Abstract
MYC family proteins are implicated in many human cancers, but their therapeutic targeting has proven challenging. MYCN amplification in childhood neuroblastoma (NB) is associated with aggressive disease and high mortality. Novel and effective therapeutic strategies are therefore urgently needed for these tumors. MYC-driven oncogenic transformation impairs cell survival under nutrient deprivation (ND), a characteristic stress condition within the tumor microenvironment. We recently identified eukaryotic Elongation Factor 2 Kinase (eEF2K) as a pivotal mediator of the adaptive response of tumor cells to ND. We therefore hypothesized that eEF2K facilitates the adaptation of MYCN amplified NB to ND, and that inhibiting this pathway can impair MYCN-driven NB progression. To test our hypothesis, we first analyzed publicly available genomic databases and tissue microarrays for eEF2K expression in NB, and for links between eEF2K, MYCN, and clinical outcome in NB. Effects of eEF2K inhibition were evaluated on survival of MYCN amplified versus non-amplified NB cell lines under ND. Finally, NB xenograft mouse models were used to confirm in vitro observations. Our results indicate that high eEF2K expression and activity are strongly predictive of poor outcome in NB, and correlates significantly with MYCN amplification. Inhibition of eEF2K markedly decreases survival of MYCN amplified NB cell lines in vitro under ND. Growth of MYCN amplified NB xenografts is markedly impaired by eEF2K knockdown, particularly under caloric restriction. In summary, eEF2K protects MYCN overexpressing NB cells from ND in vitro and in vivo, highlighting this kinase as a critical mediator of the adaptive response of MYCN amplified NB cells to metabolic stress.
The closely related MYC, MYCN, and MYCL transcription factors are implicated in many aggressive human cancers, thus representing ideal targets for cancer therapy.1, 2, 3 However, MYC family members are widely considered “undruggable”, as they lack active sites susceptible to binding by inhibitory small molecules.4 New approaches will likely require targeting of additional pathways required for their oncogenic transformation. By regulating the expression of its target genes, MYC activation results in numerous biological effects including cell cycle progression, cell growth, and metabolic reprogramming.5, 6 In addition to these pro-growth and survival effects characteristic of a classic oncogene, MYC expression also renders diverse cell types susceptible to apoptosis when cells are deprived of nutrients.7, 8, 9, 10
In the tumor microenvironment, reduced blood flow exposes cancer cells to potentially lethal stress forms, including nutrient deprivation (ND), forcing cells to adapt or die.11 Recent work suggests that a major component of stress adaptation occurs through acute changes in mRNA translation. Under acute stress, cells undergo a block in global translation to save energy,12 but with selective synthesis of key survival proteins that allow a more rapid response than through transcriptional mechanisms.13, 14 We previously reported that under ND, eukaryotic Elongation Factor 2 Kinase (eEF2K) acts as a major mediator of cell survival by phosphorylating and inhibiting its substrate eukaryotic translation Elongation Factor 2 (eEF2). The latter mediates the translocation of ribosomes along mRNAs, which is the major rate-limiting step of mRNA translation elongation.15, 16, 17, 18 The clinical relevance of eEF2K in human tumors is highlighted by our recent finding that high eEF2K expression predicts poor outcome in two central nervous system (CNS) tumors, medulloblastoma, and glioblastoma, where high eEF2K activity was detected in tumor tissues but not in normal surrounding CNS tissues.18
Neuroblastoma (NB) is the most common pediatric extracranial solid cancer,19 causing ~15% of all childhood cancer related deaths in the North America.20 High-level amplification of the chromosome 2p24.3 MYCN locus results in MYCN protein overexpression in ~20% of NB cases.21 Importantly, MYCN amplification strongly predicts poor prognosis in all stages of disease in terms of overall survival in all multivariate regression analyzes of prognostic factors.21, 22 Interestingly, MYC overexpression has been shown to impair cell viability in nutrient depleted cells.7 On the basis of this observation, we hypothesized that eEF2K activity is required for adaptation of MYCN amplified NB to ND, and that targeting this pathway can therefore impair tumor progression under metabolic stress conditions.
Here, we show that NB cell lines overexpressing MYCN are highly dependent on eEF2K to overcome acute ND in vitro. Ectopic overexpression of MYCN in a non-MYCN amplified NB cell line results in increased sensitivity to eEF2K inhibition under ND. Moreover, shRNA mediated stable genetic inactivation of eEF2K in vivo results in decreased tumor growth and massive necrosis in MYCN amplified NB, particularly under caloric restriction. Collectively, these data suggest that eEF2K inhibition may be a novel therapeutic strategy for this aggressive tumor.
Results
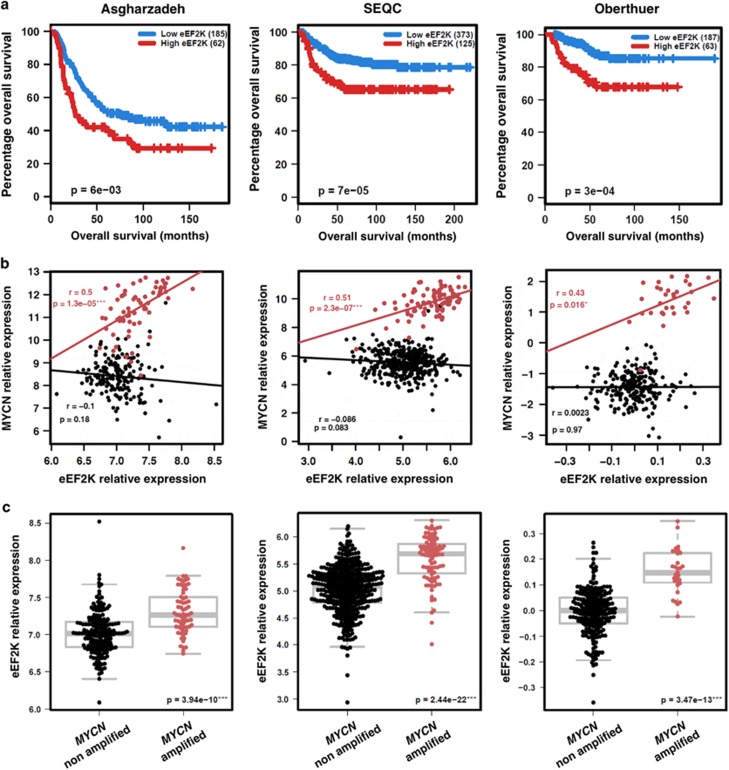
High eEF2K expression is associated with poor outcome and MYCN amplification in NB
Given the association between high eEF2K expression and poor outcome in medulloblastoma and glioblastoma,18 we first asked whether eEF2K expression levels predict outcome in NB. We therefore interrogated gene expression data from published RNAseq23 and gene expression microarray cohorts,24, 25 and found that increased eEF2K levels are strongly predictive of worse outcome in NB among four independent data sets (Figure 1a: Asgharzadeh, P=6e-03; SEQC: P=7e-05; and Oberthuer: P=3e-04, respectively; Supplementary Figure S1: Versteeg, P=4e-04). We next tested if this correlation is linked to MYCN status. Indeed, there was a strong association between MYCN and eEF2K expression in MYCN amplified NB across five independent data sets (Figure 1b: Asgharzadeh, P=1.3e-05; SEQC, P=2.3e-07; and Oberthuer, P=0.016, respectively; Supplementary Figure S1b: Versteeg, P=0.073; and Kocak, P=2.5e-07). In addition, MYCN amplified NB cases have significantly higher eEF2K mRNA expression compared to non-MYCN amplified tumors in five independent data sets (Figure 1c: Asgharzadeh, P=3.94-10; SEQC, P=2.44e-22; Oberthuer, P=3.47e-13, respectively; Supplementary Figure S1c: Versteeg, P=2.39e-07 and Kocak, P=4.11e-39). Higher levels of eEF2K expression in MYCN amplified cases, as opposed to non-MYCN amplified cases, were also observed in analyses limited to higher clinical risk tumors (stage 4, age >18 months; Supplementary Figure S1d). Taken together, these data provide evidence for high eEF2K expression in MYCN amplified NB.
Figure 1.
Clinical relevance of eEF2K mRNA expression in NB. (a) eEF2K higher expression correlates with poor outcome in NB. Survival curves by Kaplan Meier plotting, cases were split based on eEF2K expression. (b) eEF2K and MYCN expression correlates in MYCN amplified NB (r=Pearson’s correlation coefficient). (c) eEF2K is overexpressed in MYCN amplified NB compared to non-MYCN amplified NB. Cases were split based on MYCN amplification status and eEF2K expression was measured. Each analysis is performed on three different cohorts (Asgharzadeh n=247; SEQC n=493; Oberthuer n=250). *P<0.05, **P<0.01, ***P<0.001. Data obtained from the R2 platform (http://r2.amc.nl)
eEF2K is strongly activated in MYCN amplified NB cells
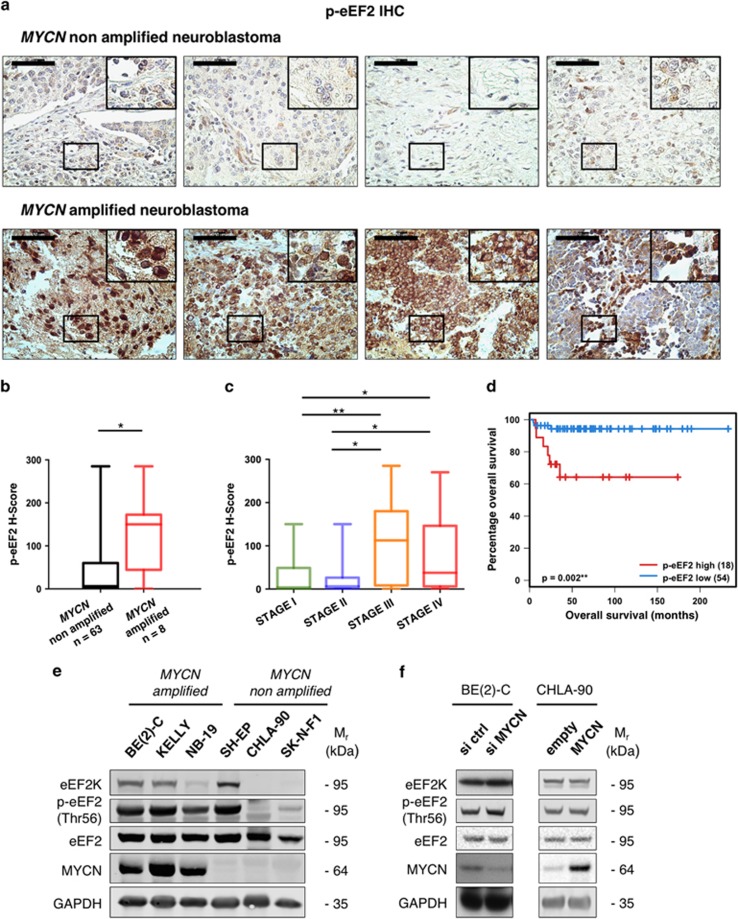
Since mRNA levels are not always predictive of the activation state of a specific biological pathway, we next wished to confirm that eEF2K is functionally activated in MYCN amplified NB. We therefore stained NB tissue microarrays (TMAs) obtained from the Children’s Hospital of Philadelphia (CHOP) by immunohistochemistry (IHC) with antibodies targeting the eEF2K specific Thr56 phosphorylation site of eEF2 (p-eEF2). This revealed higher p-eEF2 (i.e., eEF2K activation or expression) in MYCN amplified NB compared to non-MYCN amplified tumors (P=0.011; Figures 2a and b), further pointing to a role for eEF2K in this particular subset of NB. Moreover, elevated p-eEF2 levels were associated with tumors at a higher INSS clinical stage, according to the International NB Staging System, INSS, (ref. 26 and Figure 2c). In addition, high p-eEF2 levels are predictive of worse outcome in NB in a univariate survival analysis (P=0.002; Figure 2d). This was confirmed with two different manual semi-quantitative methods for IHC quantification, namely the histo-score (H-score), and the immunoreactive score (IRS; Supplementary Figure S2a; see Materials and Methods section). Of note, we found no association between the tumor proliferative index (MIB-1) and p-eEF2 immunoreactivity (Supplementary Figure S2b). Western blotting revealed that NB cell lines with MYCN amplification had overall higher eEF2K expression and activity in vitro under ambient conditions compared to non-MYCN amplified lines (Figure 2e). One exception was the non-MYCN amplified SH-EP cell line, suggesting that MYCN overexpression may not be the only factor linked to eEF2K up-regulation. To test potential MYCN-mediated transcriptional regulation of eEF2K, MYCN knockdown in MYCN amplified BE(2)-C NB cells and MYCN ectopic expression in non-MYCN amplified CHLA-90 NB cells was performed (Figure 2f). However, neither manipulation had any significant effect on eEF2K protein expression or activity (Figure 2f, Supplementary Figures S2c and S2e), or on eEF2K mRNA levels (Supplementary Figures S2d and S2f). These results indicate that, despite being highly expressed in MYCN amplified NB, eEF2K is likely not a direct transcriptional target of MYCN.
Figure 2.
eEF2K is highly active in MYCN amplified NB. (a) Representative IHC images for p-eEF2 (Thr56) performed on the neuroblastoma TMA. Every image is representative of a different patient. Scale bar=50 μm. (b) H-score based quantification of p-eEF2 (Thr56) IHC on the CHOP neuroblastoma TMA. The cohort was split based on MYCN amplification status. (c) H-score based quantification of p-eEF2 (Thr56) IHC on the CHOP neuroblastoma TMA. The cohort was split based on INSS stage. (d) Survival analysis by Kaplan–Meyer plotting on the CHOP neuroblastoma TMA based on p-eEF2 H-score. *P<0.05, **P<0.01, ***P<0.001. (e) Western blotting analysis of neuroblastoma cell lines with or without MYCN amplification. (f) Left: SH-EP neuroblastoma cells with or without stable overexpression of MYCN were lysed and analyzed by immunoblotting for the indicated proteins. Right: BE(2)-C neuroblastoma cells (with MYCN amplification) were transiently transfected with control (si ctrl) or MYCN (si MYCN) siRNAs. Cells were lysed 72 h post transfection and analyzed by immunoblotting for the indicated proteins
Inactivation of eEF2K impairs the adaptive response of MYCN amplified BE(2)-C NB cells to acute ND
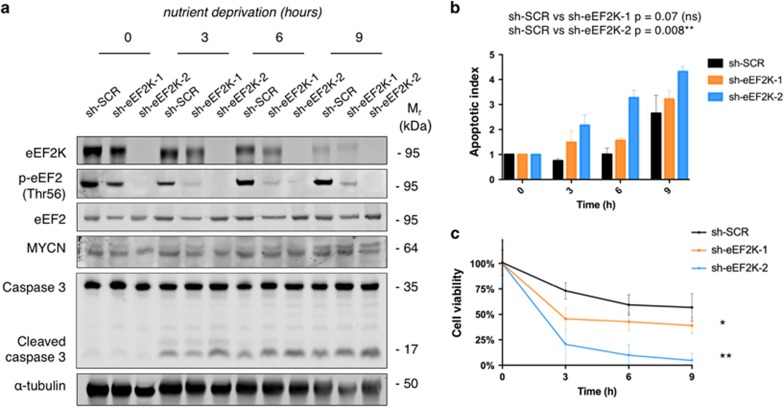
Since eEF2K confers cell survival under acute severe ND,18 and MYC overexpression impairs cell viability under ND,7 we hypothesized that eEF2K activity is particularly relevant for the adaptation of MYCN amplified NB cells to severe ND, and that targeting this pathway impairs tumor survival under metabolic stress. To test whether, we performed stable shRNA mediated knockdown of eEF2K in the well characterized MYCN amplified BE(2)-C and non-MYCN amplified SH-EP NB cell lines.27 Screening of a commercially available pool of six shRNA sequences identified two shRNAs for subsequent studies, one with strong and the other with moderate eEF2K knockdown, as confirmed by Western blotting (Figure 3a, see eEF2K panel, and Supplementary Figure S2g). As predicted by our previous findings,18 eEF2K had no effect on cell proliferation or survival under ambient conditions (i.e., complete growth media), as determined using MTT assays (Supplementary Figure S2h). We then performed a time course analysis of cell survival under acute severe ND, using media lacking glucose, amino acids and serum as described.18 Western blotting confirmed that BE(2)-C cells transduced with shRNAs targeting eEF2K were deficient in eEF2 phosphorylation under ND conditions (Figure 3a and Supplementary Figure S2i), particularly cells transduced with sh-eEF2K-2, which was more effective in knocking down eEF2K (Figure 3a and Supplementary Figure S2g). This was not associated with loss of MYCN expression in the eEF2K knockdown cell lines (Figure 3a). Phosphorylation of AMPKα, a known sensor of energy depletion,28 confirmed that cells were experiencing energy stress under ND conditions in these experiments (Supplementary Figure S2l). To evaluate the impact of eEF2K loss on cell apoptosis under ND, we performed western blotting for caspase-3 cleavage. MYCN amplified cells with eEF2K knockdown seemed more susceptible to apoptosis induced by ND compared to controls (Figure 3a; see caspase-3 panel). To confirm this result, we performed flow cytometry for PI-Annexin V staining. Knockdown of eEF2K in MYCN amplified cells led to a significant increase in apoptosis compared to controls (Figure 3b). To assess cell viability, we performed MTT and trypan blue assays over the same time course (Figure 3c and Supplementary Figure S3a), which demonstrated that eEF2K knockdown in MYCN amplified cells led to a dramatic reduction in cell number under ND. Interestingly, under similar conditions, eEF2K knockdown in non-MYCN amplified SH-EP cells increased caspase-3 cleavage and PI/Annexin V staining (Supplementary Figure S3b–c), but knockdown did not significantly decrease cell viability under ND (Supplementary Figure S3d–e). This supports the notion that high MYCN expressing NB cells may be preferentially dependent on eEF2K to fully maintain cell viability and prevent cell death under ND, compared to cells without MYCN overexpression. The mechanistic basis of this discrepancy warrants further investigation.
Figure 3.
Genetic inactivation of eEF2K impairs the adaptive response of MYCN amplified BE(2)-C cells to acute ND. (a) Western blotting analysis on the BE(2)-C cell line ±shRNA mediated eEF2K knockdown under acute ND at different time-points. Equal loading of protein samples is verified with α-tubulin. (b) PI/Annexin V flow cytometry based staining for the BE(2)C cell line ±shRNA mediated eEF2K knockdown over time of acute ND. Apoptotic index is defined as the ratio between PI-Annexin V double positive cells over PI-Annexin V double negative cells. Values are normalized for every cell line for the apoptotic index at time 0 (baseline). (c) MTT assay measuring BE(2)-C ±shRNA mediated eEF2K knockdown cells survival under acute ND at different time-points. The experiment is carried out in eight replicates
MYCN amplified NB cell lines are highly sensitive to eEF2K chemical inhibition under ND
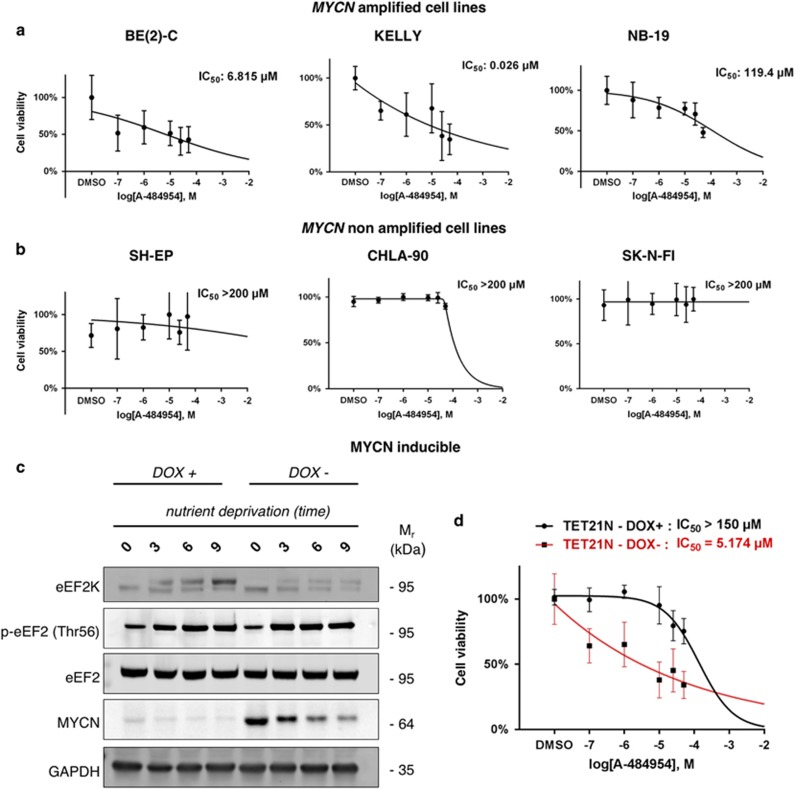
To extend our findings to additional NB cell lines and to evaluate the utility of eEF2K chemical inhibition as a therapeutic strategy, we studied effects of a known small molecule eEF2K inhibitor, A-484954 (EMD Millipore, Billerica, MA, USA;29) on NB cell lines under ND in vitro. At concentrations active against eEF2K (Supplementary Figure S4), A-484954 has minimal activity against other serine/threonine and protein tyrosine kinases and does not affect proliferation in diverse cancer cell lines.29 To study effects of eEF2K chemical inhibition on NB cells under ND, we compared survival of MYCN amplified BE(2)-C, KELLY and NB-19 NB cells with non-MYCN amplified SH-EP, CHLA-90 and SK-N-FI NB cells,30 each treated with A-484954 over a broad concentration range. A-484954 strongly decreased survival of MYCN amplified cell lines under ND (KELLY IC50=0.03 μM, BE(2)-C IC50=6.81 μM, NB-19 IC50=119.4 μM; Figure 4a), while non-MYCN cell lines were each relatively insensitive to eEF2K inhibition under the same conditions (IC50>200 μM for SH-EP, SK-N-FI and CHLA-90 cell lines; Figure 4b). Differential sensitivity in these cell lines was maintained over extended time periods, as shown for MYCN amplified KELLY compared to non-MYCN amplified CHLA-90 cells over a 72 h time course in Supplementary Figure S5a,b. Moreover, in MYCN amplified BE(2)-C cells, A-484954 more effectively reduced cell survival under ND compared to complete media conditions (Supplementary Figure S5c), similar to effects of eEF2K knockdown in the same cells (Figure 3c). In contrast, A-484954 only minimally affected survival under ND of the non-MYCN amplified CHLA-90 cell line (Supplementary Figure S5d). For both the BE(2)-C and CHLA-90 cell lines, the effect of A-484954 on cell proliferation under complete media conditions was minimal (Supplementary Figure S5c,d). These data indicate that, similar to eEF2K knockdown, A-484954 selectively inhibits survival of nutrient deprived MYCN amplified compared to non-MYCN amplified NB cell lines in vitro. To further explore whether MYCN overexpression sensitizes nutrient deprived NB cells to eEF2K chemical inhibition, we utilized the Tet21N cell line, which is a non-MYCN amplified NB cell line harboring a conditional doxycycline (DOX)-dependent MYCN repression system.31 Western blotting confirmed eEF2K activation under acute ND for Tet21N cells both with or without MYCN induction, based on increased p-eEF2 levels (Figure 4c). This is expected as eEF2 phosphorylation is a general regulatory mechanism by which eukaryotic cells control translation.17 MYCN induction had no effects on eEF2K levels in Tet21N cells ± DOX (Figure 4c), further highlighting lack of eEF2K transcriptional regulation by MYCN. However, induction of MYCN in Tet21N cells markedly increased sensitivity to A-484954 treatment under ND, compared to Tet21N cells lacking MYCN ectopic expression (Figure 4d; MYCN induced cells had an IC50=5.17 μM, while MYCN uninduced cells had an IC50>150 μM). Conversely, A-484954 treatment under complete media conditions had only a minimal effect on cell proliferation (Supplementary Figure S5e–f). These data are consistent with published studies,7 as in our studies MYCN expression renders Tet21N cells more sensitive to the effects of ND (Supplementary Figures S5g and S5h). ND strongly inhibited kinases of the mTORC1 signaling pathway involved in translation initiation (Supplementary Figure S6a), consistent with our previous results.18 To test whether MYCN expression affects eEF2K stability, we treated Tet21N cells with the translation inhibitor cycloheximide and measured eEF2K protein levels in the presence and absence of DOX. However, MYCN levels did not affect eEF2K stability, either under ambient conditions or ND (Supplementary Figure S6b–e). Taken together, these findings provide further evidence that eEF2K activity is required for the adaptation of MYCN amplified NB cells to ND in vitro.
Figure 4.
A-484954 impairs MYCN overexpressing NB cell line survival under ND. (a,b) Dose-response curve for the MYCN amplified (a) and MYCN non-amplified cell lines (b) after 48 h of ND and different concentrations of A-484954 or equal dose of DMSO. Cell viability values are normalized for the DMSO (control) average cell viability value at the specific time-point. Each MTT assay is carried out in eight replicates. (c) Western blot analysis on the Tet21N cell line under ND at different time-points. GAPDH is used to verify equal samples loading. (d) Dose-response curve for the Tet21N cell line after 48 h of ND and different concentrations of A-484954 or equal dose of DMSO. Cell viability values are normalized for the DMSO (control) average cell viability value at the specific time-point. Each MTT assay is carried out in eight replicates. The 48 h time point was chosen for the dose-response curves and to calculate the IC50 values based on the results of the time course analysis for all the cell lines, showing the most significant differences in cell viability at this specific time point
Inactivation of eEF2K induces massive cell death in MYCN amplified NB in vivo
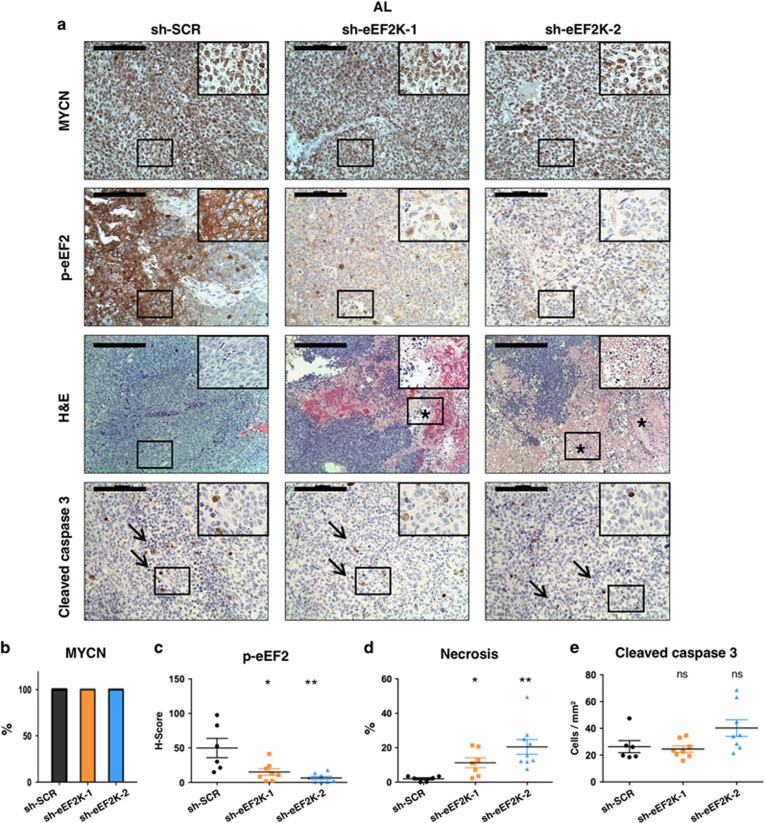
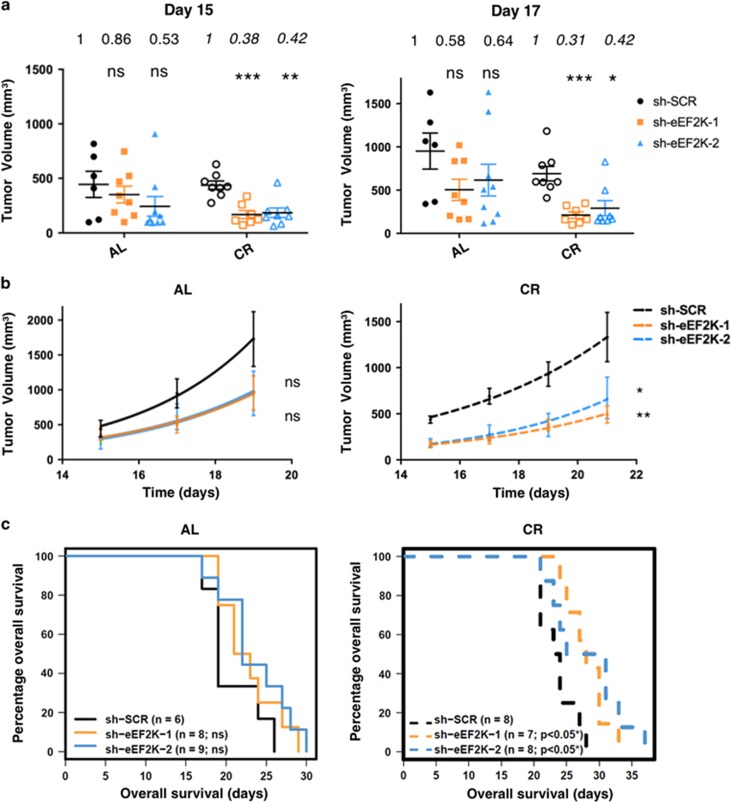
To begin to assess eEF2K inhibition as a potential therapeutic strategy for MYCN amplified NB, we tested effects of eEF2K genetic inactivation using an MYCN amplified in vivo NB xenograft model. We subcutaneously transplanted BE(2)-C cells with or without stable eEF2K knockdown into immunodeficient mice fed normal chow (at libitum; AL). This cell line is well established for in vivo NB studies due to its aggressive behavior27, 32 and high in vivo tumorigenicity.33 As expected for a MYCN amplified NB cell line, tumors from all groups showed strong nuclear staining for MYCN, confirming high expression of MYCN which was not affected by eEF2K knockdown in vivo (Figure 5a, top panels, and Figure 5b). IHC for eEF2K and p-eEF2 on tumor sections confirmed a marked decrease in eEF2K activity and expression in tumors with eEF2K knockdown (Figure 5a, see p-eEF2 panels, and 5c; P=0.020 and P=0.001 for sh-eEF2K-1 and sh-eEF2K-2 tumors compared with controls, respectively; and Supplementary Figure S7a). Notably, by hematoxylin and eosin (H&E) staining, sections from eEF2K deficient tumors revealed extensive areas of geographic coagulative necrosis compared to control tumors (Figure 5a, see H&E panels, and 5d; P=0.008 and P=4e-04 for sh-eEF2K-1 and sh-eEF2K-2, respectively, compared to controls). On the other hand, there was no significant difference in caspase-3 mediated apoptotic activity (Figure 5a, see caspase-3 panels, and 5e) or cell proliferation (Ki67 staining, Supplementary Figure S7b) in eEF2K deficient compared to control tumors, when evaluated in viable (non-necrotic) regions. Consistent with the Ki67 staining, tumor volumes did not differ significantly between eEF2K knockdown and control tumors, although there was a trend toward reduced sizes of tumors with eEF2K knockdown (Figure 6a). Together, these results indicate that eEF2K has a protective effect on MYCN amplified NB tumor growth in vivo, but that this is unlikely to be due to differential proliferative rates.
Figure 5.
Inactivation of eEF2K induces massive necrotic cell death in MYCN amplified NB in vivo. (a) Representative IHC and H&E images of subcutaneous tumor xenografts derived from the MYCN amplified BE(2)-C cell line ±eEF2K knockdown. Asterisks (*) indicate areas of necrosis and arrows (→) indicate apoptotic cells. Scale bar represents 100 μm on IHC images and 200 μm on HE images. (b) Graphic representing the quantification of percentage of MYCN positive tumors by IHC. (c) Graphic evaluating p-eEF2 H-score by IHC. (d) Graphic representing the percentage amount of necrosis on H&E sections. (e) Quantification of cleaved caspase three positive cells by IHC. The figure represents the experiment with mice fed AL. The sample size for the IHC analysis in the figure is as follows: sh-SCR n=6; sh-eEF2K-1 n=8; sh-eEF2K-2 n=9. *P<0.05, **P<0.01, ***P<0.001
Figure 6.
CR synergizes with eEF2K inactivation to inhibit growth of MYCN amplified NB xenografts in vivo. (a) BE(2)-C derived ±eEF2K knockdown tumors were measured by caliper at day 15 and 17 after tumor xeno-transplantation into mice fed with AL or CR diet. After day 17, mice killing was required as tumors in some mice exceeded humane practice guidelines. (b) Analysis of BE(2)-C derived ±eEF2K knockdown tumor growth and mice survival by Kaplan–Meyer plotting (c) in mice fed AL or with CR diet. Mice were killed when tumors reached 1,500 mm3. The sample size for the analysis in the figure is as follows: AL: as in the previous figure. CR: sh-SCR n=8; sh-eEF2K-1 n=7; sh-eEF2K-2 n=8. *P<0.05, **P<0.01, ***P<0.001
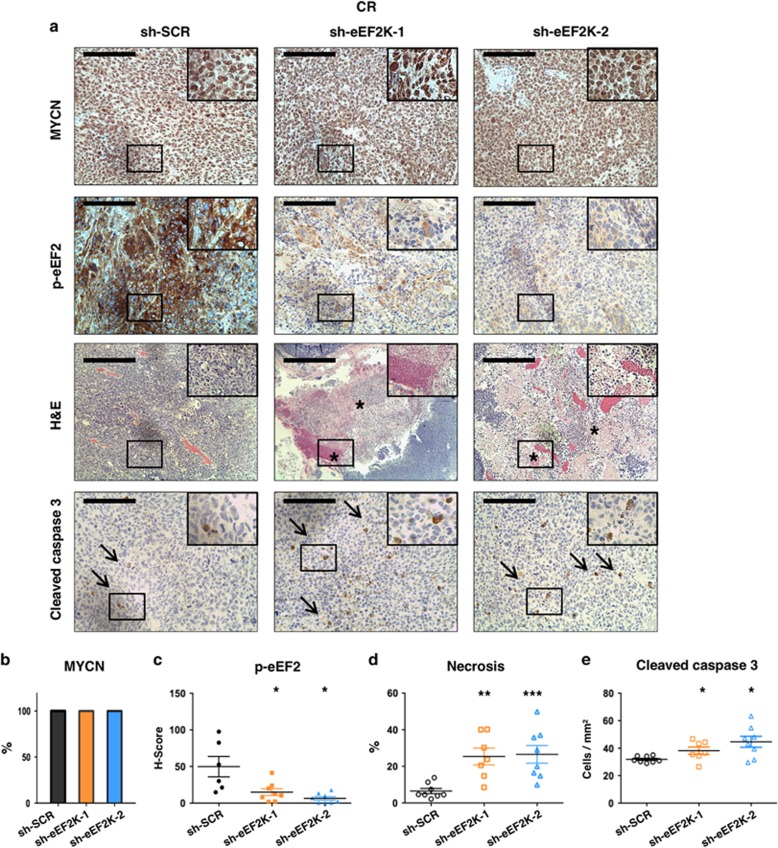
Inactivation of eEF2K restricts MYCN amplified NB xenograft growth under caloric restriction
The tumor microenvironment can limit the delivery of oxygen and other nutrients to proliferating tumor cells.11, 34 Therefore, one possibility to explain the in vivo results of the previous section is that eEF2K protects tumor cells from ND in vivo. To further explore this, we similarly transplanted BE(2)-C NB cell lines ±eEF2K knockdown as above, but instead maintained mice on a calorie restricted (CR) diet.18 Mouse weights were significantly lower than mice fed AL diets (Supplementary Figure S7c), confirming the dietary effects of CR. As opposed to mice fed AL, eEF2K deficient tumors grew significantly slower than controls under CR (Figures 6a and b). Notably, this was accompanied by improved survival of mice bearing eEF2K knockdown tumors compared to control tumors under CR (Figure 6c; P=0.013 for sh-eEF2K-1 and P=0.041 for sh-eEF2K-2 tumors, respectively, compared to controls). Effective eEF2K knockdown was confirmed by IHC (Supplementary Figure S7d). IHC analysis on tumor sections showed no effects of eEF2K knockdown on the tumor proliferative index in mice under CR (Supplementary Figure S7e). MYCN expression was also unaffected by eEF2K knockdown under CR (Figure 7a, see MYCN panels, and 7b). Moreover, while p-eEF2 IHC showed high levels of eEF2K activity in control tumors, confirming eEF2K activation in vivo under CR, p-eEF2 levels were significantly reduced by eEF2K knockdown in mice fed the same diet (Figure 7a, see p-eEF2 panels, and 7c; P=0.039 for sh-eEF2K-1 and P=0.037 for sh-eEF2K-2 tumors, compared to controls). As before, eEF2K knockdown in tumors correlated with extensive areas of coagulative necrosis compared to control tumors, but under CR the degree of necrosis was even more significantly increased in eEF2K knockdown compared to control tumors (Figure 7d; P=0.001 and P=6e-04 for sh-eEF2K-1 and sh-eEF2K-2, respectively, compared to controls). Moreover, although difficult to directly compare across distinct experiments, we observed that in mice fed CR diets, eEF2K knockdown tumors had quantitatively larger necrotic areas than counterpart eEF2K knockdown tumors in mice fed AL diets (average total % necrosis in sh-eEF2K-1: AL 11.2% versus CR 25.4% average total % necrosis in sh-eEF2K-2: AL 20.5% versus CR 26.6%). The combination of CR and eEF2K inactivation also markedly increased the number of apoptotic cells observed in non-necrotic (viable) areas, as measured by cleaved caspase-3 IHC (Figure 7a, see caspase-3 panels, and 7e; P=0.031 for sh-eEF2K-1 and P=0.020 for sh-eEF2K-2, compared to controls). The latter raises the possibility that apoptosis precludes the occurrence of geographic necrosis in eEF2K deficient tumors under CR, further arguing that eEF2K protects MYCN amplified NB against the effects of CR. Finally, comparing reductions in tumor volume between experimental animals fed either AL or CR diets showed a greater reduction upon eEF2K knockdown in mice under CR (Supplementary Figure S8). Taken together, these findings indicate that CR increases the sensitivity of MYCN amplified NB to eEF2K inhibition in vivo. Therefore, combining eEF2K inhibitors with CR mimetics warrants further investigation as a potential therapeutic approach to MYCN amplified NB.
Figure 7.
CR and inactivation of eEF2K induce massive necrotic cell death and apoptosis in MYCN amplified NB in vivo. (a) Representative IHC and H&E images of subcutaneous tumor xenografts derived from the MYCN amplified BE(2)-C cell line ±eEF2K knockdown, grown in mice under CR. Arrows (→) indicate apoptotic cells and asterisks (*) indicate areas of necrosis. Scale bar represents 100 μm on IHC images and 200 μm on H&E images. (b) Graphic representing the quantification of percentage of MYCN positive tumors by IHC. (c) Graphic evaluating p-eEF2 H-score by IHC. (d) Graph representing the percentage amount of necrosis on H&E sections. (e) Quantification of cleaved caspase three positive cells by IHC. The figure represents the experiment with mice fed with CR died. The sample size for the IHC analysis in the figure is as follows: sh-SCR n=6; sh-eEF2K-1 n=8; sh-eEF2K-2 n=9. *P<0.05, **P<0.01, ***P<0.001
Discussion
Genetic amplification of the chromosome 2p34.3 locus encoding MYCN is a major oncogenic event in a subset of highly aggressive pediatric NBs.21 However, pharmacological inhibition of MYCN has proven to be challenging, since the structure of this molecule lacks druggable moieties. Recently, several groups reported anti-cancer efficacy of molecules targeting pathways specifically activated by MYCN,33, 35, 36, 37, 38 highlighting this approach as a promising alternative strategy for MYCN-driven NB. MYC family protein overexpression is known to impair cell viability under stress conditions such as ND.7, 8, 9 Therefore we hypothesized that targeting cellular stress responses to ND in MYCN-driven NB might offer a tractable therapeutic strategy for this disease.
Solid tumors are often poorly and inefficiently vascularized, leading to intra-tumoral hypoxia and ND,11 which must be overcome for cancer cells to survive and proliferate. In this context, cancer cells typically reduce highly ATP-consuming processes such as protein synthesis,39 allowing them to effectively mantain energy balance and survive. Despite encouraging preclinical data, targeting the initiation phase of mRNA translation is still clinically challenging.40We previously reported that mRNA translational control at the elongation phase through eEF2K allows cells to adapt and survive under ND.18 The eEF2K protein is an evolutionary conserved calmodulin-dependent kinase, which phosphorylates its only known substrate, eEF2, to slow the rate of translation elongation.15 Indeed, addition of each amino acid to the elongating nascent polypeptide chain during translation utilizes one ATP and two GTP molecules per bond, which vastly exceeds the energy demands of translation initiation.41 Thus, restoring mRNA translation elongation by blocking eEF2K as a means to increase energy demands represents a promising but relatively unexplored cancer therapeutic strategy.14 Several studies have previously reported eEF2K inhibition by small molecules with varying specificities or effective doses.29, 42 Here, we use both genetic knockdown studies and chemical inhibition with A-484954 to show that the adaptive response of MYCN amplified NB to acute ND requires eEF2K activity. First, we report a highly significant association between eEF2K expression and poor outcome in NB, and that this association is predominantly due to eEF2K overexpression in MYCN amplified NB. This was validated functionally by IHC data showing a strong correlation between p-eEF2 staining (i.e., eEF2K activity) and poor outcome in MYCN amplified NB. Finally, in vitro and in vivo results show that MYCN expression sensitizes NB cells to eEF2K inhibition under ND, further suggesting a specific role for eEF2K in the adaptation of MYCN transformed NB cells to metabolic stress.
The mechanism of eEF2K activation in MYCN amplified NB remains unknown. Our data fails to support a direct transcriptional role for MYCN in promoting eEF2K gene expression. Also, given that eEF2K transcript levels are independent of MYCN in vitro, the latter is unlikely to indirectly modulate eEF2K expression through other transcription factors, through effects on eEF2K mRNA stability, or via epigenetic mechanisms. Moreover, there are no known copy number alterations (CNAs) of the eEF2K locus in NB, arguing against a genetic basis for eEF2K overexpression. Therefore an intriguing alternative model is that MYCN transformation requires the presence of intact eEF2K functional activity; i.e., there may be clonal selection of MYCN transformed cells on the basis of their basal eEF2K activity. Apart from their transcriptional activities, MYC family members are also well known to regulate mRNA translation. At the initiation phase, MYC upregulates the mRNA cap binding protein eIF4E, a pro-oncogenic translation initiation factor.5 In addition, MYC increases the selective translation of specific mRNAs, such as cyclins and cyclin-dependent kinases, by promoting methylation of their 5′-UTR caps.43 Finally, MYC promotes ribosome biogenesis and transcription of ribosomal protein encoding genes,44 further highlighting its importance in translation. In light of our data, it is tempting to speculate that cells with high MYC activity therefore also require mechanisms to rapidly alter translation elongation rates, particularly during adaptation to ND to preserve energy. Further studies are necessary to determine how loss of eEF2K might influence cellular transformation by MYC family proteins.
Our in vivo mouse data indicate that effects of eEF2K inactivation on NB xenograft growth are enhanced when combined with CR. Reducing caloric intake not only significantly reduced tumor volumes and increased survival in mice with eEF2K knockdown compared to control tumors, but also increased the degree of necrosis and apoptosis in these lesions. Therefore future therapeutic studies should aim to combine eEF2K inhibition with CR mimetics such as metformin or 2-DG, as eEF2K activity appears to be particularly critical under metabolic stress conditions. Having said this, we observed significant tumor necrosis in eEF2K knockdown tumors compared to controls even when mice were fed AL diets (Figure 5a). While this may indicate that relative ND occurs in MYCN-driven tumors even when mice are fed ambient diets, we cannot rule out the possibility that eEF2K facilitates adaptation to other microenvironmental stress forms in MYCN amplified NB. We conclude that strategies to reverse the negative regulation of eEF2K on mRNA translation elongation to increase energy demands in stressed tumor cells warrant further investigation therapeutically. This appears to be particularly relevant in pediatric tumors where MYC proteins play major oncogenic roles, such as in NB and in aggressive brain tumors including medulloblastoma45 and pediatric high grade glioma.46
Materials and Methods
mRNA expression and survival analysis
Gene expression data, MYCN amplification status and survival data were obtained from the R2 platform (http://r2.amc.nl) for five publicly available independent NB cohorts (Asgharzadeh, SEQC, Oberthuer, Verteeg, and Kocak). Technical details on gene expression analysis for each cohort can be found on the R2 platform (http://r2.amc.nl). For survival analysis, the five cohorts were split based on relative eEF2K expression, using upper quartiles as the threshold for low or high expression. Overall significance of survival differences was calculated using a log-rank test.
NB tissue microarrays
A NB tissue microarray (TMA) constructed as previously described47 with duplicate cores of 187 NB tumors was obtained from the Children’s Hospital of Philadelphia (CHOP, PA, USA). The demographic and clinical characteristics of the CHOP neuroblastoma TMA patients are summarized in Supplementary Table 1. IHC was performed using an anti-phospho-eEF2 antibody (Thr56, 1:800, Abcam ab53114, Cambridge, UK). IHC was blindly quantified by an MD with pathology training (AD) for staining intensity (0–3) and percentage of positive cells (0–100%). For each sample, the H-score was calculated as staining intensity × percentage of positive cells.48 In addition, the immunoreactive score (IRS) was calculated as previously described.49 When cores derived from the same sample showed a different score, the highest score was considered for the analysis. Only tumors with a histopathological diagnosis of NB and ganglioneuroblastoma were included in the analysis. Tumors without cores available in duplicates for the analysis were excluded. Overall survival significance differences were calculated using a log-rank test.
Cell culture
BE(2)-C and SH-EP NB cell lines were purchased from ATCC. KELLY was purchased from Sigma. Tet21N cells were a gift from Professor M. Schwab (DKFZ, Heidelberg, Germany). NB-19, SK-N-FI and CHLA-90 cells were a gift from Professor Y. DeClerck (CHLA, Los Angeles, USA). BE(2)-C cells were cultured in EMEM:F12+10% FBS. SH-EP, KELLY and Tet21N cells were cultured in RPMI 1640+10% FBS. NB-19, CHLA-90 and SK-N-FI cells were cultured in IMDM+20% FBS. MYCN repression for the Tet21N cell line was achieved with 10 ng ml–1 DOX treatment. For eEF2K stability assays, cells were treated with cycloheximide over a 9 h time course at a final concentration of 100 mg ml–1 to inhibit mRNA translation, and eEF2K protein levels were assessed by Western blotting. All cells were kept at 37 °C and 5% CO2.
siRNA transfections
BE(2)-C cells were transfected at ~30% confluency in 6-well plates with 25 nM control siRNA (1:1 mix of non-targeting siRNA (Stealth RNAi negative control duplexes, medium GC Duplex; Invitrogen, Carlsbad, CA, USA) and luciferase targeting siRNAs (Stealth RNAi siRNA Luciferase Reporter Control; Invitrogen) or with 25 nM of pooled siRNA targeted against human MYCN (ON-TARGET plus siRNA; Dharmacon, Lafayette, CO, USA) using the Lipofectamine RNAiMAX transfection reagent (Invitrogen) as per manufacturer’s instructions. Cells were replenished with new media after 24 h and collected at 72 h post transfection. CHLA-90 cells were transfected at ~30% confluency in 6-well plates with 1 μg full-length human MYCN plasmid DNA using the X-tremeGENE HP DNA Transfection Reagent (Roche, Basel, Switzerland) following the manufacturer’s instructions. Cells were replenished with new media after 24 h and collected at 72 h post transfection.
RNA isolation and quantitative RT-PCR
For RNA isolation in mammalian cells, after indicated treatments cells were lysed and total RNA was extracted using RNeasy RNA extraction kit (Qiagen, Hilden, Germany). RNA concentrations were determined by nanodrop and equal amounts of RNA were used to generate cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). cDNA and Power SYBR Green PCR master mix (Applied Biosystems) were mixed with primers, as previously described,14 and ran in an Applied Biosystems Quant Studio 6 Flex RT-PCR system. As an internal control, GAPDH or β-actin was amplified.
Lentiviral transduction
HEK293T cells were infected with the pLKO-1 vector (Sigma-Aldrich, St. Louis, MO, USA) containing two different eEF2K targeting shRNA sequences, sh-eEF2K-1 and sh-eEF2K-2, respectively:
CCGGCGATGAGGAAGGTTACTTCATCTCGAGATGAAGTAACCTTCCTCATCGTTTTT
CCGGCCAAAGGATTTGATTACTTACCTCGAGGTAAGTAATCAAATCCTTTGGTTTTTG or a non-targeting shRNA (Sigma) using OptiMEM, according to the manufacturer’s instructions. Viral particle producing media was then added to the BE(2)-C and SH-EP target NB cell lines with the addition of Polybrene (Santa Cruz Biotech, Santa Cruz, CA, USA). Cells successfully transduced were finally selected with the appropriate concentration of puromycin.
Nutrient deprivation
ND was performed with 50% subconfluent cultures in Hanks buffered saline solution (HBSS) as described:18 HEPES (CaCl2•2H2O, 0.185 g l1; MgSO4•7H2O, 0.2 g l–1; KCl, 0.4 g l–1; KH2PO4[anhydrous], 0.06 g l–1; NaHCO3, 0.35 g l–1; NaCl, 8 g l–1; Na2HPO4•7H2O, 0.09 g l–1; 20 mM HEPES, pH 7.4) containing no glucose, for the indicated times. As (HBSS)-HEPES buffer did not allow addition of DMSO without affecting cell viability, ND studies using A-484954 were performed with serum-free DMEM lacking glucose, sodium pyruvate, HEPES, or L-glutamine, for the indicated times.
Western blot analysis
Western blotting was carried out on cell lysates as previously described,18 using antibodies against phospho-eEF2 (Thr56), eEF2, eEF2K, caspase-3, cleaved caspase-3, AMPK, phospho-AMPKα (Thr172), GAPDH (all from Cell Signaling Technology, Danvers, MA, USA), MYCN (NCM-II, Thermo Fisher Scientific, Waltham, MA, USA), α-Tubulin (Sigma). Western blotting densitometric quantification was achieved using Adobe Photoshop 13.0.6.
MTT assays
To assess cell number at defined time-points, cell media was aspirated and MTT-supplemented media was added to the cells, which were subsequently incubated for 3 h and lysed with DMSO before measuring absorbance at 570 nm with a plate reader. Specifically, for MTT assays measuring cell proliferation, cells were plated in a 48-well plate at 20,000 cells per well in 200 μl media. Cells were allowed to attach overnight and an absorbance measure was obtained as a baseline for normalization of the following measures (i.e., time point 0 h), followed by the indicated time-points and treatments. For MTT assays measuring cell survival, cells were plated in a 48-well plate at 40,000 cells per well in 200 μl media, and the same procedures measuring absorbance as described above were performed.
Trypan blue assay
Cells were plated in 6-well plates at 1 × 106 cells per well in 2 ml media and were allowed to attach overnight. At the indicated time-points, cells were detached with 200 μl trypsin before blocking the reaction with 2 ml media. Cell samples were placed on a hemocytometer and pictures were taken from four grids. Viable cells were finally manually counted from the images.
Flow cytometry
Cells ±eEF2K knockdown were treated with ND media for the indicated time. Subsequently, cells were trypsinized, centrifuged (1,100 r.p.m., 4 °C, 5 min) and resuspended in PBS containing 20% FBS and stained for Annexin V and propidium iodide (PI) with a Dead Cell Apoptosis Kit, Thermo Fisher, according to the manifacturer’s instructions. Cells were analyzed on a FACSCalibur-Tangerine flow cytometry instrument. Unstained cells from all conditions were used to detect the background fluorescence signal.
Mouse studies
Aliquots of 2 × 106 cells were resuspended in 200 μl of 1:1 mixture of PBS and Matrigel and injected subcutaneously into the flanks of 5–6-week old female NOD SCID gamma immunodeficient mice using standard procedures. After day 17, mice klling was required when tumors exceeded humane practice guidelines (1,500 mm3). Mice were evaluated for tumor growth periodically until the experimental endpoints. Tumor volumes were estimated using the following formula: tumor length × (tumor width)2 × π/6 mm3(ref. 50). For CR experiments (i.e., mice receiving 70% of the daily food intake of their ad libitum counterparts), mice were fed a 30% calorie restricted irradiated rodent chow diet (70% caloric Restriction 2020, Envigo, Huntingdon, UK) from the day after cell injection until the experimental endpoints. Body weights and tumors size were recorded every 2 days. The CR diet was similar in consistency to the ad libitum diet but was fortified with vitamins and minerals to prevent deficiency in these nutrients. All animal experiment underwent ethical approval from the Animal Care Committee of the University of British Columbia.
Histopathology, immunohistochemistry, and tissue microarrays (TMAs)
For histopathology of mouse tissues, samples were formalin-fixed for 48 h, paraffin embedded, and sectioned at 5 μm. For each sample, sections were stained using standard hematoxylin and eosin (H&E) protocols. Extent of necrosis was manually quantified using Adobe Photoshop on 5 pictures (× 10 magnification, total surface: 1.61 mm2) randomly acquired on each HE section. Areas of structurally damaged tissue with no viable tumor cell nuclei were considered necrotic. For TMAs, four 1 mm diameter cores for each sample were punched from the paraffin block in non-necrotic areas and used for a TMA construction. The recipient block was subsequently sectioned at 5 μm thickness and IHC was performed using the following antibodies: p-eEF2 (Thr56, 1:800, Abcam ab53114), MYCN (1:100ss, Abcam ab198912), cleaved Caspase 3 (1:200ss, Cell Signaling 9664), and Ki67 (1:300, Thermoscientific RM-9106). IHC was blindly quantified by an MD with pathology training (AD) for staining intensity (0–3) and percentage of positive cells (0–100%). For each sample, the H-score was calculated as staining intensity × percentage of positive cells. When cores derived from the same sample showed a different H-score, the highest score was considered for the analysis.
Statistical analysis
Statistical differences between groups of data not normally distributed or with unknown distribution were calculated with the non-parametric Mann–Whitney U-test, unless otherwise specified. Repeated measures two-way ANOVA was performed for experiments with multiple time-points. Data are represented as mean+s.e.m., unless if box plots were used as indicated. Statistical analysis was performed using GraphPad Prism 6. Survival analysis and mRNA expression analysis were performed using R 3.3.1.
Acknowledgments
The work was supported in part by a Canadian Cancer Society Research Institute (CCSRI) Impact Grant (grant #703205) to PHS. The authors acknowledge Professor M. Schwab (DKFZ, Heidelberg) and Professor Y. DeClerck (CHLA, Los Angeles) for kindly providing cell lines.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
The authors also thank Amy Li, Melvin Pan, Jordan Cran, Saki Konomura, and Sylvia Lee for technical assistance. The authors have no conflicts of interest to disclose.
Supplementary Material
References
- Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F et al. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell 1983; 35(2 Pt 1): 359–367. [DOI] [PubMed] [Google Scholar]
- Schwab M, Varmus HE, Bishop JM, Grzeschik KH, Naylor SL, Sakaguchi AY et al. Chromosome localization in normal human cells and neuroblastomas of a gene related to c-myc. Nature 1984; 308: 288–291. [DOI] [PubMed] [Google Scholar]
- Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 1985; 318: 69–73. [DOI] [PubMed] [Google Scholar]
- Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev 2010; 20: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA 1999; 96: 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer 2002; 2: 764–776. [DOI] [PubMed] [Google Scholar]
- Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci USA 1998; 95: 1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 2007; 178: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 2013; 3: a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 1992; 69: 119–128. [DOI] [PubMed] [Google Scholar]
- Wek RC, Staschke KA. How do tumours adapt to nutrient stress? EMBO J 2010; 29: 1946–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell 2010; 40: 228–237. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet 2011; 12: 833–845. [DOI] [PubMed] [Google Scholar]
- Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochim Biophys Acta 2015; 1849: 845–860. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG, Ward MD, Mendola CE, Pavur KS, Dorovkov MV, Wiedmann M et al. Identification of a new class of protein kinases represented by eukaryotic elongation factor-2 kinase. Proc Natl Acad Sci USA 1997; 94: 4884–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Regulation and roles of elongation factor 2 kinase. Biochem Soc Trans 2015; 43: 328–332. [DOI] [PubMed] [Google Scholar]
- Ryazanov AG, Shestakova EA, Natapov PG. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature 1988; 334: 170–173. [DOI] [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 2013; 153: 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM. Recent advances in neuroblastoma. N Eng J Med 2010; 362: 2202–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer 2013; 13: 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Eng J Med 1985; 313: 1111–1116. [DOI] [PubMed] [Google Scholar]
- Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 2009; 27: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012; 483: 589–593. [DOI] [PubMed] [Google Scholar]
- Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst 2006; 98: 1193–1203. [DOI] [PubMed] [Google Scholar]
- Oberthuer A, Hero B, Berthold F, Juraeva D, Faldum A, Kahlert Y et al. Prognostic impact of gene expression-based classification for neuroblastoma. J Clin Oncol 2010; 28: 3506–3515. [DOI] [PubMed] [Google Scholar]
- Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol 1993; 11: 1466–1477. [DOI] [PubMed] [Google Scholar]
- Rosen N, Reynolds CP, Thiele CJ, Biedler JL, Israel MA. Increased N-myc expression following progressive growth of human neuroblastoma. Cancer Res 1986; 46: 4139–4142. [PubMed] [Google Scholar]
- Wu CA, Chao Y, Shiah SG, Lin WW. Nutrient deprivation induces the Warburg effect through ROS/AMPK-dependent activation of pyruvate dehydrogenase kinase. Biochim Biophys Acta 2013; 1833: 1147–1156. [DOI] [PubMed] [Google Scholar]
- Chen Z, Gopalakrishnan SM, Bui MH, Soni NB, Warrior U, Johnson EF et al. 1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) induces phosphorylation of eukaryotic elongation factor-2 (eEF2): a cautionary note on the anticancer mechanism of an eEF2 kinase inhibitor. J Biol Chem 2011; 286: 43951–43958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari V, Borriello L, Turcatel G, Shimada H, Sposto R, Fernandez GE et al. MYCN-dependent expression of sulfatase-2 regulates neuroblastoma cell survival. Cancer Res 2014; 74: 5999–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 1996; 13: 803–812. [PubMed] [Google Scholar]
- Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res 1989; 49: 219–225. [PubMed] [Google Scholar]
- Puissant A, Frumm SM, Alexe G, Bassil CF, Qi J, Chanthery YH et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov 2013; 3: 308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P. Metabolic microenvironment of tumor cells: a key factor in malignant progression. Exp Oncol 2010; 32: 125–127. [PubMed] [Google Scholar]
- Ham J, Costa C, Sano R, Lochmann TL, Sennott EM, Patel NU et al. Exploitation of the apoptosis-primed state of MYCN-amplified neuroblastoma to develop a potent and specific targeted therapy combination. Cancer Cell 2016; 29: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 2012; 21: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson WC, Meyerowitz JG, Nekritz EA, Chen J, Benes C, Charron E et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 2014; 26: 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogen D, Wei JS, Azorsa DO, Ormanoglu P, Buehler E, Guha R et al. Aurora B kinase is a potent and selective target in MYCN-driven neuroblastoma. Oncotarget 2015; 6: 35247–35262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6: 318–327. [DOI] [PubMed] [Google Scholar]
- Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab 2014; 19: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. GTP hydrolysis in protein synthesis: two for Tu? Science 1993; 259: 1264–1265. [DOI] [PubMed] [Google Scholar]
- Arora S, Yang JM, Kinzy TG, Utsumi R, Okamoto T, Kitayama T et al. Identification and characterization of an inhibitor of eukaryotic elongation factor 2 kinase against human cancer cell lines. Cancer Res 2003; 63: 6894–6899. [PubMed] [Google Scholar]
- Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 2007; 27: 2059–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005; 7: 311–318. [DOI] [PubMed] [Google Scholar]
- Pfister S, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J et al. Outcome prediction in pediatric medulloblastoma based on DNA copy-number aberrations of chromosomes 6q and 17q and the MYC and MYCN loci. J Clin Oncol 2009; 27: 1627–1636. [DOI] [PubMed] [Google Scholar]
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 2013; 3: 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C, Pawel B, Seiser E, Zhao H, Raabe E, Wang Q et al. Neural cell adhesion molecule (NCAM) isoform expression is associated with neuroblastoma differentiation status. Pediatr Blood Cancer 2008; 51: 10–16. [DOI] [PubMed] [Google Scholar]
- Detre S, Saclani Jotti G, Dowsett M. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995; 48: 876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaemmerer D, Peter L, Lupp A, Schulz S, Sanger J, Baum RP et al. Comparing of IRS and Her2 as immunohistochemical scoring schemes in gastroenteropancreatic neuroendocrine tumors. Int J Clin Exp Pathol 2012; 5: 187–194. [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X et al. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci USA 2007; 104: 18636–18641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.