bar represents 25 μm
The level of GLI1 was regulated by H3.3
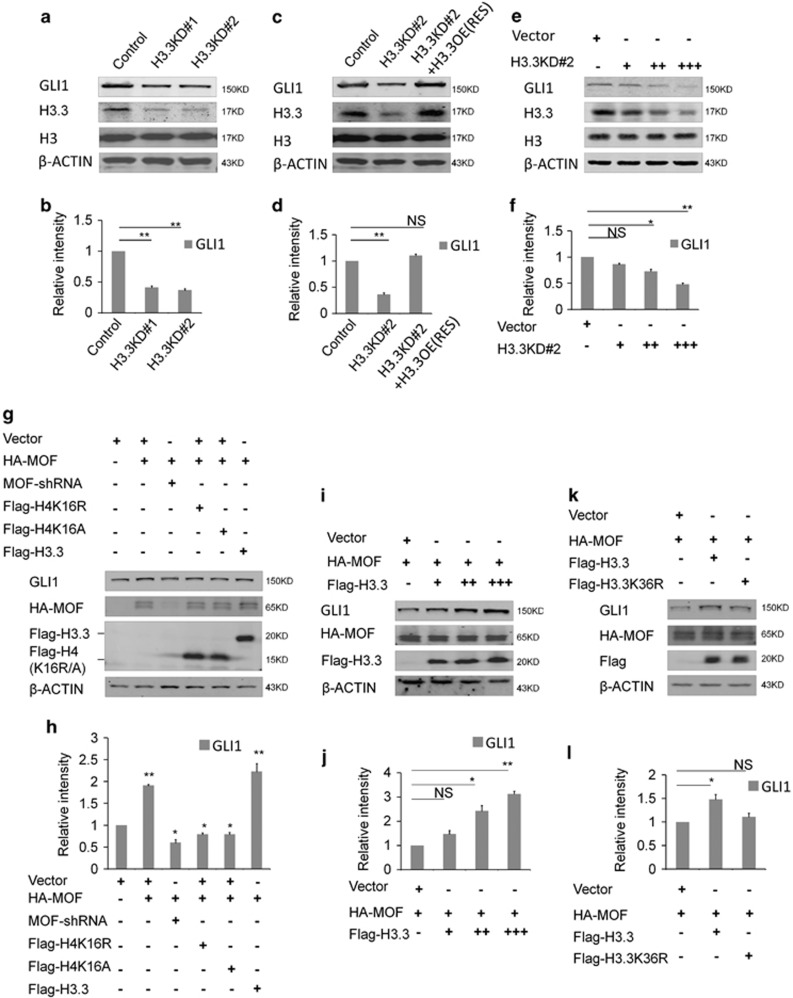
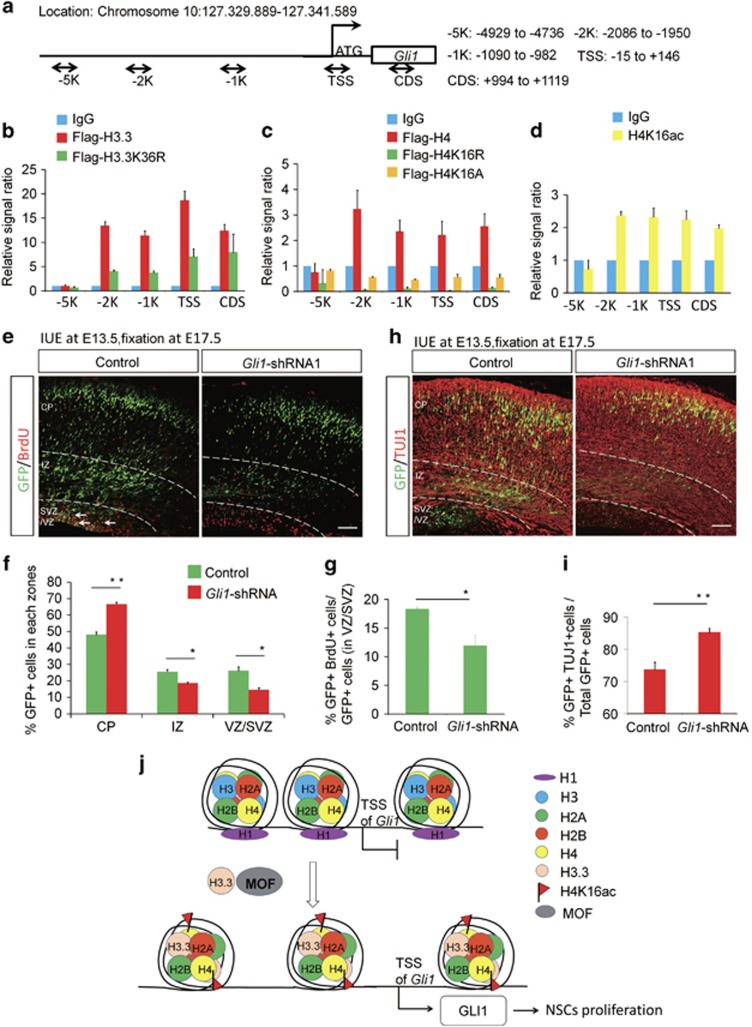
Previous study has proved that GLI1 acts as a strong transcriptional activator.24 Therefore, we want to know whether GLI1 regulated by histone H3.3 promotes the activation of transcription sequentially in early brain development. The results showed that the level of GLI1 was decreased when the H3.3 was knockdown in NSCs (Figures 9a and b). And we found that H3.3(h3f3a) overexpression could also rescue the reduction of the GLI1, which was caused by H3.3 knockdown in NSCs (Figures 9c and d). Further study showed that there was a dose–response relationship between H3.3 and GLI1. When H3.3 was gradually decreased by different dose of shRNA, the protein level of the GLI1 was reduced as well (Figures 9e and f). We also found the collaborative role between H3.3 and MOF and they both promoted the expression of the GLI1 significantly than the MOF itself. When MOF was knockdown, the expression of the GLI1 was reduced. H4K16R and H4K16A could both reduce the expression of the GLI1 (Figures 9g and h). When H3.3 was gradually increased by different dose of H3.3WT, the protein level of the GLI1 was augmented as well (Figures 9i and j). The western result showed that H3.3K36R could not promote the expression of GLI1 in NSC (Figures 9k and l). The ChIP-realtime PCR showed that H3.3, H4, and H4K16ac could bind to the Gli1 promoter in NSCs, but the binding of H3.3K36R, H4K16R, and H4K16A mutants to the Gli1 promoter was reduced in NSCs (Figures 10a–d). These results suggest the H3.3 and MOF regulate the expression of the GLI1 in a collaborative way.
Figure 9.
H3.3 regulates the expression of the GLI1. (a,b) Protein level of GLI1 is decreased in the H3.3 knockdown NSCs versus the control (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; β-ACTIN served as loading control). (c,d) Protein level change of GLI1 can be rescue by H3.3 overexpression (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; β-ACTIN served as loading control). (e,f) The level of GLI1 protein was gradually reduced by gradually increased dose of H3.3KD shRNA-mediated interference in NSC (n=3 independent experiments; bar represents mean±S.E.M; *P<0.05, **P<0.01; β-ACTIN served as loading control). (g,h) H3.3 and MOF promote the expression of the GLI1 in the collaborative role in NSCs. The indicated viruses were delivered into NSC cells, and the protein level of GLI1 was detected (n=3 independent experiments; bar represents mean±S.E.M; *P<0.05, **P<0.01; β-ACTIN served as loading control). (i,j) H3.3 and MOF promote the expression of the GLI1 in the collaborative dose-response role. The indicated viruses were delivered into NSC cells, and the protein level of GLI1 was detected (n=3 independent experiments; bar represents mean±S.E.M; *P<0.05, **P<0.01; β-ACTIN served as loading control). (k,l) H3.3K36R mutant overexpression failed to promote expression of the GLI1. The indicated viruses were delivered into NSCs, and the protein level of GLI1 was detected (n=3 independent experiments; bar represents mean±S.E.M; **P<0.01; β-ACTIN served as loading control)
Figure 10.
GLI1 participates in the process of the embryonic neurogenesis. (a) Sketch map shows the different region selected for ChIP- realtime-PCR. (b) Flag-H3.3 binds to the Gli1 promoter region. However, the binding of Flag-H3.3K36R to each region was decreased versus the Flag-H3.3 in the NSCs (n=3 independent experiments; bars represent mean±S.E.M). (c) Flag-H4 binds to the Gli1 promoter and the gene body. However, the binding of Flag-H4K16R and Flag-H4K16A to each region was decreased versus the Flag-H4 in the NSCs (n=3 independent experiments; bars represent mean±S.E.M). (d) H4K16ac binds to the region of the Gli1 promoter in the NSCs (n=3 independent experiments; bars represent mean±S.E.M). (e,f) GLI1 knockdown causes Gli1-shRNA-GFP cell positioning changes in utero. The empty shRNA control or GLI1 knockdown plasmids was electroporated into E13.5 embryonic mouse brains, and the phenotype was analyzed at E17.5. The percentage of Gli1-shRNA-GFP or Control-GFP cells in each region is shown (n=3 independent experiments; error bars represent mean±SEM; *P<0.05). (g) The bar graph shows the BrdU incorporation is reduced in GLI1 knockdown sections. The mouse brain was electroporated at E13.5 and BrdU (50 mg/kg) was injected 2 hours prior to killing at E17.5. The white arrows indicate GFP and BrdU double-positive cells. The bar graph shows the percentage of BrdU and GFP double-positive cells relative to the total GFP-positive cells in the VZ/SVZ (n=3 independent experiments; *P<0.05, bars represent mean±SEM). The scale bar represents 50 μm. (h,i) GLI1 knockdown increases neuronal differentiation. E17.5 brain sections were stained for TUJ1 after the electroporation of the control or GLI1 knockdown plasmid into the brain at E13.5. The quantification of the percentage of GFP and TUJ1 double-positive cells relative to the total GFP-positive cells is shown as a bar graph (n=3 independent experiments; *P<0.05; bars represent mean±S.E.M). The scale bar represents 50 μm. (j) Model of how H3.3 regulates the level of H4K16ac and GLI1 expression
GLI1 Is participated in the process of the embryonic neurogenesis
To investigate the role of the GLI1 in the early embryonic neurogenesis, we constructed the GLI1 knockdown and overexpression plasmids for further study. We employed IUE to introduce gli1 shRNA into neural progenitor cells of the developing cortex in E13.5 mouse embryos, and brains were collected at E17.5 for phenotypic analysis. We found that the knockdown of GLI1 resulted in a similar phenotype consistent with the H3.3 knockdown in cell distribution compared to controls. There was a significant loss of GFP-positive cells in VZ/SVZ and the GFP-positive cells in the IZ were also reduced. There was augmentation of GFP-positive cells in the CP. To test if the reduction of GFP-positive cells from the VZ/SVZ was due to decreased neural progenitor cells proliferation, pregnant dams were injected with BrdU 2 h prior to brains collection at E17.5. Knockdown of Gli1 resulted in an obvious decrease in BrdU and GFP double-positive cells (Figures 10e–g). And the quantification of GFP-TUJ1 double-positive cells revealed that GLI1 knockdown led to an obvious increase in the percentage of GFP-labeled cells that were also positive for TUJ1 (Figures 10h and i). We determined that GLI1 knockdown could decrease the expression of neural stem cell markers PAX6, and increase the expression of neurogenesis marker TUJ1. And GLI1 overexpression had the opposite phenotype (Supplementary Figure 6). These results demonstrated that GLI1 knockdown resulted in an increase in neuronal differentiation. Finally, we found that the H3.3-WT, MOF-WT, and GLI1-WT could rescue the abnormal phenotype caused by H3.3 knockdown, but the H3.3K36R could not (Supplementary Figures 7a and 7b). And the proportion of the BrdU+GFP+ could also be rescued by H3.3-WT, MOF-WT, GLI1-WT, but not H3.3K36R (Supplementary Figures 7c and 7d). These results suggest that GLI1 acts as the downstream of the H3.3 and participates in the process of the embryonic neurogenesis.
Discussion
Histone variants, including H2A.Z, H2A.X, macroH2A, and H3.3 have been reported to be associated with organ bioprocess.25, 26, 27 Among them, whether H3.3 plays a role in early cortical development arouses our interest. H3.3 protein was encoded by h3f3a and h3f3b genes simultaneously. Previous studies demonstrate that h3f3a28 or h3f3b29 knockout cause serious developmental disorders, and the knockout embryos die. It suggests that H3.3 is required for embryonic development. Previous study has shown that H3.3 is enriched in active chromatin with some covalent modification,30 and H3.3 regulates the biological process with a DNA synthesis dependent or independent way.31, 32
We demonstrate that the expression of the H3.3 is dynamic in the embryonic brain. We report here that the histone variant H3.3 is required for the proliferation of the PAX6-postive NSCs in vivo. H3.3 knockdown causes the abnormal GFP-positive cells distribution via IUE experiment. The ratio of BrdU-positive proliferative NSCs was decreased and the percentage of TUJ1 or MAP2 positive neurons is increased. H3.3 could also regulate the neuronal development in early cortical development. The crosstalk of the histone modifications is frequent.33 Here we report in this study that there is crosstalk between H3.3 and H4K16ac bridged by MOF, which is required for the embryonic cortical development.
In addition, a causal relationship between H3.3 knockdown and the reduction of the H4K16ac has been demonstrated in our study. The H4K16ac has been shown to be associated with many bioprocesses.34 However, the function H4K16ac in the embryonic neurogenesis is unclear. We report that the expression of the H4K16ac is also consistent with the expression of H3.3, PAX6, and PCNA. It is co-localized with the NSCs markers of NESTIN and SOX2. The collaborative role between H3.3 and MOF by promoting the acetylation on the H4K16 provides a new view of the histone crosstalk. Since H3.3 and H4K16ac are critical epigenetic codes and they are conserved in evolution, the crosstalk between H3.3 and H4K16ac bridged by MOF might be applicable to different stem cells. Our study has established a functional link among H3.3, MOF, and H4K16ac in NSCs. This result is supported by experiments showing that the H3.3-WT and MOF overexpression could rescue the abnormal phenotype caused by H3.3 knockdown in vivo and in vitro.
In our study, RNA-seq result shows the downregulated genes in H3.3 knockdown NSCs have a significant enrichment in biological processes related to NSCs proliferation. The upregulated genes exhibit a significant enrichment in biological processes related to NSCs differentiation. Through in-depth analysis, we find the GLI1, a member of the GLI zinc finger family, is regulated by H3.3. GLI1 itself has been shown to be a transcriptional factor in previous study, which may have similar function with H3.3 as a transcription regulator. However, the downstream of the GLI1 requires further study.
Taken together, we report that GLI1 knockdown decreases the proliferation of the NSCs and promotes the augmentation of the differentiated neurons (Figure 10j). The findings about the histone crosstalk in NSCs will strengthen our understanding of the function of epigenetic regulation in the developing brain.
Materials and Methods
Mice
ICR pregnant mice were purchased from Vital River. The mice were maintained in standard conditions (12 h light/dark cycles) with food and water provided. All of them were taken care according to the Guide for the Care and Use of Laboratory Animals. All animal experiments were approved by the Animal Committee of the Institute of Zoology, Chinese Academy of Sciences.
In utero electroporation
The detail experimental protocols have been described in our previously study.35 In brief, the ICR pregnant mice (E13.5) were anesthetized by given an injection of pentobarbital sodium, and then the uterine horns were exposed for experiment. 1.5 μl recombinant plasmid (final concentration 2 mg/ml) mixed with Venus-GFP at a 3:1 mol ratio and fast green (2 mg/ml; Sigma, St. Louis, MO, USA) was microinjected into the fetal brain ventricles. And the mixed plasmid was electroporated into the brain ventricle cells by using an electroporator (BTX ECM830). After electroporation, the pregnant mice were killed at different time point for phenotype analysis. The fetal brains were fixed in 4% paraformaldehyde (PFA) overnight and then dehydrated in 30% sucrose at 4 °C. Different sub-regions of the cortex were identified by cell density and visualized with DAPI nuclear staining. In IUE experiments, brains from at least three independent experiments were collected. Brain sections (at least nine sections) were analyzed by confocal microscopy, and the GFP-positive cell number or multi-marker co-labeled cells in different regions were counted by the confocal microscopy software (ZEISS Zen 2010, Zeiss, Oberkochen, Germany) and Photoshop CS4 software (Adobe Systems Inc., New York, NY, USA). For morphological and quantitative analysis of cultured neurons and IUE brain slides, the number of branches and branch length of the neurons (at least eleven cells) were calculated using confocal microscopy software (ZEISS Zen 2010).
Co-Immunoprecipitation
Cells were washed three times with PBS and suspend with cold lysis buffer with protease inhibitors. The samples were centrifuged at 4 °C, and the supernatants were transferred to another tube. The BCA method was used for measuring the concentration of protein. 200 μg protein sample was incubated with anti-Flag-tag (or anti-HA-tag) magnetic beads at 4 °C overnight, and the same amount of sample was incubated with anti-IgG magnetic beads for negative control. The supernatant was removed by magnetic rack, and the beads were washed by cold washing buffer three times. After that, the magnetic beads were suspended with 1X protein running buffer, and boiled for 2 min. Each sample was analyzed by Western blot and 20 μg (10%) protein sample was used as In-put.
ChIP
The NSCs were isolated from E12.5 cortex and infected with lentivirus for 8 hours with 2 μg/ml polybrene. Twelve hours later, the proliferation medium was changed into a differentiation medium and cultured for 4 days. After that, 1% formaldehyde was used to cross link the chromosome and protein for 15 min at room temperature, and 2.5 M glycine was added to stop the reaction. Cells were rinsed three times with cold PBS and collected in lysis buffer 1 (50 mM HEPES KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton, and protease inhibitor). Then sample was resuspended in lysis buffer 2 (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitor). After centrifugation, the sample was resuspended and sonicated in lysis buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, and protease inhibitor). After that, each lysate sample was divided into two parts: one was incubated with anti-Flag-tag (or anti-HA-tag) magnetic beads; the other one was incubated with anti-IgG magnetic beads at 4 °C overnight. After washing six times with washing buffer, the beads-antibody-DNA complex was incubated at 65 °C to reverse the covalent histone–DNA bonds. The DNA was extracted and then analyzed by realtime-PCR. The data was analyzed as previous described.36
Immunostaining
Immunostaining for cultured cells or brain slices was performed according to the following procedure: the samples were washed with PBST (1%Triton X-100 in 1 M PBS), fixed in 4%PFA, blocked by 5%BSA (in 1% PBST), incubated with primary antibodies overnight at 4 °C, and visualized using fluorescence-labeling secondary antibodies.
Antibodies
Anti- beta III Tubulin (MAB1637, Millipore), Anti- beta III Tubulin (T2200, Sigma), anti-NeuN (MAB377), anti-PCNA (SC7907, SANTA CRUZ), anti-PAX6 (AB2237, Millipore, Darmstadt, Germany), anti-H3.3 (MABE872, Millipore), anti-Tri-Methyl-Histone H3(Lys4) (05-1339, Millipore), anti-Histone H3 (4499, Cell Signaling Technology, Beverly, MA, USA), anti-H4K16ac (07-329, Millipore), anti-β-ACTIN (20536-1-Ap, Proteintech, Rosemont, IL, USA), anti-NESTIN (MAB353, Millipore), anti-SOX2 (3728, Cell Signaling Technology), anti-SOX2 (MAB2018, R&D, Minneapolis, MN, USA), anti-Flag (F1804, Sigma), anti-phospho-Histone H3(Ser10) (3377, Cell Signaling Technology), anti-HA (M20003, Abmart, Shanghai, China), anti-Tri-Methyl-Histone H3(Lys36) (9763, Cell Signaling Technology), anti-BrdU (AB6326, Abcam, Cambridge, UK), anti-GLI1 (SC20687, SANTA CRUZ, Delaware Avenue, CA, USA), IgG (bs0295P, Bioss, Beijing, China), and anti-Caspase3 (9662, Cell Signaling Technology) anti-Lamin A/C (bs1446, bioworld, Nanjing, China).
Cell culture
293FT were cultured in DMEM medium that contained 10% FBS, NEAA, and penicillin/streptomycin.
Lentiviral package DNA and the core DNA was transfected into HEK293FT by GenEscortTMI (Wisegen, Nanjing, China). The supernatant containing the virus was collected at 24, 48, and 72 h post-transfection, and the cell debris was removed by centrifugation at 3000 rpm for 5 min.
NSCs were isolated from E12.5 cortex and seeded in plates, the plates were coated with Poly-D-Lysine (10 μg/ml, Sigma) and Laminin (10 μg/ml, Invitrogen, Carlsbad, CA, USA). Approximately 1.5 million cells per well were seeded for a 6-well-plate, while 20 000 cells per well were seeded for a 48-well-plate. After growing in the proliferation medium for 12 h, which consisted of 50% DMEM/F12 (Invitrogen), 50% Neurobasal medium (Invitrogen), 0.5% GlutaMAX (Invitrogen), 1% nonessentialamino acids, bFGF (10 ng/ml, Invitrogen), EGF (10 ng/ml, Invitrogen), and 1% penicillin-streptomycin (Invitrogen). Cells were infected with lentivirus by the addition of 2 μg/ml polybrene to improve the efficacy of infection. Twelve hours later, the proliferation medium was changed into a differentiation medium, which consisted of low glucose DMEM (Gibco, Grand Island, NY, USA), 2% B27, and 1% fetal bovine serum (Invitrogen).
BrdU labeling
For NPC proliferation analysis, BrdU (50 mg/kg) was administered to pregnant female mice for 2 h before collecting the embryonic brains. For the birth-dating experiment, BrdU (50 mg/kg) was administered to pregnant mice 24 h after electroporation, and the embryonic brains were collected three days later for phenotype analysis. For the cell-cycle exit experiment, BrdU (50 mg/kg) was administered to pregnant mice for 24 h before the mouse brains were separated and processed. Then, the brains were co-stained with anti-Ki67 and anti-BrdU antibodies for analysis.
Western blotting
Cells were lysed with RIPA with protease inhibitor on ice for 5 min and the cell debris was eliminate by centrifuged at 4 °C for 5 min. The BCA method was used for measuring the concentration of protein. The protein samples were loaded onto SDS-PAGE gel for electrophoresis, and the bands were transferred to nitrocellulose or polyvinylidene fluoride membranes. The membranes were blocked in 5% nonfat milk in PBS-T (PBS with 0.1% Tween-20) for 1 h at room temperature, and incubated with primary antibody at 4 °C. The different secondary antibodies were used to visualize the bands. In the western data analysis, the gray density of the different protein band is measured using the Odyssey software. The gray density of the target protein band was divided by that of the internal reference protein of the corresponding sample (Lamin A/C or β-ACTIN) to make the corresponding statistical data.
Realtime-PCR
The total RNA was extracted by using the TRIzol (Invitrogen) according to the instructions. FastQuant RT Kit (with DNase, TIANGEN, Beijing, China) was used to get first-strand cDNA. Realtime-PCR was performed on the realtime-PCR machine (ABI 7500, Life Technologies, Singapore).
Confocal imaging and statistical analysis
All images were got by using Zeiss 780 laser scanning confocal microscope and analyzed with Photoshop CS4 (Adobe).
Statistical analyses were performed using T-test (*P<0.05, **P<0.01). All bar graphs in the figures are shown as the means±S.E.M.
Primers
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (2015CB964500 and 2014CB964903), the National Science Foundation of China (31371477 and 31621004), and K.C.Wong Education Foundation. The GEO accession numbers for the Control and H3.3-Knockdown transcriptome sequencing data reported in this paper is GSE92720.
Author contributions
WX and JJ designed the research; WX performed the research and analyzed the data; and WX and JJ wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by Y Shi
The authors declare no conflict of interest.
Supplementary Material
References
- Durak O, Gao F, Kaeser-Woo YJ, Rueda R, Martorell AJ, Nott A et al. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat Neurosci 2016; 19: 1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol 2010; 28: 1079–1088. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell 2014; 53: 631–644. [DOI] [PubMed] [Google Scholar]
- Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X et al. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 2014; 158: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 2005; 6: 777–788. [DOI] [PubMed] [Google Scholar]
- McConnell SK. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron 1995; 15: 761–768. [DOI] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O'Connell DJ et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and Polycomb gene repression. Dev Cell 2013; 26: 223–236. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin Regulates Cadherin Function via Dab1/Rap1 to Control Neuronal Migration and Lamination in the Neocortex. Neuron 2011; 69: 482–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LL, Wang J, Hong F, Spector DL, Fang YD. Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes. Proc Natl Acad Sci USA 2011; 108: 10574–10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 2010; 11: 264–275. [DOI] [PubMed] [Google Scholar]
- Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 2013; 27: 2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 2005; 19: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet 2009; 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaev N, Keohane AM, Turner BM. Differential underacetylation of histones H2A, H3 and H4 on the inactive X chromosome in human female cells. Hum Genet 1996; 97: 573–578. [DOI] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 2012; 11: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem 1999; 274: 8143–8152. [DOI] [PubMed] [Google Scholar]
- Zahreddine HA, Borden KL. Molecular pathways: gli1-induced drug glucuronidation in resistant cancer cells. Clin Cancer Res 2015; 21: 2207–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Grund EM, Silva HM, Lafaille JJ, Fishell G, Salzer JL et al. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature 2015; 526: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palle K, Mani C, Tripathi K, Athar M. Aberrant GLI1 activation in DNA damage response, carcinogenesis and chemoresistance. Cancers 2015; 7: 2330–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal NK, Kim CH, Kunkalla K, Konno H, Tjendra Y, Kwon D et al. Active IKKbeta promotes the stability of GLI1 oncogene in diffuse large B-cell lymphoma. Blood 2016; 127: 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 2013; 140: 3624–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J 2007; 26: 4974–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126: 3915–39241999. [DOI] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2013; 12: 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Johnson MR, Ke Z, Chen L, Welte MA. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol 2014; 24: 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, Leroy G et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun 2013; 4: 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet 1999; 8: 2489–2495. [DOI] [PubMed] [Google Scholar]
- Bush KM, Yuen BT, Barrilleaux BL, Riggs JW, O'Geen H, Cotterman RF et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics & chromatin 2013; 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci USA 2004; 101: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–612004. [DOI] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 2010; 328: 94–98. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell 2008; 135: 604–607. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xing ZB, Wang SQ, Chen S, Liu YK, Li YH et al. MDM2-MOF-H4K16ac axis contributes to tumorigenesis induced by Notch. FEBS J 2014; 281: 3315–3324. [DOI] [PubMed] [Google Scholar]
- Xia W, Liu Y, Jiao J. GRM7 regulates embryonic neurogenesis via CREB and YAP. Stem Cell Reports 2015; 4: 795–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 2006; 1: 179–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.