Abstract
Circular RNAs (circRNAs) are a subclass of noncoding RNAs widely expressed in mammalian cells. We report here the tumorigenic capacity of a circRNA derived from angiomotin-like1 (circ-Amotl1). Circ-Amotl1 is highly expressed in patient tumor samples and cancer cell lines. Single-cell inoculations using circ-Amotl1-transfected tumor cells showed a 30-fold increase in proliferative capacity relative to control. Agarose colony-formation assays similarly revealed a 142-fold increase. Tumor-take rate in nude mouse xenografts using 6-day (219 cells) and 3-day (9 cells) colonies were 100%, suggesting tumor-forming potential of every cell. Subcutaneous single-cell injections led to the formation of palpable tumors in 41% of mice, with tumor sizes >1 cm3 in 1 month. We further found that this potent tumorigenicity was triggered through interactions between circ-Amotl1 and c-myc. A putative binding site was identified in silico and tested experimentally. Ectopic expression of circ-Amotl1 increased retention of nuclear c-myc, appearing to promote c-myc stability and upregulate c-myc targets. Expression of circ-Amotl1 also increased the affinity of c-myc binding to a number of promoters. Our study therefore reveals a novel function of circRNAs in tumorigenesis, and this subclass of noncoding RNAs may represent a potential target in cancer therapy.
Circular RNAs (CircRNAs) are a large group of noncoding transcripts that form covalently closed continuous loop where the 3′ and 5′ RNA ends are joined together.1, 2, 3 CircRNAs can be generated from exons, producing exonic circRNAs,4, 5 or from introns, producing intronic circRNAs.6 They can also be generated from intron-containing exons, producing exon–intron circRNAs.7 Recent studies have shown that a wide array of endogenous circRNAs are expressed in animal cells,3, 8, 9 while certain circRNAs are highly specific to cell type and/or developmental stage.8, 10, 11 Genome-wide analyses have revealed high levels of abundance and evolutionary conservation of circRNAs across species.12, 13, 14, 15 One mode of action for circRNAs may be the miRNA sponge activity of some circRNAs.16 We have also reported that circ-Foxo3 functions as a sponge to interact with a number of microRNAs.17 There is evidence suggesting that some circRNAs may bind to RNA-binding proteins to form RNA–protein complexes and regulate their activities.18, 19 Levels of circRNAs can also be regulated by microRNA-dependent silencing involving Ago2-mediated cleavage.20 Our studies have shown that circ-Foxo3 interacts with different proteins in related signal pathways and regulated cell activities.15, 21 This study was designed to explore the potential role of circ-Amotl1 in breast cancer tumorigenesis.
Results
Effect of circ-Amotl1 on tumor cells
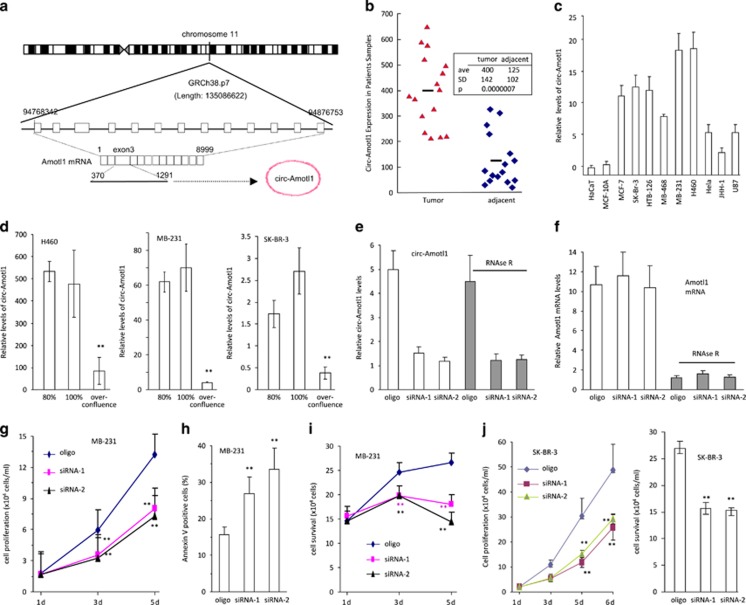
To explore the roles of circRNAs in cancer development, we examined circRNA expression in a number of cancer and non-cancer cell lines, based on previous sequencing information.14, 22 We confirmed the endogenous expression of a few circRNAs, including the circRNA generated from angiomotin like1 (circ-Amotl1, hsa_circ_0004214). This circRNA was generated from human angiomotin-like 1 gene.14, 22, 23, 24 Amotl1 is located in chromosome 11 and circ-Amotl1 is generated from Exon-3, the largest exon of Amotl1 (Figure 1a). We then measured circ-Amotl1 levels in breast carcinoma biopsies and in adjacent tissues and found that the levels of circ-Amotl1 in tumor specimen were significantly higher than those in the adjacent tissues (Figure 1b). The expression of circ-Amotl1 was determined in a number of cancerous and non-cancerous cell lines. We observed that the levels of circ-Amotl1 were significantly higher in cancer cell lines than in non-cancer cell lines (Figure 1c). The levels of circ-Amotl1 decreased significantly when the cells were maintained to over-confluence (Figure 1d), suggesting a role of circ-Amotl1 in cell proliferation.
Figure 1.
Circ-Amotl1 increased cell survival and colony formation. (a) Relationship of Amotl1 genomic DNA, mRNA and circRNA. (b) Total RNAs were isolated from the specimens of patients with breast carcinoma and subject to real-time PCR. Tumor samples showed significantly higher levels of circ-Amotl1 than the adjacent tissues. (c) Expression of circ-Amotl1 was analyzed in a variety of cell lines by real-time PCR. Nine cancer cell lines expressed higher levels of circ-Amotl1 relative to two non-cancer cell lines HaCaT and MCF-10A. (d) H460, MDA-MB-231, and SK-BR-3 were maintained at 80 and 100% or cultured to over-confluence, when some cells began to detach. RNAs were isolated and subject to real-time PCR to measure circ-Amotl1 levels. Asterisks indicate significant differences. (e and f) Total RNA extracted from the control oligo- or circ-Amotl1 siRNA-transfected MDA-MB-231 cells was incubated with or without RNAse R followed by real-time PCR. RNAse R treatment did not affect circ-Amotl1 levels but decreased linear Amotl1 mRNA levels. (g) Silencing circ-Amotl1 expression in MDA-MB-231 cells decreased ability of cell proliferation compared with the control. **P<0.01. Error bars, S.D. (n=4). (h) Silencing circ-Amotl1 expression in MDA-MB-231 cells increased cell apoptosis (n=4). (i) Silencing circ-Amotl1 expression in MDA-MB-231 cells decreased ability of cell survival. (n=4). (j) Left, Silencing circ-Amotl1 in SK-BR-3 cells decreased rates of cell proliferation (left) and viability (right) relative to the control. **P<0.01, n=4
To test this possibility, we designed two siRNAs (Supplementary Figure S1a) to specifically target circ-Amotl1 (Figure 1e), but not linear Amotl1 mRNA (Figure 1f). Following this, we used RNAse-R to digest linear Amotl1 mRNA but not circ-Amotl1. We examined the phenotypic effects of these siRNAs and found that silencing circ-Amotl1 decreased cell proliferation (Figure 1g) but increased cell apoptosis (Figure 1h,Supplementary Figure S1b). Silencing circ-Amotl1 subsequently decreased cell survival (Figure 1i). Circ-Amotl1 siRNA impeded the ability of MDA-MB-231 cells to form colonies (Supplementary Figure S1c). We confirmed that the siRNA did not show off-target effects on Amotl1 protein expression (Supplementary Figure S1d). In addition, we silenced circ-Amotl1 in SK-BR-3 cells and observed similar results (Figure 1j).
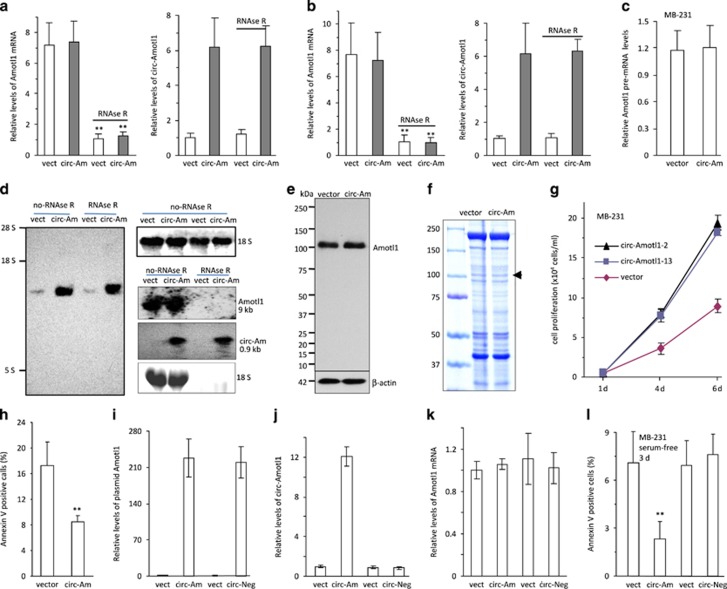
We generated a circ-Amotl1 expression construct (Supplementary Figures S1e and f) and confirmed that ectopically expressed circ-Amotl1 was resistant to RNAse-R digestion, as measured by real-time PCR and northern blotting (Supplementary Figure S1g). RNAse-R treatment decreased linear mRNA but not circ-Amotl1 (Figure 2a). Ectopic addition of mouse total RNA to the digested samples did not affect significantly the results (Figure 2b,Supplementary Tables S1 and S2). We confirmed that ectopic circ-Amotl1 did not affect pre-mRNA levels (Figure 2c). By northern blotting, a probe specific to circ-Amotl1 could detect the predicted circ-Amotl1 band (Figure 2d left). We also designed a probe specific for a shared sequence between circ-Amotl1 and Amotl1 (Figure 2d right) and found that transfecting circ-Amotl1 did not affect the translation of linear Amotl1 mRNA (Figure 2e). Transfection of circ-Amotl1 did not alter the expression of total proteins (Supplementary Figure S1h) nor the interaction of Amotl1 with its partners (Figure 2f). Functionally, we found that expressing circ-Amotl1 in MDA-MB-231 cells conferred a higher proliferative capacity (Figure 2g) and resistance to serum-free growth conditions (Supplementary Figure S1i). The circ-Amotl1-transfected cells also displayed decreased rates of apoptosis (Figure 2h,Supplementary Figure S1j).
Figure 2.
Expression of circ-Amotl1 increased cell proliferation. (a) Total RNA extracted from mock- or circ-Amotl1-transfected MDA-MB-231 cells was incubated with or without RNAse R followed by real-time-PCR. Although RNAse R treatment decreased Amotl1 linear RNA levels, it did not affect circ-Amotl1 levels. **P<0.01. Error bars, S.D. (n=4). (b) After RNAse R digestion, the samples were subject to RNA spike-in with mouse total RNA followed by real-time PCR. Similar results were obtained. (c) Real-time PCR using two primers located in the intron and exon showed that transfection with circ-Amotl1 did not affect pre-mRNA levels of Amotl1. n=4. (d) Northern blotting showing circ-Amotl1 expression in vector (vect)- and circ-Amotl1 (circ-Am)-transfected MDA-MB-231 cells. RNAse R digestion did not affect circ-Amotl1 levels (left) but degraded linear RNAs (right). (e) circ-Amotl1 transfection did not affect linear Amotl1 expression. (f) Cell lysates were precipitated with anti-Amotl1 antibody followed by SDS-PAGE and Coomassie blue staining. Transfection with circ-Amotl1 did not affect interaction of Amotl1 with its partners. (g and h) Cell proliferation was assayed in vector- and circ-Amotl1-transfected MAD-MB-231 cells. Transfection with circ-Amotl1 increased proliferation (g) but decreased apoptosis (h) compared with the control. (i) Cells transfected with the circ-Amotl1 construct without an acceptor intron (circ-Neg) produced the same levels of transcript as cells transfected with circ-Amotl1 (n=4). (j) Transfection with circ-Neg did not produce circ-Amotl1 (n=4). (k) Transfection with circ-Neg had no effect on Amotl1 mRNA (n=4). (l) Circ-Neg had no effect on repressing cell apoptosis relative to circ-Amotl1 (n=4)
To test the effect of circularization, we removed the acceptor intron from the circ-Amotl1 construct producing circ-Neg. As a consequence, the construct circ-Neg could express the fragment of circ-Amotl1 (Figure 2i), but the fragment could not circularize (Figure 2j). Transfection with circ-Neg, similar to circ-Amotl1, had no effect on the expression of endogenous Amotl1 mRNA (Figure 2k). As a result, the inhibitory effect of circ-Amotl1 on cell apoptosis was abolished owing to lack of circularization (Figure 2l). Transfection with circ-Amotl1 (Supplementary Figure S2a) also increased proliferation in other cell lines, including HepG2 (Supplementary Figure S2b), NIH3T3 (Supplementary Figure S2c), and YPEN (Supplementary Figure S2d). Colony formation was assayed in the cancerous cell lines, and we detected larger colonies formed in the circ-Amotl1-transfected MDA-MB-231 cells (Supplementary Figure S2e) and HepG2 cells (Supplementary Figure S2f).
Circ-Amotl1 in nude mice xenografts
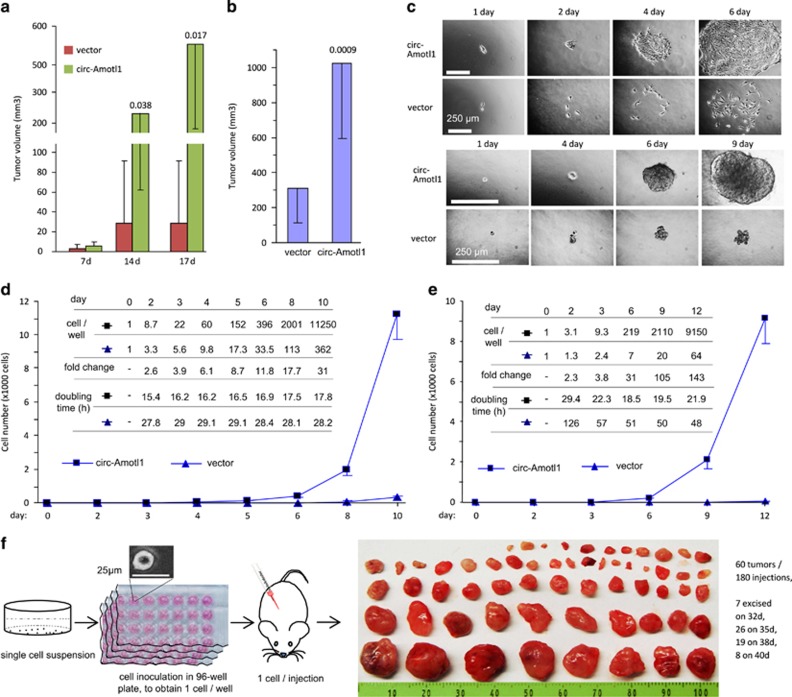
Similar to our in vitro findings, the tumor volume of both MDA-MB-231 and HepG2 xenografts increased significantly, compared with controls (Figures 3a and b). MDA-MB-231 tumors were excised 17 days postinjection and analyzed histologically. In addition to the gross difference in tumor volume (Supplementary Figure S2g), we found significant adhesion between the tumor mass and stromal tissues. H&E staining showed extensive invasion between tumor cells and muscle and between tumor cells and bone (Supplementary Figure S2h). In vitro, cell invasion assays demonstrated that the circ-Amotl1 transfection increased the invasive capacity of MDA-MB-231 cells through Matrigel (Supplementary Figure S2i). Sections from the MDA-MB-231 tumors were subject to TUNEL staining, where we detected a reduced number of apoptotic cells in circ-Amotl1 xenografts, relative to control (Supplementary Figures S3a and b). Immunohistochemical (IHC) staining of CD34 further revealed increased tumor endothelium in the circ-Amotl1 tumors (Supplementary Figure S3c). Interestingly, in mice with circ-Amotl1-transfected xenografts, we also observed invasion of tumor cells and tumor vasculature into bone, resulting in bone degradation (Supplementary Figure S3d).
Figure 3.
Expression of circ-Amotl1 enhanced tumor formation. (a) MDA-MB-231 cells transfected with circ-Amotl1 or the vector were injected subcutaneously into nude mice. Tumor growth was monitored for 17 days. Expression of circ-Amotl1 increased tumor growth. (b) HepG2 cells transfected with circ-Amotl1 or the vector were injected subcutaneously into nude mice for tumor growth assay. Expression of circ-Amotl1 increased tumor growth. (c) Upper, cells were inoculated in 96-well plates to obtain one cell per well. Typical photos are shown for up to 6 days. Lower, cells were inoculated in 96-well plates to obtain one cell per well. Colony formation was monitored for up to 12 days. Typical photos are shown for up to 9 days. (d) Single-cell growth curve for up to 10 days. The numbers of cells per well are shown as the inset. To the end, the circ-Amotl1 cells grew 30-fold faster than the control with doubling time 17.8 h. (e) Colony growth curve for up to 12 days. The numbers of cells per colony are shown as the inset. To the end, the circ-Amotl1 cells grew 142-fold faster than the control with doubling time 21.9 h. (f) Single-cell suspension was distributed into 96-well plates. After confirming the presence of one cell per well, each cell was injected subcutaneously. Tumors were isolated on days 32, 35, 38 and 40
To understand how circ-Amotl1 functioned in tumorigenesis, we selected uniform cell populations by isolating cells from the single colonies formed in agarose. Although the total population of circ-Amotl1-transfected cells showed an elongated phenotype as compared with the control, the uniform circ-Amotl1 cells not only maintained this elongated phenotype but also displayed a greater capacity in cell–cell adhesion (Supplementary Figure S4a). By northern blotting, we confirmed that circ-Amotl1 was highly expressed in circ-Amotl1-transfected cells (Supplementary Figure S4b).
To further characterize the highly tumorigenic circ-Amotl1-transfected cells, we then performed single-cell cultures. Within 6 days, large spread colonies, 1 mm in diameter, had formed (Figure 3c, upper). A single circ-Amotl1-expressing cell grew to a population of 11 250 cells in 10 days, which represented a 30-fold increase over the vector-transfected cells (Figure 3d). The doubling time of the circ-Amotl1-expressing cells was between 15 and 18 h, a growth rate much faster than any known wild-type cancer cell (Figure 3d,Supplementary Figure S4c). It is reported that wild-type MDA-MB-231 cells have a doubling time of 1.1 days and the GFP-transfected MDA-MB-231 cells have a doubling time of 1.4 days.25 In a population culture, we confirmed that the doubling time for the vector-transfected MDA-MB-231cells was 26.64 h, whereas the doubling time for the circ-Amotl1-transfected cells was 15.94 h (Supplementary Figure S4d). We also found that individual circ-Amotl1 cells could form colonies significantly larger than those from the vector-transfected cells (Figure 3c,Supplementary Figure S4e). Twelve days after inoculation, we detected a 142-fold increase in the number of circ-Amotl1-transfected cells compared with the control (Figure 3e).
We performed single-cell culture in agarose gel for 6 days. These colonies of approximately 219 cells were then individually injected into nude mice. Mice were also injected with colonies derived from vector-transfected cells. Each of the 219 cell colonies that had originated from one circ-Amotl1 cell began to form tumors within 10 days. By 14 days, some tumors caused skin ulcerations and were discarded. All mice were killed on day 28. Seventeen tumors were obtained from 18 injections (Supplementary Figure S5a). Excluding one technical incident, this represented a nearly 100% tumor-take rate (Supplementary Figure S5b). No tumor formation was observed from control cells, as 107 vector-transfected cells were needed to form a tumor in 28 days.
We then repeated the procedure using 3-day-old colonies of approximately nine cells. Twenty-eight days postinjection, some tumors began to cause skin ulcerations and the mice were killed. By day 31, all mice were killed. Seventeen tumors were obtained from 20 injections (Supplementary Figure S5c). In the controls, mice were injected with different number of cells, none of which produced tumor. Considering an 85% success rate of colony harvest and transfer (Supplementary Figure S5d), this represented 100% rates of tumor formation injected from the 3-day colonies.
Finally, we injected mice with single cells. Some animals formed visible tumors by 20 days after injection. Some tumors were excised by 32 and 35 days after injection. All mice were killed by day 40, and we obtained 60 tumors from 180 injections (Figure 3f). Considering 80% success rates of cell harvest and transfer (Supplementary Figures S5e and f), this represented a 41% tumor-take rate using single-cell subcutaneous injections.
Interaction of circ-Amotl1 with c-myc
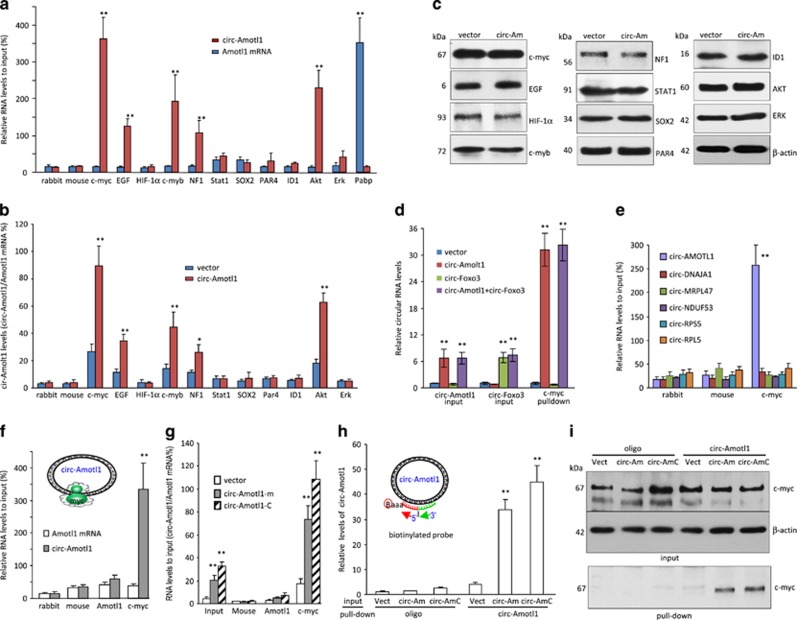
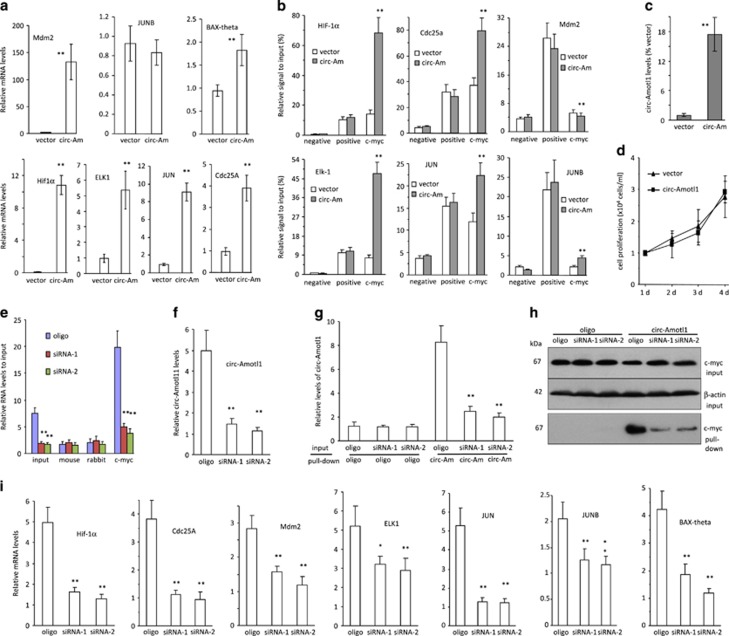
To study how circ-Amotl1 could trigger this highly tumorigenic phenotype, we tested potential interactions of circ-Amtol1 with oncogenic proteins. Cell lysis prepared from MDA-MB-231 cells were subject to immunoprecipitation. It showed that circ-Amotl1 was pulled down by antibodies against c-myc, EGF, c-myb, NF1, and AKT, which did not pull down linear Amotl1 mRNA (Figure 4a). Circ-Amotl1 also interacted with other oncogenic proteins (Supplementary Figure S6a). In addition, we found that only antibodies against c-myc, EGF, c-myb, NF1, and AKT were able to pull down significantly higher levels of circ-Amotl1 from the circ-Amotl1-transfected cells than those from the vector-transfected cells (Figure 4b). It was noted that c-myc showed the greatest binding capacity to circ-Amotl1, suggesting a critical role of c-myc in mediating circ-Amotl1 functions. By western blotting, we showed that these proteins were equally expressed (Figure 4c).
Figure 4.
circ-Amotl1 interacted with c-myc protein. (a) MDA-MB-231 cell lysate was incubated with antibodies as indicated. Anti-c-myc, EGF, c-myb, NF1, and AKT antibodies pulled down circ-Amotl1 but not linear Amotl1 mRNA. **P<0.01. Error bars, S.D. (n=4). (b) Lysates prepared from vector- or circ-Amotl1-transfected MDA-MB-231 cells were incubated with antibodies as above, followed by real-time PCR. Anti-c-myc, EGF, c-myb, NF1, and AKT antibodies pulled down significantly higher levels of circ-Amotl1 in the circ-Amotl1-transfected cells relative to the control. *P<0.05, **P<0.01. Error bars, S.D. (n=4). (c) Equal amounts of proteins were detected in the vector- and circ-Amotl1-transfected cells. (d) The lysates were also incubated with antibodies as indicated, followed by real-time PCR. Anti-c-myc antibody pulled down circ-Amotl1 but not circ-Foxo3. (n=4). (e) Cell lysates prepared from MDA-MB-231 cells were subjected to immunoprecipitation, followed by real-time PCR. Antibodies against c-myc pulled down circ-Amotl1 but not the other circRNAs. (n=4). (f) MDA-MB-231 cell lysate was incubated with different antibodies followed by real-time PCR. Anti-c-myc antibody pulled down circ-Amotl1 but not linear Amotl1 mRNA. (n=4). (g) Lysates prepared from vector- or circ-Amotl1-transfected (including circ-Amotl1-m and circ-Amotl1-c) cells were incubated with antibodies as above, followed by real-time PCR. Anti-c-myc antibody pulled down circ-Amotl1 but not Amotl1 mRNA. (n=4). (h) The mixes were incubated with streptavidin beads to pull down circ-Amorl1. Real-time PCR showed that the probe of circ-Amotl1 pulled down significantly higher levels of circ-Amotl1 in cells transfected with circ-Amotl1 than in the control cells. (n=4). (i) Protein lysates prepared from panel (h) were subject to western blotting. The probe complementary to circ-Amotl1 pulled down more c-myc than the control
We then prepared cell lysates from MDA-MB-231 cells transfected with circ-Amotl1, an empty vector, an unrelated circ-Foxo3, or both circ-Amotl1 and circ-Foxo3.21, 26 Although we showed that both circRNAs were equally expressed, anti-c-myc antibody precipitated circ-Amotl1 only and not circ-Foxo3 (Figure 4d). We also examined the interaction between c-myc with other circRNAs. The pull-down experiment revealed that the antibody against c-myc only precipitated circ-Amotl1 but did not pull down the other circRNAs (Figure 4e).
We examined the interaction between circ-Amotl1 and c-myc in detail. MDA-MB-231 cell lysate was incubated with an antibody against c-myc, followed by real-time PCR. We found that anti-c-myc antibody pulled down circ-Amotl1 but not the linear Amotl1 mRNA (Figure 4f). Anti-c-myc antibody also pulled down significantly higher levels of circ-Amotl1 from the circ-Amotl1-transfected cells than those from the control cells (Figure 4g). We tested whether the probe used for northern blotting had the capacity to pull down circ-Amotl1. Although the total inputs of circ-Amotl1 and the probe were similar (Supplementary Figure S6b), streptavidin beads pulled down significantly higher levels of circ-Amotl1 compared with the control (Figure 4h). Using this probe, higher levels of c-myc were pulled down (Figure 4i).
Circ-Amotl1 regulates c-myc function and translocation
It has been reported that c-myc can regulate up to 15% of gene expression.27 We analyzed the expression of some known c-myc targets in our circ-Amotl1- and vector-transfected cells and found that a number of c-myc targets were upregulated in the circ-Amotl1-transfected cells cultured at sub-confluence (Figure 5a) and full confluence (Supplementary Figure S6c). As some cells might start to undergo apoptosis, we performed all of our experiments using sub-confluent cultures. We analyzed the effect of circ-Amotl1 on the interaction between c-myc and different promoter regions. We found that ectopic circ-Amotl1 enhanced the binding affinity of c-myc to the promoters of HIF-1α, Cdc25a, ELK-1, and JUN (Figure 5b), using primers amplifying these promoters (Supplementary Figure S6d). To validate the effect of c-myc in mediating circ-Amotl1 functions, we transfected the c-myc null cells HO15.1928, 29 with either circ-Amotl1 or a control vector. Although the transfected c-myc null cells now expressed circ-Amotl1 (Figure 5c), there was no change in cell proliferation (Figure 5d), confirming the role of c-myc in mediating circ-Amotl1 functions. To validate these results, we performed silencing assays. Transfection with the siRNAs targeting circ-Amotl1 decreased the levels of precipitated c-myc (Figure 5e). With circ-Amotl1 siRNA knockdown (Figure 5f), the probe that specifically targeted circ-Amotl1 was shown to pull down significantly lower levels of circ-Amotl1 mRNA (Figure 5g,Supplementary Figure S6e) and c-myc protein (Figure 5h). Finally, silencing circ-Amotl1 significantly downregulated c-myc targets compared with the control oligo (Figure 5i).
Figure 5.
circ-Amotl1 promoted the expression of c-myc targets. (a) The circ-Amotl1- and vector-transfected MDA-MB-231 cells were maintained to 80% confluence followed by RNA extraction and measurement of c-myc targets. The circ-Amotl1 cells expressed significantly higher levels of c-myc targets than the control. (b) Chromatins of vector- and circ-Amotl1-transfected MDA-MB-231 cells were isolated, digested and immunoprecipitated with rabbit IgG (negative control) and antibodies against Histone H3 (positive control) or c-myc, followed by PCR with specific primers flanking a piece of DNA sequence at the promoter regions of HIF-1α, Cdc25a, Mdm2, ELK1, JUN, and JUNB. circ-Amotl1 expression enhanced c-myc binding to the promoters of HIF-1α, Cdc25a, ELK1, and JUN. (c) The c-myc null cells HO15.19 were transfected with circ-Amotl1 or a control vector followed by real-time PCR to confirm overexpression of the construct. (d) Cell proliferation was tested in the HO15.19 cells transfected with circ-Amotl1 or the vector. Little difference was detected. (e) Lysates prepared from control oligo- or circ-Amotl1 siRNA-transfected cells were subject to anti-c-myc antibody precipitation assay. PCR showed that circ-Amotl1 silencing decreased precipitation by anti-c-myc antibody. **P<0.01. Error bars, S.D. (n=4). (f) Cells transfected with the siRNAs expressed significantly lower levels of circ-Amotl1 than the control. (n=4). (g) The mixes of input and probe as indicated were incubated with streptavidin beads to pull down circ-Amorl1. Real-time PCR showed that the probe of circ-Amotl1 pulled down significantly lower levels of circ-Amotl1 in cells transfected with siRNAs than in the control cells. (n=4). (h) Protein lysates prepared from panel (e) were subject to western blotting probed with antibodies as indicated. The probe complementary to circ-Amotl1 pulled down less c-myc in the siRNA-transfected cells than the control. (i) The siRNA-transfected cells expressed significantly lower levels of c-myc targets than the control. *P<0.05, **P<0.01. Error bars, S.D. (n=4)
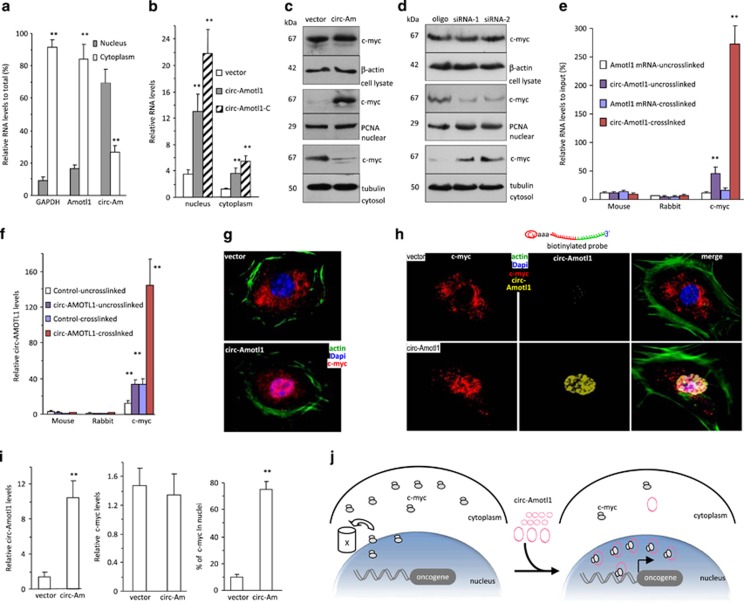
It has been reported that exonic circRNAs are distributed mainly in cytoplasm.2, 16 We isolated total RNA from cytoplasm and nuclei and measured the levels of circ-Amotl1. Higher levels of circ-Amotl1 were detected in nuclei than in cytoplasm (Figure 6a). Significantly higher levels of the ectopic circ-Amotl1 were also detected in the nuclei relative to the cytoplasm (Figure 6b). Western blotting using fractionated samples showed a similar distribution of c-myc (Figure 6c). Silencing circ-Amotl1 produced an opposite pattern of c-myc distribution (Figure 6d). To examine whether circ-Amotl1 bound to c-myc in nuclei, we performed a crosslinking assay and found that the anti-c-myc antibody precipitated circ-Amotl1 but not linear Amotl1 mRNA (Figure 6e). Ectopic expression of circ-Amotl1 increased the levels of the pulled down product (Figure 6f).
Figure 6.
circ-Amotl1 promoted nuclear translocation of c-myc. (a) RNAs extracted from cytoplasm and nuclei of MDA-MB-231 cells were subject to real-time PCR. GAPDH and Amotl1 were mainly expressed in the cytoplasm, while significantly higher levels of circ-Amotl1 were detected in the nuclei than in cytoplasm. **P<0.01. Error bars, S.D. (n=4). (b) RNAs extracted from cytoplasm and nuclei of MDA-MB-231 cells transfected with circ-Amotl1 or the vector were subjected to real-time PCR. Significantly higher levels of circ-Amotl1 were detected in the nuclei than in the cytoplasm. (n=4). (c) Upper showing equal levels of c-myc in the cell lysate. Middle showing higher level of c-myc was detected in the nucleus fraction in the circ-Amotl1-transfected cell. Lower showing lower level of c-myc was detected in the cytoplasm. (d) Western blottings revealed equal levels of c-myc in the cell lysates. Lower levels of c-myc were detected in the nucleus fraction, but higher levels were detected in the cytoplasm in the cells transfected with circ-Amotl1 siRNAs relative to control oligo. (e) MDA-MB-231 nuclear lysate was subjected to immunoprecipitation, followed by real-time PCR. RIP assays indicated that anti-c-myc antibody pulled down more circ-Amotl1 than linear RNA (n=6). (f) RIP assays indicated anti-c-myc antibody pulled down more circ-Amotl1 from cells transfected with circ-Amotl1 than from control (n=6). (g) The mock- and circ-Amotl1-transfected MDA-MB-231 cells were stained with phalloidins (green), DAPI (blue), and c-myc (red). High intensity of c-myc was detected in the nucleus of the circ-Amotl1-transfected cells. (h) The overexpressed circ-Amotl1 was found co-localized with c-myc in the nucleus in the circ-Amotl1-transfected cells. (i) Quantitation of circ-Amotl1 expression and nuclear translocation. (j) Diagram showing c-myc nuclear translocation facilitated by circ-Amotl1
By confocal microscopy, we detected mainly cytoplasmic c-myc in the vector-transfected cells and nuclear distribution of c-myc in the circ-Amotl1-transfected cells (Figure 6g, full panels provided in Supplementary Figure S6f). Using the probe, we detected co-localization of circ-Amotl1 and c-myc in the nuclei of the circ-Amotl1 cells (Figure 6h, full panels provided in Supplementary Figure S7a). Scanning the expression levels confirmed circ-Amotl1 overexpression alongside nuclear c-myc in circ-Amotl1-transfected cells (Figure 6i,Supplementary Figure S7b). Silencing circ-Amotl1 facilitated cytoplasmic distribution of c-myc (Supplementary Figure S7c). Taken together, our study showed that the ectopic expression of circ-Amotl1 induced nuclear translocation of c-myc (Figure 6j). It has been reported that nuclear c-myc is readily degraded.30, 31 In our study, high levels of c-myc were translocated to the nucleus, but we did not detect decreased level of total c-myc. This suggested that c-myc was co-localized with circ-Amotl1, preventing degradation.
Identification of circ-Amotl1-binding site in mediating c-myc function
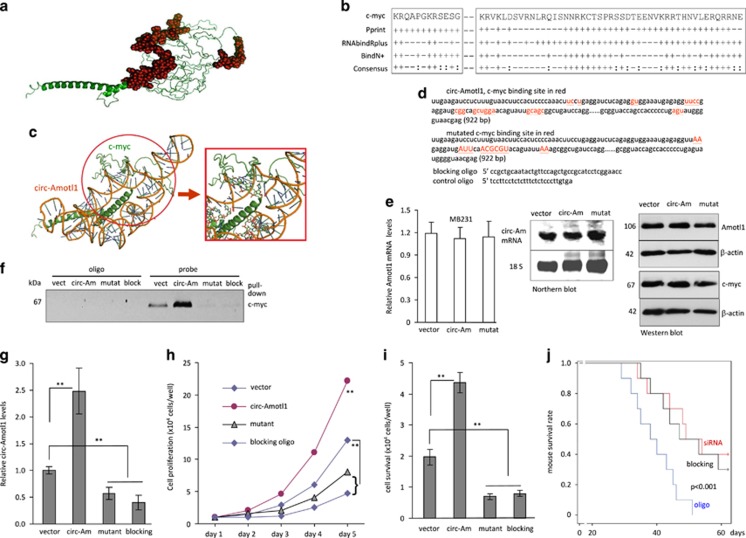
As the structure of full-length c-myc has yet to be elucidated, a three-dimensional (3D) model was built using I-TASSER (Iterative Threading ASSEmbly Refinement).32 Five models of c-myc were computationally generated using the I-TASSER algorithm with C-scores ranging from −3.80 to −1.26, the TM score was 0.56±0.15 and the estimated root-mean-square deviation was 9.9±4.6 Å. The confidence scores ranged from −5 to 2, with higher scores representing higher confidence in the model. We thus derived a 3D structure of c-myc (Figure 7a) to analyze potential circ-Amotl1-binding sites. Using RNABindRPlus,33 BindN+,34 and Pprint,35 we conducted a computational screen of c-myc for RNA-binding residues. We found that c-myc shared consensus sequences with other RNA-binding proteins (Figure 7b).
Figure 7.
Identification of circ-Amotl1 binding on c-myc. (a) Three-dimensional model of c-myc. (b) Prediction of c-myc RNA-binding residues was performed using Pprint, RNABindRPlus, and BindN+. Plus signs indicate the predicted RNA-binding residues. (c) NPDock graphical representation of the three-dimensional docking models of circ-Amotl1 and c-myc. (d) Circ-Amotl1 sequence labeling c-myc-binding site (red) and the mutated nucleotides (red). (e) MB-231 cells transfected with the mutant construct produced similar levels of Amotl1 mRNA, measured by real-time PCR (left) and northern hybridization (middle) relative to controls. They also expressed similar protein levels of Amotl1 and c-myc (right). (f) The probe could not precipitate c-myc following transfection with either the circ-Amotl1 siRNA or the blocking oligo. (g) Anti-c-myc antibody precipitated reduced circ-Amotl1 following transfection with circ-Amotl1 siRNA or the blocking oligo. (h and i) Delivery of circ-Amotl1 siRNA or the blocking oligo decreased cell proliferation (h) and cell survival (i). (j) Mice were injected intraperitoneally with MDA-MB-231 cells. Kaplan–Meier survival curves show that injection with circ-Amotl1 siRNA or the blocking oligo prolonged mouse survival significantly
The secondary structure of circ-Amotl1 was predicted by analyzing its thermodynamic properties using the formula ΔG=ΔH–TΔS, ΔG=−52.2 kcal/mol at 37 °C, ΔH=−515 kcal/mol, and ΔS=−1492.1 cal/(K·mol), where T (K) is the absolute temperature and ΔG, ΔH, ΔS, denote the change in free energy, enthalpy, and entropy, respectively. NPDock was used to perform in silico molecular docking of circ-Amotl1 with c-myc. Based on these predictions and the electrostatic surface potential calculation studies, c-myc was modeled with the circ-Amotl1 (Figure 7c). It appeared that the RNA-binding residues were concentrated in the central region and in the C-terminal domain of c-myc. Our in silico simulation demonstrated that circ-Amotl1 could dock with c-myc. The contact map (Supplementary Figure S8a), residue-level resolution contact maps (Supplementary Figure S8b), MC score (Supplementary Figure S8c), contact distance (Supplementary Table S3), accessible surface area (Supplementary Table S4), and interaction overview (Supplementary Table S5) all supported our conclusion that circ-Amotl1 could bind to c-myc.
We mutagenized the c-myc interaction site and designed blocking oligos complementary to the binding sites (Figure 7d). Mutation of the c-myc-binding site did not affect expression of Amotl1 and c-myc (Figure 7e). Lysates prepared from MDA-MB-231 cells transfected with circ-Amotl1, the vector, the mutant construct, or the blocking oligo were subjected to pull-down assay. The circ-Amotl1 probe could no longer precipitate c-myc when the cells were transfected with the mutant construct or the blocking oligo (Figure 7f). As well, the levels of circ-Amotl1 precipitated by the anti-c-myc antibody were significantly lower in the cells transfected with the mutant or the blocking oligo (Figure 7g). As a result, cells transfected with the mutant or the blocking oligo displayed decreased levels of proliferation (Figure 7h) and survival (Figure 7i).
We then examined the blocking oligo in vivo. As the efficiency of delivering the blocking oligo into solid tumors was relative low, formation of ascites tumors was performed in nude mice by injecting MDA-MB-231 cells intraperitoneally. This was followed by control oligo, circ-Amotl1 siRNA, or blocking oligo treatment. Mouse survival was monitored until day 63. We found that siRNA and blocking oligo treatment both prolonged the survival of mice with tumor xenografts, relative to mice that were administered the control oligo (Figure 7j).
Discussion
In this study, we found that ectopically expressed circ-Amotl1 reprogrammed the tumorigenic capacity of MDA-MB-231. The circ-Amotl1-expressing cells displayed a 30-fold increase in single-cell culture and a 142-fold increase in colony formation. Tumor-take rate in nude mice reached almost 100%. This appeared to have occurred through facilitating translocation of c-myc. Although c-myc was mainly distributed in cytoplasm normally, ectopic expression of circ-Amotl1 increased c-myc retention in the nucleus and promoted c-myc stability. The interaction of circ-Amotl1 with c-myc was tested computationally and its function was validated by blocking experiments in vitro and in vivo.
The tumor xenografts from circ-Amotl1-transfected cells displayed a marked increase in proliferation and tumor-forming capacity. Although MDA-MB-231 cells are malignant, these cells typically do not exhibit this phenotype. We observed non-uniform tumors in mice. This suggested some degree of heterogeneity within the cells, with certain populations growing much more rapidly than others. We then selected uniform cell population by isolating cells from the single colonies formed in agarose. This would reduce the heterogeneity of the cell populations.
We found that every circ-Amotl1-transfected cell could efficiently form a colony, which differs markedly from a normal cancer cell population. Through subcutaneous injection of 3-day colonies formed from circ-Amotl1-transfected cells, we derived a 100% rate of tumor formation. These results suggested that every circ-Amotl1-transfected cell had the potential to form an individual tumor. In parental MDA-MB-231 cells, 1 × 107 cells are typically required to form a detectable tumor 30 days postinjection. Our results suggest that the tumorigenicity of MDA-MB-231 cells had been reprogrammed by circ-Amtol1 overexpression. By single-cell culture, we found that each cell formed a robust and spread colony. It should be noted that, in population culture, the difference in proliferation rates between the circ-Amotl1-transfected cells and the vector-transfected cells was approximately one-fold increased. In single-cell culture, we detected a 30-fold increase in cell number after 10 days. By colony-formation assay, we detected a 142-fold increase in the number of circ-Amotl1-transfected cells compared with control at 12 days.
To study how circ-Amotl1 could trigger this highly tumorigenic phenotype, we examined potential interactions between circ-Amotl1 and c-myc, which is a transcription factor regulating oncogene expression.36, 37, 38 Myc has been shown to selectively regulate gene expression to control cellular growth.39, 40, 41 It has also been shown that reduced c-myc expression increases longevity in mice.42 Our results demonstrated that circ-Amotl1 is highly expressed in breast cancer biopsies and cell lines. We then systematically show that this circRNA triggers potent tumorigenicity in vitro and in vivo through the nuclear translocation of c-myc. This leads to upregulation of multiple known c-myc targets. It is known that nuclear c-myc is readily degraded.30 In our study, large levels of c-myc were translocalized to the nucleus, but we did not detect decreased level of total c-myc. This suggests that c-myc was localized to the nucleus, where it co-localized with circ-Amotl1, preventing degradation. Crosslinking assays confirmed the interaction of circ-Amotl1 with c-myc within the nucleus.
It has been reported that Amotl1 is highly expressed in human breast cancer tissues and promotes metastasis.43 This seems correlated with our finding that circ-Amotl1 is highly upregulated in breast cancer and promotes cancer growth. It has been known that circRNAs can function as sponges for regulating miRNA activities,44, 45 and we found that circ-Foxo3 could promote Foxo3 expression and function serving as a sponge for a number of miRNAs.17 In the current study, we detected approximately equal levels of Amotl1 mRNA and protein. This suggests that circ-Amotl1 does not function as a sponge for binding miRNAs. Indeed, to serve as a sponge, the circRNA must stay in cytosol, and we observed that circ-Amotl1 interacted with c-myc and translocated to nucleus. These results suggest that Amotl1 and circ-Amotl1 function by different mechanisms in promoting cancer progression. This awaits further investigation.
Materials and methods
Circ-Amotl1 construct generation
Generation of circ-Amotl1: A Basic Sequence was synthesized, which contained the 3′ half-intron-enox-II fragment (splice acceptor or SA) of bacteriophage T4 td gene, a Small Space Sequence, a exon-I splice donor 5′ half-intron segment. The Basic Sequence, driven by a CMV promoter, was cloned into the multiple cloning sites of pEGFP-N1, which allowed generation of circRNA and GFP by replacing Small Space Sequence with the circular Amotl1, producing circ-Amotl1. The control plasmid was inserted with a non-related sequence instead of the human circ-Amotl1 sequence. Both sequences of circ-Amotl1 and the control vector are provided in Supplementary Figures S1e and f.
Generation of mutated circ-Amotl1: Two primers, cir-Amo-FMut and cir-Amo-RMut, were designed to generate mutation in the c-myc-binding site on circ-Amotl1 using similar approach as above. Generation of circ-Neg1: There were two restriction enzyme sites HindIII and NotI in circ-Amotl1, which were cut and subcloned into pEGFE-N1 plasmid producing circ-Neg. In circ-Neg construct, the acceptor intron was removed, abolishing the activity of circularization of circ-Amotl1. The anti-circ-Amotl1 siRNAs, DNA oligo probes against endogenous or ectopic expression of circ-Amotl1, labeled with biotin or Cy5, were obtained from Integrated DNA Technologies, Inc. (Coralville, IA, USA).
Single-cell proliferation and colony formation
MDA-MB-231 cells were inoculated in Petri dishes in DMEM containing 10% FBS, which allowed the cells to attach but not spread as tissue culture plates did. The next day, cells were treated with trypsin/EDTA and quantified by serial dilutions. Fresh medium (DMEM containing 10% FBS) was added to obtain a density of 1 cell per 100 μl followed by distribution into 96-well tissue culture plates at 100 μl per well. The plates were examined carefully under a light microscope. The wells that contained one single cell were marked. Some of these wells were excluded if more than one or none growing colonies were detected within 2–3 days. The growing colonies were monitored and photographed, and cell count was quantified daily.
Following quantification, the single-cell suspension was mixed with low melting agarose gel in the density of 1 cell per 100 μl. The mixture was immediately distributed into 96-well plates, which had been coated with 0.66% agarose, at the amount of 100 μl per well. After solidification, the plates were examined carefully under a light microscope. The wells that contained single cells were marked. Some of these wells were excluded if more than one or none growing colonies were detected within 2–3 days. The growing colonies were monitored and photographed, and the cell number was determined daily. Owing to the difficulty in obtaining single-cell suspension, the large colonies were treated with trypsin/EDTA extensively before cell counting.
Single colony tumor formation
After quantification, the single-cell suspension was mixed with low melting agarose gel in DMEM containing 10% FBS (reaching a final concentration of 0.25% agarose) at 1 cell per 100 μl. The mixture was distributed into 96-well plates, which had been coated with 0.66% agarose, at 100 μl per well. After solidification, the plates were examined carefully under a light microscope. The wells that contained one single cell were marked. Some of these wells were excluded if more than one or none growing colonies were detected within 2–3 days. The growing colonies were monitored for 6 days.
Some colonies were used for single colony injection into nude mice subcutaneously. In all, 10 μl of Matrigel were diluted with 90 μl medium and added to each well before injection. Each colony was carefully collected with a syringe and re-examined by a dissecting microscope prior to injection. The other colonies were used to estimate the efficiency of colony transfer and injection. In brief, the colonies in the 96-well plates in agarose gel were transferred, one-by-one in the same manner as mouse injection. After transfer, the number of colonies in the new plates was examined under a light microscope (Axiovert 200, Oberkochen, Germany) to estimate the efficiency of colony transfer. As controls, mice were injected with 500, 5000, or 50 000 vector-transfected cells (n=5).
For the 3-day colony injection, the procedure was similar except the colony was too small to be seen under a dissecting microscope. For estimating the transfer and injection efficiency, another set of the 3-day colonies, after transfer to a new plate, were allowed to grow for an additional 3 days, so that the size of each colony could be seen easier under a light microscope. As controls, mice were injected with 500, 5000, or 50 000 vector-transfected cells (n=5).
Single cell tumor formation
For single-cell injection, MDA-MB-231 cells were inoculated in Petri dishes 1 day before injection. After obtaining single-cell suspension, the cell number was determined. Medium (DMEM containing 10% FBS) was added to obtain a density of 1 cell per 100 μl followed by immediate distribution into 96-well tissue culture plates at the amount of 100 μl per well. The plates were examined carefully under a light microscope. The wells that contained one single cell were marked. Some wells were used for single-cell injection into nude mice subcutaneously. In all, 10 μl of Matrigel were diluted with 90 μl medium (DMEM containing 10% FBS) and added to each well before injection. Each cell in the well was collected with a syringe, followed by mouse injection. The other wells were used to estimate the efficiency of single-cell transfer and injection. In brief, the cell in the 96-well plates were transferred, one-by-one in the same manner as mouse injection, to a well of a new plate. It is important that all these procedures were finished before cell attachment to the plates. After transfer, the cell was allowed to grow for 3–4 days to form spreading colonies from single cells, so that the size of each spreading colony could be seen easier under a light microscope.
Routine in vitro and in vivo assays
We performed in vitro experiments as described, including cell transfection and colony formation,46 western blotting,47 flow cytometry, IHC and immunofluorescence microscopy,48, 49 real-time PCR using U6 as an internal control,50 protein pull down by RNA,21 and northern blotting.17 Given the different sizes of circ-Amotl1 and linear Amotl1 mRNA, the samples were separated on a 1.5% formaldehyde agarose gel for circ-Amotl1 and a 0.6% gel for Amotl1 mRNA by using voltages of 50 and 90 V, respectively. Tumor-formation assay was described previously.15 In the preparation of nuclear proteins, cells were lysed in 1 ml nuclear extract buffer (20 mM Hepes, pH 7.2, 10 mM KCl, 2 mM MgCl2, and PI) and homogenized with a prechilled Dounce homogenizer with 20 strokes. Cell lysis was centrifuged at 4200 rpm for 5 min. The pellet was washed with PBS 3 times and resuspended in 100 μl lysis containing 0.5 M NaCl. After centrifugation at 13 200 rpm for 10 min, supernatants containing nuclear extract were used for western blotting analysis.
Statistical analysis
All experiments were performed at minimal in triplicate, and numerical data were subjected to independent sample t-test. The levels of significance were set at *P<0.05 and **P<0.01.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (PJT-149083) to BBY, who is the recipient of a Career Investigator Award (CI 7418) from the Heart and Stroke Foundation of Ontario. WWD is supported by a Postdoctoral Fellowship from the Breast Cancer Foundation of Ontario.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
The authors declare no conflict of interest.
Supplementary Material
References
- AbouHaidar MG, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA 2014; 111: 14542–14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res 2016; 44: D196–D202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 2016; 44: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 2016; 44: D209–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Fang L, Yang X, Sheng W, Yang BL, Seth A et al. The role of versican in modulating breast cancer cell self-renewal. Mol Cancer Res 2013; 11: 443–455. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22: 256–264. [DOI] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 2016; 44: 1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH et al. Circular intronic long noncoding RNAs. Mol Cell 2013; 51: 792–806. [DOI] [PubMed] [Google Scholar]
- Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res 2016; 45: 4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res 2016; 44: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YZ, Yang F, Tan W, Li X, Jiao C, Huang R et al. The anti-cancer components of Ganoderma lucidum possesses cardiovascular protective effect by regulating circular RNA expression. Oncoscience 2016; 3: 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher JP et al. Circular RNAs and their associations with breast cancer subtypes. Oncotarget 2016; 7: 80967–80979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 2017; 24: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 2016; 35: 3919–3931. [DOI] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73: 1019–1030. [DOI] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012; 7: e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30: 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2017; 38: 1402–1412. [DOI] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9: e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20: 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Adwan H, Bauerle T, Najajreh Y, Elazer V, Golomb G, Berger MR. Decreased levels of osteopontin and bone sialoprotein II are correlated with reduced proliferation, colony formation, and migration of GFP-MDA-MB-231 cells. Int J Oncol 2004; 24: 1235–1244. [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016; 44: 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart J, Pashos EE, Prasad MK. Pluripotency redux—advances in stem-cell research. N Engl J Med 2007; 357: 1469–1472. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ 1997; 8: 1039–1048. [PubMed] [Google Scholar]
- Oster SK, Marhin WW, Asker C, Facchini LM, Dion PA, Funa K et al. Myc is an essential negative regulator of platelet-derived growth factor beta receptor expression. Mol Cell Biol 2000; 20: 6768–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 2003; 11: 1189–1200. [DOI] [PubMed] [Google Scholar]
- Li S, Jiang C, Pan J, Wang X, Jin J, Zhao L et al. Regulation of c-Myc protein stability by proteasome activator REGgamma. Cell Death Differ 2015; 22: 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemege KE, Hickey JM, Lovell S, Battaile KP, Zhang Y, Hefty PS. Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes. J Bacteriol 2011; 193: 6517–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia RR, Xue LC, Wilkins K, El-Manzalawy Y, Dobbs D, Honavar V. RNABindRPlus: a predictor that combines machine learning and sequence homology-based methods to improve the reliability of predicted RNA-binding residues in proteins. PLoS ONE 2014; 9: e97725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Huang C, Yang MQ, Yang JY. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst Biol 2010; 4(Suppl 1): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Gromiha MM, Raghava GP. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins 2008; 71: 189–194. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M et al. Non-transcriptional control of DNA replication by c-Myc. Nature 2007; 448: 445–451. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kim SH, Jin X, Kim H. KCTD2, an adaptor of Cullin3 E3 ubiquitin ligase, suppresses gliomagenesis by destabilizing c-Myc. Cell Death Differ 2017; 24: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wu JJ, Huang L. Nanoparticles targeted with NGR motif deliver c-myc siRNA and doxorubicin for anticancer therapy. Mol Ther 2010; 18: 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phesse TJ, Myant KB, Cole AM, Ridgway RA, Pearson H, Muncan V et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ 2014; 21: 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SB, Seo KW, So AY, Seo MS, Yu KR, Kang SK et al. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ 2012; 19: 534–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JW, Zhao X, De Cecco M, Peterson AL, Pagliaroli L, Manivannan J et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell 2015; 160: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc C, Boin A, Fuhrmann L, Vincent-Salomon A, Mandati V, Kieffer Y et al. AMOTL1 promotes breast cancer progression and is antagonized by merlin. Neoplasia 2016; 18: 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016; 37: 2602–2611. [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Kay MA. The expanding repertoire of circular RNAs. Mol Ther 2013; 21: 1112–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ et al. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res 2013; 41: 9688–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chang L, Du WW, Gupta S, Khorshidi A, Sefton M et al. Anti-microRNA-378a enhances wound healing process by upregulating integrin beta-3 and vimentin. Mol Ther 2014; 22: 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gupta S, Du WW, Yang BB. MicroRNA-17 inhibits tumor growth by stimulating T-cell mediated host immune response. Oncoscience 2014; 1: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol 2009; 11: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Rutnam ZJ, Du WW, Yang W, Yang X, Yang BB. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat Commun 2014; 5: 2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.