Abstract
Although human embryonic stem cells (hESCs) were first derived almost 20 years ago, it was only recently acknowledged that they share closer molecular and functional identity to postimplantation lineage-primed murine epiblast stem cells than to naïve preimplantation inner cell mass-derived mouse ESCs (mESCs). A myriad of transcriptional, epigenetic, biochemical, and metabolic attributes have now been described that distinguish naïve and primed pluripotent states in both rodents and humans. Conventional hESCs and human induced pluripotent stem cells (hiPSCs) appear to lack many of the defining hallmarks of naïve mESCs. These include important features of the naïve ground state murine epiblast, such as an open epigenetic architecture, reduced lineage-primed gene expression, and chimera and germline competence following injection into a recipient blastocyst-stage embryo. Several transgenic and chemical methods were recently reported that appear to revert conventional human PSCs to mESC-like ground states. However, it remains unclear if subtle deviations in global transcription, cell signaling dependencies, and extent of epigenetic/metabolic shifts in these various human naïve-reverted pluripotent states represent true functional differences or alternatively the existence of distinct human pluripotent states along a spectrum. In this study, we review the current understanding and developmental features of various human pluripotency-associated phenotypes and discuss potential biological mechanisms that may support stable maintenance of an authentic epiblast-like ground state of human pluripotency.
Keywords: : naive human pluripotency, inner cell mass, human embryonic stem cell, hESC, blastocyst, epiblast
Developmental Capacities of the Murine and Human Preimplantation Embryo
The concept of totipotency was first introduced by Driesch in the 1890s to define the potency of the first two cleavage cells in echinoderms [1] and refers to the capacity of a (single) cell to develop into a complete organism. This potency includes not only differentiation into all embryonic lineages but also the developmental competence to form an organized embryo [2]. Totipotency was first experimentally demonstrated in 1942 in rats through full-term embryo development of isolated single blastomeres (2-cell stage) or fused zygotes following transfer into foster females [3].
In most mammals, totipotency sensu stricto is limited to the zygote and to 2-cell blastomeres (although there have been successful reports of functional totipotency from 4- or 8-cell blastomeres) [2]. The cleavage and blastula stages of development mark the loss of totipotency and the subsequent specification of the epiblast, which is a transient embryo-forming structure that undergoes species-specific morphogenetic reorganization before gastrulation [4] (Fig. 1).
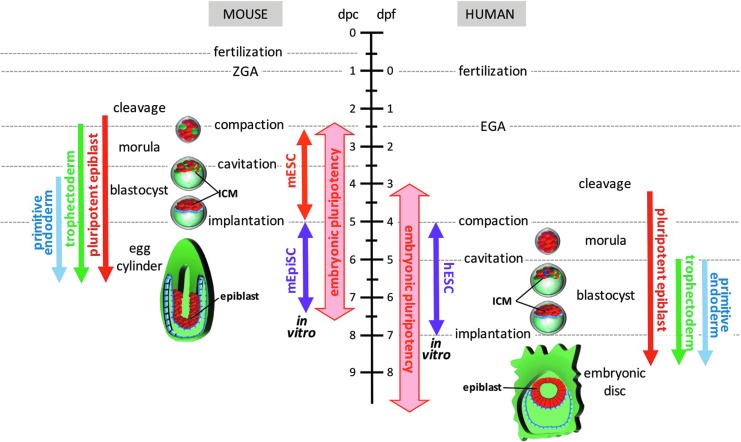
FIG. 1.
Embryonic pluripotency in early mouse and human embryonic development. Left: Pluripotent cells arise in the murine embryo during the cleavage stage, following loss of totipotency. Functional capacity to form all three germ layer lineages is retained up to the postimplantation egg cylinder epiblast. Two categories of PSCs have been isolated from murine embryos: mESCs and mEpiSCs. mESC lines can be isolated from postcleavage preimplantation embryos and model the ground state of pluripotency in the ICM of the blastocyst. In contrast, mEpiSC lines can be isolated from postimplantation epiblasts and mimic the continuum of lineage-primed developmental states that proceed to gastrulation. Right: Human embryonic pluripotency follows slower developmental kinetics than the mouse, but can be classified by analogous morphogenetic changes. Similar to mESCs, hESCs have been isolated from postcleavage preimplantation embryonic ICMs, but hESC lines share closer phenotypic and functional similarity with mEpiSCs than mESCs. hESCs may represent an equivalent of the developmentally more advanced human embryonic disc rather than the preimplantation epiblast cells they originate from. Red: pluripotent cells, green: trophectoderm, blue: primitive endoderm. hESCs, human embryonic stem cells; ICM, inner cell mass; mESCs, mouse embryonic stem cells; mEpiSCs, mouse epiblast-derived stem cells; PSCs, pluripotent stem cells.
Following zygotic activation, embryonic development follows defined rapid successions of ontogenetic phases that can be classified through standardized systems (eg, Carnegie or Hamburger–Hamilton stages) (Fig. 1). The morulae and early blastocyst stages of preimplantation development (up to the fifth cleavage division in the mouse [5]) conserve pluripotent capacity for differentiation into most, if not all, lineages. However, their capacity to self-organize into an integrated body plan is limited and has been accomplished only through artificial methods such as multicellular aggregation or tetraploid complementation [2]. The term pluripotency was originally employed by Haecker in 1914 [6] as the potential for several different developmental options [7].
The rodent preimplantation inner cell mass (ICM) (Fig. 1) transiently embraces a naïve ground state of pluripotency phenotype that is captured in vitro by ICM-derived self-renewing embryonic stem cells (ESCs) [8]. In contrast, the mouse postimplantation epiblast and its derivatives [eg, epiblast-derived stem cells (EpiSCs)] adopt primed pluripotent states with variable degrees of lineage commitment [9] and defective chimeric contribution following injection into recipient blastocysts, although limited contribution can be achieved using postimplantation embryos [10].
Current consensus dictates that putative pluripotent (pluripotential) cells should demonstrate, at a minimum, a differentiation capacity in all three germ layers (although this may extend to differentiation capacity in some or all extraembryonic tissues); although without requirement for competence of self-organization into a coherent embryo. The most widely utilized assay to validate the functional pluripotency of pluripotent stem cells (PSCs) remains teratoma formation, which is a method that was originally developed using single embryonal carcinoma cells [11].
This assay detects differentiation in all germ layers following the subcutaneous, intramuscular, intrarenal, or intratesticular injection of putative pluripotent cells into mice. However, pluripotency is more rigorously validated through potency for chimera formation and germline incorporation following morula aggregation or injection of PSC test cells into a blastocyst-stage embryo. This assay was first described following the injection of murine teratocarcinoma [12] or murine ICM [13] into mouse blastocysts or interspecifically between rat ICMs into mouse blastocysts [14]. Unlike teratoma formation, the capacity for functional chimeric incorporation into a murine blastocyst is lost by murine blastocyst ICM cells following embryo implantation [15]. Thus, this divergence in functional chimera-forming capacity broadly represents a critical delineation of at least two functional classes of pluripotent cells in early rodent embryos [16].
A critical distinction between mouse and human postimplantation embryos is revealed by the progression of the human ICM into an embryonic disc, which contrasts with the developmental structure of the well-described mouse egg cylinder (Fig. 1) [4]. However, the general nonaccessibility of implanted human embryos restricts detailed in vivo studies of this process. Recent descriptions of in vitro systems for ex utero culture and development of human embryos may provide information about human-specific cues governing human epiblast development, epithelialization, and proamniotic cavity formation throughout these poorly accessible early postimplantation phases [17,18]. However, although determination of human functional pluripotency in pre- and postimplantation embryos is limited by ethical and availability constraints, it can be extrapolated from nonhuman primate studies.
For example, using nonhuman primate embryonic cells as surrogates for human PSCs, whole rhesus ICMs and rhesus ESCs both failed to robustly chimerize with rhesus monkey host embryos with the ease routinely observed with rodent PSCs [19]. Interestingly, these studies revealed that rhesus ICMs generated reproducible chimerism only in the extraembryonic compartment and a limited engraftment in fetal liver and spleen that possibly reflected blood cell exchange through placental perfusions [19]. In contrast, monkey chimeras were efficiently generated from totipotent cleavage-stage 4-cell embryos [19], suggesting that preimplantation epiblast pluripotency may follow different functional kinetics in primates and rodents.
Distinct Molecular and Functional Pluripotencies of the Rodent Epiblast
Pre- and postimplantation epiblast cells both possess the capacity to form all three germ layers in most species, and rodent PSC lines can be successfully derived from both developmental stages. Mouse ESCs (mESCs) were originally derived as ICM-derived explants that were expanded over mitotically inactivated mouse embryonic fibroblast (MEF) feeder cells in undefined culture systems (eg, employing specific lots of fetal bovine serum (FBS) [20] or conditioned media from teratocarcinoma cultures [21]). mESC lines were subsequently revealed to exploit gp130/LIF/STAT3 [22–25], WNT [26], and bone morphogenetic protein (BMP) [27] signaling for their self-renewal. In contrast, EpiSCs derived from the postimplantation epiblast of murine egg cylinders were stably propagated through FGF2/MEK/ERK [28–30] and WNT-β-catenin pathway [31] signals.
Serum-based cultures of mESCs produced heterogeneous populations of lineage-primed subsets [32], and a more stringent culture system was ultimately developed using small-molecule inhibition to sustain a more primitive self-renewal [33]. This system utilized two small molecules (2i) that augmented WNT/β-catenin activation while simultaneously diminishing extracellular signal-regulated kinase (ERK) signaling [via GSK3β and mitogen-activated protein ERK (MEK) inhibition, respectively] [33]. This 2i culture system proved sufficient for stably maintaining a naïve pluripotent state in mESCs [34,35] that was biologically akin to the ground state of pluripotency of the murine preimplantation ICM [36,37].
mESCs and EpiSCs are both derived from embryonic cells separated by only several cell divisions. However, they reproduce distinct pluripotent states (ie, naïve and primed) representing major peri-implantation transcriptomic, epigenomic, and metabolic transitions of the pluripotent epiblast (Figs. 2 and 3) [8,16]. Indeed, while both mESCs and EpiSCs share a similar core pluripotency molecular network [38,39] and can differentiate into derivatives of the three germ layers in teratoma or directed differentiation assays [29,30], they retain distinct molecular and functional characteristics.
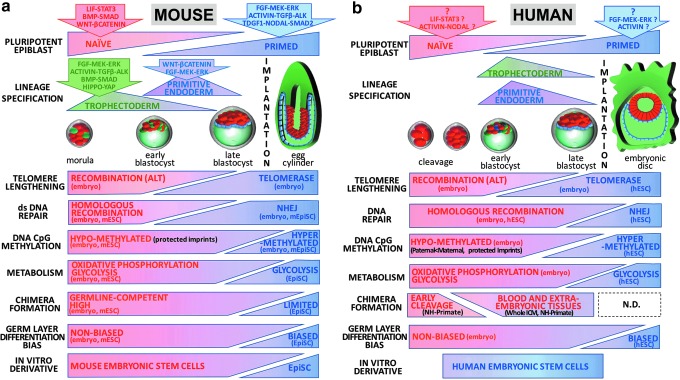
FIG. 2.
Functional phenotypes of primed and naïve pluripotent states. (a) Functional shifts in the peri-implantation mouse embryo. The mouse pluripotent epiblast progresses from a naïve ground state (red) to a primed lineage-biased state (blue) following implantation. Naïve and primed states exploit distinct signaling pathways and their transition is accompanied by the sequential specification of trophectoderm (green) and primitive endoderm (violet) lineages. Known signaling pathways directing trophectoderm and primitive endoderm are indicated. In the mouse embryo, naïve and primed states can be distinguished by differing telomere lengthening and DNA repair strategies, levels of global repressive epigenetic marks (eg, DNA CpG methylation), and usage of metabolic pathways. Both states also display nonequivalent functional pluripotencies, with only the naïve state showing capacity for germline-competent chimera formation. In contrast, postimplantation epiblast cells have a partially committed lineage bias. In vitro expansion of mouse naïve epiblast cells generates mESC lines, while the postimplantation epiblast can generate lineage-primed mEpiSC lines. Functional capacities that have been demonstrated in vivo (embryo) or using in vitro surrogates (mESC, mEpiSC) are indicated. (b) Functional shifts in the human peri-implantation embryo. Similarly to the mouse, the human pluripotent epiblast is believed to recapitulate a steady progression from a naïve preimplantation state (red) to postimplantation primed lineage-biased states (blue). The signaling pathways that are essential for human naïve and primed states remain a subject of debate and have been extrapolated from hESC or single-cell RNA sequencing of preimplantation human embryos. The progression of human pluripotency is accompanied by the specification of trophectoderm (green) and primitive endoderm (violet) lineages, although the kinetics for emergence of extraembryonic lineages diverge between both species. The human naïve and primed states can also be distinguished by differing telomere lengthening and DNA repair mechanisms, global levels of repressive epigenetic marks, and metabolic pathway usage. The chimeric contribution of the postimplantation epiblast of nonhuman primates remains undetermined. However, nonhuman primate (NH-Primate) studies indicate that chimera formation may be restricted to early cleavage embryos, with possible low engraftment capacity for later preimplantation stages demonstrated by whole ICM transfer experiments. Functional capacities that have been demonstrated in vivo (embryo) or using in vitro surrogates (hESCs) are indicated.
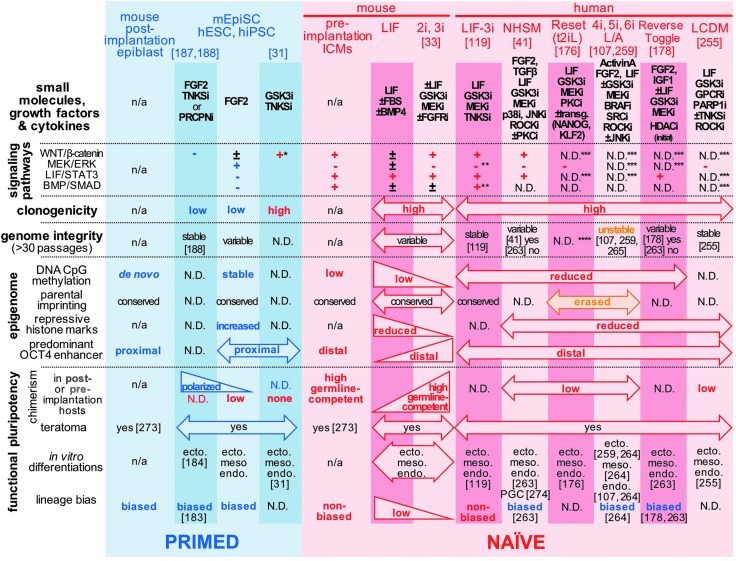
FIG. 3.
Summary of epigenetic and functional phenotypes that are detected in distinct human and mouse pluripotent states. Select human and mouse PSC culture systems are presented with their downstream outcomes on WNT/β-catenin, FGF2/MEK/ERK, LIF/STAT3, and BMP/SMAD circuitries. + and − indicate signaling activities that have been verified to be, respectively, up- and downregulated using the aforementioned cocktails of small molecules, growth factors, and cytokines. The figure lists a series of epigenomic and functional hallmarks that have been associated with and distinguish between primed and naïve pluripotent cell populations. *Non-nuclear β-catenin only, **unpublished data (MEK/ERK) or subject to interline variability (BMP/SMAD), ***directly targeted by culture conditions, but nonverified, ****normal chromosome preparations were only verified between 5 and 17 passages. n/a, not applicable; N.D., not determined. ecto., meso., endo., PGC, and TE indicate reported detections of neuroectoderm, mesoderm (ie, cardiac, hemato-vascular), definitive endoderm, primordial germ cell, and trophectoderm lineages in directed differentiation assays. BMP, bone morphogenetic protein; ERK, extracellular signal-regulated kinase; LIF, leukemia inhibitory factor; MEK, mitogen-activated protein ERK.
For example, EpiSCs exhibited higher levels of epigenetic repressive marks (eg, increased CpG promoter DNA methylation [40,41] and bivalent/repressive histone marks [41,42]) that were partially erased following reversion to naïve culture [34,35,41]. The reduced levels of classical repressive chromatin marks in naïve PSCs may potentiate a distinct epigenetic regulation of transcriptional activities [43,44] that includes seed enhancers [45], miRNA networks, RNA-induced silencing complex-mediated control of chromatin [46], and post-transcriptional regulators (eg, PARsylation by PARP1 and PARP7 [47], YAP/TAZ signaling [48–50], and regulation of retrotransposon elements [51,52]).
The postimplantation epiblast (and thus EpiSCs [53–55]) is characterized by increased lineage-primed gene expression relative to the naïve ground state [9,41,54] that also correlates with a functional lineage differentiation bias [9,54]. This functional discrepancy between primed and naïve pluripotencies in the mouse is revealed by their chimeric contribution capacity in pre- or postimplantation embryos (Fig. 2a). ICM-derived mESCs show robust capacity for contribution to chimeric animals when aggregated with morulae or injected into blastocysts, including efficient contribution to germline lineages. This chimera-forming capacity is further reinforced with LIF-2i-cultured mESCs [56]. In contrast, mEpiSCs expanded under standard culture conditions are not capable of significant chimeric contribution when injected into preimplantation embryos [29,30].
However, this deficiency can be improved by injection into stage-matched postimplantation epiblasts [10]. Even though EpiSCs may be artificially conditioned for engraftment into preimplantation ICM [57], their baseline inefficiency of contribution to the germline supports the notion that naïve reversion of EpiSCs is necessary for such contribution. Thus, functional and molecular pluripotencies may overlap between the two states and may not necessarily be stringently compartmentalized. For example, specific EpiSC subsets have been reported to retain naïve-like phenotypes, including chimera contribution [58]. Additionally, hybrid EpiSC culture systems using FGF2, Activin, and leukemia inhibitory factor (LIF) [59] or alternatively FGF2, Activin, and a GSK3β inhibitor [60] were shown to produce pluripotent stem cells that retained capacity for chimeric germline contribution.
Importantly, primed mEpiSCs can be successfully reverted into a naïve-like pluripotent state by exposing them to LIF/STAT3 signaling [61] or transgenic expression of key naïve inducers (eg, E-Cadherin [62], Esrrb [63], the Krüppel-like factors Klf2 (in synergy with Prdm14) [64], Klf4 [65,66] or Klf5 [67], Mbd3 [68], cMyc [66], Nanog [69], or the nuclear receptors Nr5a1/Nr5a2 [70]). Discrepancies in reversion efficiencies have been attributed to either advanced developmental progression of the starting primed state [9] or genetic background [66].
Interestingly, although LIF/STAT3 activation may be sufficient to revert specific EpiSC lines [71,72], poor reversion efficiencies or strain-specific requirements of other lines may be circumvented by employing chemical WNT modulation by the ATP-competitive cyclin and GSK3β inhibitor kenpaullone [66], the tankyrase inhibitor XAV939 [62], inhibition of the histone H3K4 methyltransferase MLL1 using MM-401 [73], nonspecific histone deacetylase (HDAC) inhibition with sodium butyrate [74], or a combination of WNT, MEK, FGFR, and TGFβ pathway inhibitions and epigenetic erasure involving inhibition of histone demethylase LSD1 [75].
The multiplicity of mouse pluripotency states captured in vitro appears to correspond to a spectrum of dynamic shifts in molecular and cellular identities in vivo that naturally progress within epiblast cells during the peri-implantation period (Fig. 2a). Pluripotency briefly persists through the developmental progression of the ICM by continuous expression of core pluripotency regulators. Thus, maintenance of a stable naïve pluripotent state in vitro may similarly require sustained reinforcement of WNT, BMP4, and LIF/STAT3 signaling [27,76–84]. Such reinforcement likely requires a stable orchestration of events that incorporate repressive and bivalent epigenetic marks and subsequent downstream expression of epiblast lineage specifiers.
For example, this epigenetic transition is known to involve dynamic reorganization of chromatin enhancer signatures for regulating developmental factors [45,85–87], most notably a shift of distal to proximal OCT4 enhancer usage [29,88]. Furthermore, the ICM undergoes dramatic metabolic transitions, including an interruption of its use of oxidative phosphorylation, and exclusive alternate use of glycolysis for sustaining its energy expenditures [89–91]. Additional developmental shifts include changes in activities responsible for DNA repair [92,93] and telomere maintenance [94,95]. For example, before implantation, the embryo relied predominantly on homologous recombination for DNA repair, whereas in the subsequent postimplantation period, there is a transition toward increase of more error-prone, but more efficacious, nonhomologous end joining for double-stranded DNA repair [96].
Determinants of Human Molecular and Functional Pluripotency
Human ESCs (hESCs) were originally derived from human blastocysts in FBS-containing medium on mitotically inactive MEF feeders and human LIF [97] in conditions similar to those sustaining mESC self-renewal. However, unlike mESCs [24,25], LIF was found to not be essential for sustenance of self-renewal of human ICM cells, including in the absence of feeders [97]. Similarly, while BMP4 and LIF/STAT3 pathways synergized to support clonal growth of mESCs, supplementation of hESC cultures with BMP4 led to trophoblast lineage differentiation [98]. Subsequent hESC culture conditions adopted supplementation with FGF2, serum replacer (ie, knockout serum replacer), and mitotically inactive MEF feeders or MEF-conditioned medium for more uniform propagation and expansion of undifferentiated hESCs [99]. These culture conditions were further optimized using feeder-free and more defined medium formulations (eg, mTESR [100] and E8 [101]).
Differences in morphology, gene expression, cell cycle regulation, telomerase activity, and functional performance between mouse and human PSCs were originally attributed to species-specific attributes [102–104]. However, subsequent isolation of EpiSCs from postimplantation mouse epiblasts [29,30] revealed that despite being a derivative of the human preimplantation ICM, human PSCs shared greater molecular, epigenetic, and functional pluripotency similarities with mEpiSCs than with mESCs (Fig. 3) [29,30,105].
In the absence of an ethically conceivable human chimera assay, the functional pluripotency of conventional (primed) human PSCs was extrapolated from surrogate nonhuman primate PSC experiments (Fig. 2b). Conventional (primed) cultures of rhesus monkey ESCs failed to participate in chimera formation when injected into rhesus blastocysts [19]. Moreover, cross-species chimera studies using host mouse blastocysts revealed that even though injected conventional monkey ESCs could transiently associate with the mouse ICM, they did not significantly contribute to developing murine fetal tissue [106]. Similar interspecies chimera approaches with human cells confirmed that even though conventional, primed hPSC cultures could not survive injection into mouse preimplantation blastocysts [41,107], limited, but measurable, integration occurred within injected gastrula-stage mouse embryos [108].
Although functional pluripotency can be validated by teratoma or directed differentiation across germ layers, interline genetic variability between conventional hESC lines [109] can result in differentiation lineage bias and skewing in response to microenvironmental cues [110,111]. Indeed, marked differentiation disparities have been documented extensively between hESC lines [112–115]. Furthermore, hESC lines comprise heterogeneous populations [116] with epigenetically distinct coexisting subsets showing variable differentiation capacities [117].
One study involving a cohort of 20 independent hESC lines revealed that discrepancies in functional pluripotency reflected variations in both epigenetic and transcriptional profiles, including a high disparity in genes regulating development and differentiation [118]. Our own studies revealed disparities between conventional human PSCs based on highly variable lineage-primed gene expression that directly impacted functional pluripotency [119]. These studies supported the notion that conventional, primed human PSCs embrace diverse states of primed pluripotency in a manner similar to mEpiSCs [9].
Various factors among conventional human PSCs have been hypothesized to contribute to this functional variability; these include genetic background [120], acquisition of mutations in key developmental genes, and differences in derivation and culture methodologies [109]. For example, derivation of hESCs under physiological oxygen concentration may result in acquisition of naïve-like X chromosome activation, which may reinforce ground state pluripotency by suppressing spontaneous differentiation [121].
Yamanaka's discovery of transcription factor-mediated cellular reprogramming [122] for generating human induced pluripotent stem cells (hiPSCs) [123,124] revolutionized the study of pluripotency and regenerative medicine. However, hiPSC reprogramming further accentuated the variability observed in functional pluripotency between conventional PSC lines. Most hiPSC lines were noted to display more augmented lineage skewing [119,125–128] than standard hESC lines despite their strong overlap of transcriptional and epigenetic signatures with conventional hESCs [118,129,130]. While optimization of differentiation methods partially erased these functional discrepancies in directed differentiation [131–137], epigenomic aberrations were identified in a number of hiPSC lines that included retention of donor cell-specific somatic memory and reprogramming errors [138–141] and were shown to be transmitted to differentiated progenies [138].
Some studies argued that such reprogramming errors resulted in differentiation bias toward their respective cell of origin lineage in mouse [142–145], human [146–150], and dog [151] iPSCs. It may be worth noting that most of these studies involved genome-integrating methods that introduced reprogramming factor transgenes through retroviral [142,144–147,150,151] or lentiviral [148,149] vectors (including a lentiviral doxycycline-inducible secondary system [143]). Such viral reprogramming methods are now known to promote transcriptional and epigenetic errors [141] that were not detected using somatic cell nuclear transfer [141] or nonintegrative episomal derivation methods [119,120].
Transgene-integrating reprogramming methods may also have potentiated an increased frequency of genomic aberrations in established hiPSC clones [152–154], which likely compromised functional pluripotency [155] through transgene reactivation in differentiated cells [156–158].
Factors Determining the Quality of the Pluripotent State in Reprogrammed hiPSCs
A series of nonintegrative reprogramming strategies have been developed (eg, Sendai virus, episomal, and mRNA) to avoid the risks associated with viral transgene integration, but with notable disparities of their aneuploidy rates, reprogramming efficiency, reliability, and workload that have been discussed by Schlaeger et al. elsewhere [159]. We previously demonstrated that optimized episomal reprogramming was uniquely superior in activated myeloid progenitors (MPs) and could consistently achieve bulk reprogramming at high efficiencies across variable donor genetic backgrounds. Moreover, human MP-iPSC lines possessed hESC-like transcriptomes closely with significantly fewer reprogramming errors than hiPSC lines obtained from standard episomally reprogrammed adult skin fibroblasts [119,160].
This myeloid reprogramming method exploited a stromal priming activation step that delivered various signals (eg, Toll receptor/NFκB, JAK/STAT3 signaling) responsible for decreasing reprogramming efficiency barriers [119,160]. Analogous interactions with the mesenchyme may contribute to aberrant reprogramming of tumor cells toward invasive cancer phenotypes [161,162] and have also been shown to induce epigenetic changes that favor cellular reprogramming using retroviral vectors [163]. Such deterministic reprogramming generated hiPSC clones with stable genomes and reduced lineage bias [119,160] that translated into the generation of highly functional progenitors across germ layers in independent studies (ie, hemato-vascular [164], cardiac [132], and photoreceptors [165]).
Importantly, the specialized cell populations obtained from such directed differentiation assays displayed enhanced functional capacities with lower senescence, superior DNA repair capacity, or improved long-term engraftment [164], underlying a correlation between terminal differentiation and the initial pluripotency state. As such, functional pluripotency requires evaluation of terminally differentiated progenies, even though most studies limit their characterization to intermediate progenitors. To this end, we and others have developed a battery of in vitro and in vivo directed differentiation assays that were included for our group hematopoietic [131,166] (eg, macrophages [167]), vascular [164], cardiac [132,168,169], or retinal [165] lineages.
A number of comparative strategies were also employed between isogenic hiPSC lines reprogrammed from distinct cell types [170–173] or from isogenic donor hESCs/iPSCs [120,174]. These studies have suggested that donor-specific genetic background rather than cell of origin or reprogramming system plays a more dominant role on the differentiation capacities of hiPSC lines [170,171,174]. These studies employed both integrative and nonintegrative derivation methods for cellular reprogramming (eg, Sendai virus [120,170–172,174], episomal plasmids [171,173], lentiviruses [174], or retroviruses [171,172,174]).
A publication from the Progenitor Cell Biology Consortium analyzed a large repertoire of 58 hiPSC lines from 10 independent laboratories and reported a segregation of DNA methylation profile signatures based on their cell type of origin, but these differences could not be directly attributed to somatic donor memory [130]. Taken together, these studies have revealed that multiple complex determinants collectively impact the differentiation potency of conventional, primed human PSCs [119,120].
Conventional mEpiSC-Like Human PSCs Can Be Chemically Reverted to Highly Variable Naïve Preimplantation Epiblast-Like Pluripotent States
Several groups have developed various culture systems to revert EpiSC-like conventional human PSCs [41,48,107,119, 175–183] or derive de novo hESCs [41,107,178,184] to pluripotent states resembling the human preimplantation epiblast (Fig. 3). These studies revealed that the classical mESC 2i cocktail of inhibitors targeting MEK and GSK3β [33] was insufficient for stable sustenance of a human ICM-like state.
LIF/STAT3 signaling, a critical self-renewal signaling pathway in mESCs [22–25], can promote naïve reversion of both mEpiSCs [61] and hESCs [181]. However, such reversion of mEpiSCs in 2i was relatively inefficient even with transgenic STAT3 reinforcement (ie, ∼1%–2% efficiency) [61], and a number of EpiSC lines required sustained transgenic STAT3 expression in 2i culture [65]. Despite LIF supplementation, forced transgenic expression of STAT3 was essential for achieving naïve reversion of both human and mouse primed pluripotent states [61,181].
In mEpiSCs, several factors were shown to potentiate LIF-dependent STAT3 responsiveness and naïve reversion. These variables included colony size [72], increased BMP/SMAD signaling [185], and FGF/ERK inhibition [28,66], as well as stimulated [66,186] or, alternatively, reduced [62,187–189] WNT signaling. Interestingly, these determinants were previously reported to impact lineage priming and differentiation [27,28,31,72,187,190–194]. These studies also exposed an interline molecular and functional variability among mEpiSC lines [28,66] that impacted naïve reversion efficiencies in LIF-2i [9,186]. Our studies similarly observed a similar pattern when reverting a cohort of variably lineage-primed human PSC lines to an LIF/STAT3-dependent naïve pluripotent state in LIF-3i [119].
Four Main Molecular Axes Intersect to Balance the Maintenance and Exit of Molecular Pluripotency
The BMP/SMAD, LIF/STAT3, FGF2/MEK/ERK, and WNT/β-catenin pathways have all been recognized to regulate the self-renewal of pluripotent stem cells (Fig. 4). These molecular axes are strongly intermingled and not only control the pluripotency states but also initiate differentiation if their balanced circuitry is altered. Even though these signaling pathways regulate multiple independent downstream transcriptional targets, they also converge to a few shared effectors (Fig. 4).
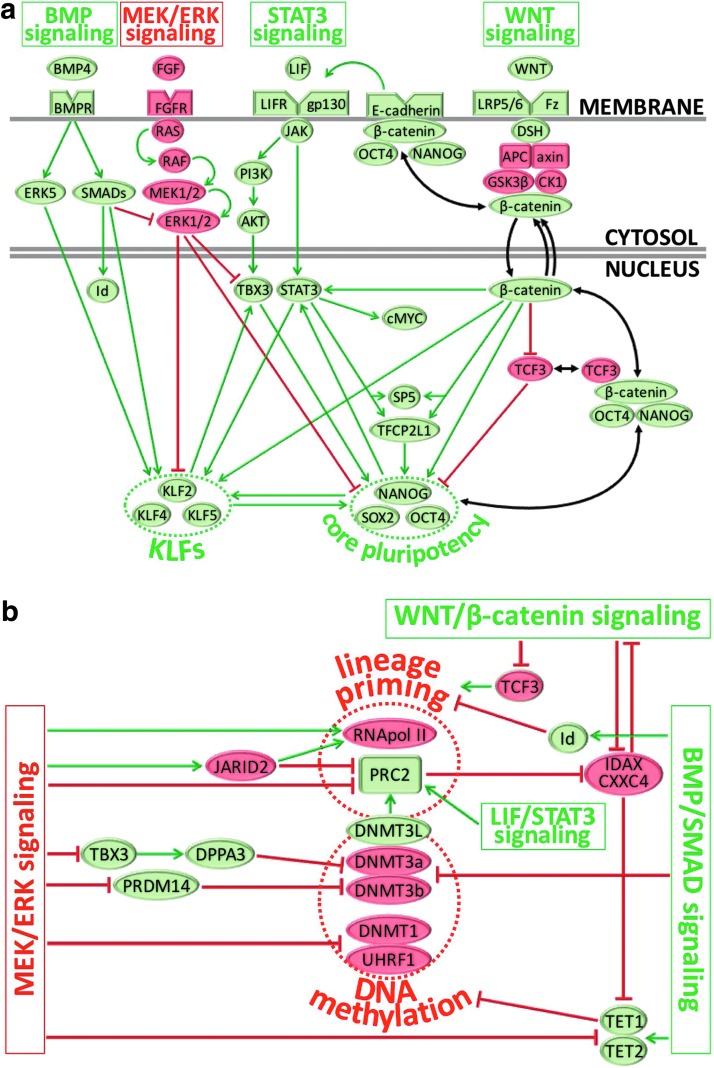
FIG. 4.
Schematic summary of four main signaling pathways that regulate the naïve pluripotent state. (a) The BMP/SMAD, LIF/STAT3, FGF2/ERK, and WNT/β-catenin pathways are the four main molecular axes regulating naïve pluripotency. These circuits not only share a few common transcriptional effectors but also act separately to reinforce the core pluripotency network through mechanisms that can involve the KLF circuitry. Fluctuating subcellular distribution of the WNT pathway effector β-catenin may regulate accessibility of the core factors, OCT4 and Nanog, by facilitating their functions in either the nucleus or at the cell membrane (eg, to reinforce E-Cadherin strengthening of STAT3 signaling). (b) Downstream signaling of BMP/SMAD, WNT/β-catenin, LIF/STAT3, or MEK/ERK suppression results in marked reductions of genome-wide chromatin repressive marks (ie, reduced DNMT3a/DNMT3b levels and impaired DNMT1 recruitment following UHRF1 downregulation) as well as downregulation of lineage priming at developmental promoters by mechanisms that involve RNA-pol II pausing and accessibility to the PRC2. Green arrows: activation. Red blunt line: inhibition. Proteins known to reinforce (green) or destabilize (red) naïve pluripotency are shown. KLF, Krüppel-like factor; PRC2, polycomb repressor complex 2.
For instance, the LIF/STAT3 and MEK/ERK pathways antagonistically regulate TBX3 [76], a known transcriptional activator of the pluripotency-associated genes Nanog and DPPA3/Stella [195,196]. The LIF/STAT3 and WNT/β-catenin pathways also synergistically converge onto the trans-acting protein 5 (Sp5), a transcription factor that closely relates to the Klf gene family and can reprogram mEpiSCs to the naïve state [197]. Another transcription factor named Tfcp2l1 is also similarly and independently reinforced by STAT3 and WNT signaling [77,198] and facilitates EpiSC reprogramming to naïve pluripotency by interacting with KLFs and Nanog [198] (Fig. 4).
The BMP/SMAD pathway stimulates ESC self-renewal by endogenously inhibiting MEK/ERK signaling [82], recruiting multiple Kruppel-like factors [83], or suppressing differentiation by stimulating expression of the inhibitor of differentiation (Id) genes [27] (Fig. 4). While BMP4 signals cooperate with the LIF/STAT3 axis to stabilize the naïve state [27], they can also promote differentiation of primed pluripotent stem cells under control of WNT signaling [191].
LIF/STAT3 activity is essential for maintenance of the mouse naïve state [22–25] and promotes transition from primed to naïve pluripotency [61,181]. STAT3 signaling regulates multiple pluripotency-associated targets (Fig. 4), including reinforcement of the core pluripotency factors Nanog and SOX2 through activation of Tbx3 and Klf4 in two parallel circuitries [76], or augmented transcriptional expression of Rex1, Stat3 itself, and the epigenetic modifiers, Lin28, Ezh2, and Mbd3 [84]. Nanog expression has been shown in return to amplify STAT3 and KLF4 activities to reinforce naïve pluripotency [199]; conversely, STAT3 activity may suppress mesoendodermal differentiation in cooperation with Nanog [200].
Forced chemical rewiring of MEK/ERK and WNT/β-catenin has been shown to bypass at least partially LIF/STAT3 and BMP/SMAD signaling and sustain the naïve ground state in mESCs [33]. MEK inhibition promotes naïve ESC self-renewal by blocking lineage priming toward primitive endoderm and differentiation in naïve cells [201] through dynamic remodeling of polycomb repressors and H3K27me3 repressive marks [202]. While suppression of the ERK1/2 pathway stabilizes mESCs [203], it will promote differentiation of mEpiSCs and hESCs [8,16]. Moreover, MEK/ERK signaling may be indispensable for self-renewal of primed pluripotent cells [28,204].
Unlike the pivotal role of ERK signaling in naïve-to-primed transition, the role of WNT/β-catenin is more ambiguous since either reinforcement [66,186] or inhibition [62,187–189] of WNT signals can augment naïve reversion of mEpiSCs. In addition, β-catenin targets have been involved in promoting both self-renewal [26,78,194,205–207] and differentiation [187,188,191,208,209] of primed and naïve PSCs. This ambiguity may reflect the importance for a synergetic balance between the different pathways and is highly context specific. For example, in the absence of LIF and without ERK inhibition, WNT stimulation will prime mESCs toward the primitive endoderm [210].
In addition, the availability of β-catenin for interaction with distinct factors directly affects the balance between propluripotency or differentiation cues. As such, the accessibility of β-catenin to the transcription factor TCF1 in the nucleus antagonizes long-term self-renewal and functional pluripotency of mESCs [211].
MEK/ERK Inhibition Suppresses Differentiation and Reinforces Naïve Pluripotency
In mouse cells, ERK inhibition potentiates naïve reversion in cooperation with WNT and LIF/STAT3 signaling (Fig. 4) [28]. MEK/ERK inhibition also antagonizes primitive endoderm differentiation of naïve cells [201], while FGF stimulation promotes differentiation [191]. In contrast, FGF promotes pluripotency in primed cells, partially by inhibiting neuroectodermal commitment [28]. The involvement of ERK inhibition in suppressing neural commitment is not clear since ERK-mediated effects have also been shown to direct differentiation toward primitive endoderm, but not neural, lineages [201]. In this study, ERK inhibition was actually reported to reinstate neural capacity of a differentiation-compromised EpiSC line [201].
Direct isolation and expansion of mouse naïve ICM using MEK/ERK inhibition support the idea that MEK signaling blockade replicates the signaling circuitry in the mouse preimplantation epiblast [37]. Furthermore, ERK-mediated phosphorylation of Nanog was shown to promote differentiation by inhibiting Nanog transactivation and compromising Nanog stability [212]. ERK also negatively regulates the Kruppel-like factors, including KLF2 [213] and KLF4 [214], which regulate maintenance of ground state pluripotency by reinforcing core pluripotency signaling [215,216] (Fig. 4). MEK inhibition protects KLF2 phospho-degradation in mouse naïve cells and cooperates with the GSK3/TCF3 cascade to establish ground state pluripotency [213].
MEK/ERK inhibition also cooperates with GSK3β inhibition in establishing global DNA hypomethylation in mESCs to reach levels that are similar to early embryos [217]. Maintenance of a hypomethylated epigenome in mESCs due to forced MEK blockade has been shown to rely on molecular mechanisms that are redundant to those exploited by primordial germ cells and early embryos: transcriptional repression of Dnmt3A and Dnmt3B [34,217], recruitment of the polycomb repressive complex 2 (PRC2) complex [218] and the ten-eleven translocation (TET)-mediated base excision repair pathway [219], and impaired recruitment of DNMT1 due to low levels of the E3 ubiquitin-protein ligase UHRF1 [220] (Fig. 4).
ERK1/2 also contributes to establishing lineage priming in mESCs by binding to DNA sequence motifs at developmental genes that are typically accessed by polycomb repressors [221]. ERK inhibition directly interferes with PRC2 promoter occupancy and contributes to decreased phosphorylation of RNA polymerase II (RNApol II) at lineage commitment genes [221]. These ERK-mediated activities mirror epigenomic features that were previously detected in 2i cultures (ie, reduced H3K27Me3 repressive marks, fewer bivalent domains and RNApol II pausing at developmental genes, and reduced lineage priming in mouse naïve ESCs) [35]. ERK1/2 activity also regulates access of the PRC2 repressor JARID2 to developmental promoters [221]. Overall, these studies support that ERK signaling inhibition may directly or indirectly play an essential role in repression of developmental genes within a naïve epigenome to maintain pluripotency.
Subcellular WNT/β-Catenin Fluctuations May Orchestrate the Naïve Pluripotency Molecular Network
The WNT/β-catenin pathway has been linked to multiple mechanisms that ensure the maintenance of naïve pluripotency in mESCs (Fig. 4). These mechanisms include upregulation of Stat3 mRNA levels [205], augmented expression of Klf2 and Tfcp2l1 [222], and downregulation of Tcf3 to suppress neuroectodermal differentiation [223]. The canonical WNT pathway also directly affects DNA methylation by regulating TET proteins through the TET-negative regulator IDAX/CXXC4 [224]. This activity is regulated in a feedback loop inhibition since IDAX can also repress WNT by binding to DVL [225].
The involvement of WNT signaling in mediating pluripotency states is complex and may depend on its synergy with other pathways, including TGFβ/Activin [186], BMP/SMAD [191], and FGF/ERK [28]. Alternatively, intrinsic regulation of the WNT pathway itself by altering β-catenin subcellular distribution may regulate pluripotent states [31,119]. Reinforcement of WNT signaling by inhibition of GSK3β not only reinforces naïve pluripotency in mESCs [192] but also promotes acquisition of naïve features in some EpiSC lines [60,66]. Conversely, the use of inhibitors of the WNT pathway facilitates derivation of EpiSCs [188,226] and reinforces primed pluripotency in EpiSCs with [31] or without [187,226] the presence of a GSK3β inhibitor (Fig. 3). GSK3β is the kinase that initiates the cascade of phosphorylation targeting β-catenin that will ultimately lead to β-catenin proteolysis.
Stabilization of members of the destruction complex of β-catenin, while preventing β-catenin phosphorylation, can alter subcellular localization of β-catenin [227–230], but not impede its expression [31], and will particularly reinforce β-catenin levels in the cytoplasm [230,231]. As a result, diverse WNT signal responses will be determined by the cellular distribution of β-catenin and the Axin/APC/GSK3β complex [230]. For instance, at a high nuclear β-catenin concentration in the on-state of the pathway, Axin/APC/GSK3β shuttling can maximize the response to WNT signaling by reducing fluctuations [230].
Interestingly, a membrane-associated β-catenin/OCT4 complex marks the mouse ground state of pluripotency [232] and supports the notion that a subcellular reorganization of β-catenin may participate in stabilizing the naïve state in EpiSCs. Further investigation will be needed to clarify the role of subcellular β-catenin levels in modulating naïve versus primed pluripotencies.
β-Catenin fluctuations were also observed to tightly correlate with Nanog expression levels in LIF/serum mESC cultures [233]. These fluctuations were retained in LIF-2i naïve conditions [233], presumably in a Nanog-independent manner since Nanog expression is homogenized in the latter system [35]. WNT stimulation was also shown to mitigate Nanog expression variability in mESCs [232], and β-catenin upregulated Nanog expression through its interaction with OCT4 [78]. Since shuttling of β-catenin participates in reinforcing the WNT response, continuous β-catenin fluctuations may be inherent to the naïve pluripotency state and may dynamically orchestrate the stabilization of the core pluripotency factors, OCT4 and Nanog. OCT4 participates in a shuttling complex with β-catenin and Axin that typically potentiates β-catenin degradation in the absence of GSK3β inhibition [234].
The detection of small amounts of both Nanog and OCT4 in mESC membrane containing fractions supports a possible regulation of these pluripotency factors through complexes with β-catenin [232], although further studies are still required to elucidate the role of these intricate interactions across subcellular compartments. These complexes at the membrane involve E-cadherin, β-catenin, Nanog, and OCT4 and are believed to be specific to the ground state [232]. Several other protein complexes comprising OCT4, Nanog, β-catenin, and TCF3 were also detected in the nucleus and were proposed to stabilize the mouse naïve ground state mainly by regulating the amount of free OCT4 [235]. Dynamic rearrangements of these complexes between β-catenin and core pluripotency factors may be induced by fluctuations of β-catenin shuttling and were reported to be augmented when mESCs were cultured in LIF-2i [235].
The subcellular distribution of the mESC proteome is complex, and even though the interactome of the three core pluripotency factors concentrates onto chromatin-bound factors, it also extends to a variety of non-nuclear targets, suggesting regulatory mechanisms involving rearrangements between compartments [236]. For example, subcellular relocalization of proteins between naïve and primed pluripotent states has been described for Tfe3, an important bHLH transcription factor that regulates Esrrb expression in mESCs and that relocates to the cytoplasmic compartment upon exit from ground state pluripotency [237].
Interestingly, by using a cocktail of small inhibitors targeting MEK, GSK3β, and tankyrase, our group (Zimmerlin et al.) achieved a rearrangement of activated β-catenin in human primed PSC lines [119]. The tankyrase inhibitor XAV939 stabilizes Axin, the presumptive limiting factor of the β-catenin destruction complex [238]. In the LIF-3i culture system, simultaneous exposure to GSK3β and tankyrase inhibitors permitted simultaneous stabilization of β-catenin and Axin and paradoxically reinforced active β-catenin levels in both nuclear and cytosolic compartments [119].
A similar approach involving GSK3β and tankyrase inhibition (but without concomitant MEK inhibition) was employed to stabilize hESC and mEpiSC lines in a primed pluripotent state [31] and also augmented cytoplasmic (at the expense of nuclear) levels of β-catenin. Simultaneous exposure to CHIR99021 and XAV939 permitted clonal propagation of mouse and human primed cells, although without any detectable acquisition of a naïve phenotype (ie, no upregulation of naïve pluripotency genes, switch of OCT4 enhancer usage, or blastocyst chimera potential) [31]. Interestingly, the effects of the CHIR99021/XAV939 inhibition combination in this study appeared to be independent of E-Cadherin since an E-cadherin-depleted EpiSC line could still be propagated in culture [31].
The CHIR99021/XAV939-induced primed state may primarily benefit from nuclear exclusion of β-catenin. Indeed, while promoting self-renewal [26,239] or derivation [240] of mESCs, transcriptional activity of β-catenin promotes emergence of lineage-specified progenitors in EpiSCs and hESCs [187,191,241,242].
Thus, although molecular rewiring in the 2i condition has been shown to be sufficient for maintaining the naïve ground state in mESCs [33], a customized tuning of the WNT/β-catenin signaling output may be required to sustain naïve molecular pluripotency in human PSCs. Such WNT regulation has been achieved in distinct mouse genetic backgrounds by further reinforcing [66,186] or attenuating [62,187–189] WNT signals. This may potentially be accomplished through marked reduction (eg, 3–10-fold) of the standard 3 μM concentration of the GSK3β inhibitor CHIR99021 in 2i-based cultures, which may prevent the spontaneous differentiation of rat ESCs [243,244] or human PSCs [107,176,178,184] possibly resulting from excessive amounts of nuclear β-catenin.
E-Cadherin May Regulate Naïve Pluripotency by Regulating the Intracellular Levels of β-Catenin
As outlined above, the balance between self-renewal and differentiation may be regulated through subcellular levels of β-catenin, and this shuttling between cytoplasmic and nuclear compartments may be controlled at the level of the actin cytoskeleton (Fig. 4). At the membrane, β-catenin promotes intercellular adhesion by complexing with E-cadherin and facilitating the binding of cadherins to the actin cytoskeleton. In contrast, in the nucleus, β-catenin serves as a transcriptional cofactor to activate target genes of the canonical WNT signaling pathway either through repression of members of the TCF protein family [192,245,246] or through TCF-independent mechanisms that involve direct targeting of pluripotency factors such as Nanog [78], OCT4 [81], and KLF4 [247].
At the membrane, β-catenin promotes cadherin-mediated intercellular adhesions by binding the cytoplasmic domain of E-cadherin and linking the adherens junction to actin filaments through interaction with α-catenin. β-Catenin may first bind E-cadherin within the endoplasmic reticulum (ER) membrane, which initially protects it from proteolytic degradation [248] and subsequently facilitates its exit from the ER before its transport toward the cell membrane [245]. Maternal E-cadherin and β-catenin are present until early morula stages in mouse embryos and promote blastomere adhesion and morula compaction [249].
The observation that the absence of maternal E-cadherin can restore the developmental deficit induced by a truncating β-catenin mutant suggests that the interactions between E-cadherin and β-catenin at the membrane may directly regulate the availability of nuclear β-catenin during embryonic genome activation [249]. A similar interdependence between adhesive and cotranscriptional roles of β-catenin has also been implied in cancer cells [250]. Moreover, E-cadherin is required for proper activation of LIF receptor/gp130 signaling and STAT3 phosphorylation in mESCs [251]. This E-Cadherin-mediated STAT3 signaling has been shown to contribute to elevated Nanog expression [80] and stabilizes naïve pluripotency.
The importance of β-catenin-induced E-Cadherin reinforcement at the membrane in safeguarding functional pluripotency was highlighted in studies exploiting a TCF/LEF signaling defective β-catenin variant that independently restored β-catenin-mediated adhesion [252]. These β-catenin-deficient mESCs exhibited impaired mesendoderm formation and neuronal differentiation, and introduction of a β-catenin variant without TCF-mediated nuclear activities partially rescued adhesion and endoderm (although not mesoderm) formation as well as neuronal differentiation [252].
Other functional data substantiating the importance of E-cadherin levels to support naïve pluripotency have been obtained from genetic manipulation of primed EpiSCs. For example, ectopic E-cadherin expression in mEpiSC lines enhanced chimerism efficiency in blastocyst injection experiments [253], although without germline contribution. In contrast, disruption of E-cadherin at the membrane following either genetic loss of β-catenin or through tankyrase inhibition (XAV939) augmented a biased integration of primed EpiSCs into postimplantation embryos [187].
Functional Validation of Human and Nonhuman Primate Naïve Pluripotent States: A Work in Progress
Conventional human PSCs were recently reverted to a highly variable spectrum of naïve-like pluripotent states that partially replicated the molecular circuitry of mESCs and human preimplantation embryos (Fig. 3). These culture systems generally not only relied on utilization of classical mouse naïve 2i conditions (GSK3β and MEK inhibition), but also required additional chemical modulation for stabilizing inherently unstable or metastable human naïve states.
These methods included (1) hybrid culture systems that costimulate primed pluripotency circuitry with exogenous FGF2 [41,107,177,178,182,254], Activin/TGFβ [41,107,177, 254], or the BMP inhibitor dorsomorphin [177]; (2) forced transgene expression of OCT4, SOX2, and KLF4 [175], OCT4 and KLF4 [175], KLF2 and KLF4 [175], NANOG and KLF2 [107,176], YAP [48], or STAT3 [181]; (3) global epigenetic erasure using HDAC inhibitors [178]; (4) chemical rewiring of antiapoptotic signaling pathways (eg, activation of adenylyl-cyclase [48,175] and/or YAP [48] or inhibition of B-RAF [107], JNK [41,107], p38 [41] PKC [41,176], ROCK [41,107,176,177,180,181], and SRC [107]); and (5) multipathway biochemical and epigenomic rewiring of uncharacterized naïve stabilizing pathways through tankyrase [119] or PARP1 [255] signal inhibition.
Many of these strategies stemmed from previous efforts of mEpiSC naïve reversion or potentiation of mESC derivation conditions. For example, (1) hybrid EpiSC culture systems (eg, FGF2, Activin and LIF [59], FGF2, Activin, and inhibition of GSK3β [60]); (2) forced transgenic expression of E-Cadherin [62], Esrrb [63], Klf2 [64], Klf4 [65,66], Klf5 [67], Mbd3 [68], cMyc [66], Nanog [69], Nr5a1 [70], Nr5a2 [70], or Stat3 [61]; (3) global epigenetic remodeling using the H3K4 methyltransferase MLL1 inhibitor MM-401 [73], the HDAC inhibitor sodium butyrate [74], or the histone demethylase LSD1 [75]; and (4) reinforcement of naïve pluripotency molecular circuitry by augmentation of LIF/STAT3 [61], BMP/SMAD [185], WNT/β-catenin [66,186], or inhibiting FGF/ERK [28,66] signaling.
Similar to mEpiSCs, conventional human PSCs have demonstrated measurable, but limited, capacity for engraftment into postimplantation mouse developing embryos [108], yet only limited chimerization in mouse blastocysts [256]. The chimeric contribution of human and nonhuman primate cells into preimplantation embryos was assessed for naïve-reverted cells obtained using the naïve human stem cell medium (NHSM) method developed by Hanna's team [41,254,257–259], variants of the 5i/L/A cocktail from the Jaenisch's group [107,259] and the t2iL technique [254,259] developed in Austin Smith's laboratory [176], and more recently the FGF2, Activin, CHIR990211 (FAC) [254] and LIF, CHIR99021, DiM, MiH (LCDM) [255] methods, which produce less clearly defined pluripotent states. These studies reported that injection of variably derived naïve-like PSC populations into mouse [41,107,255,257,259] or monkey [258] morulae or ungulate blastocysts [254] resulted in extremely limited human and monkey cell chimerism in developing embryos.
Reliable detection of transgenic green fluorescent protein (GFP) within murine embryos was technically challenging with such reduced levels of chimerism [107,254,259], and several groups employed sensitive polymerase chain reaction (PCR)-based methods (ie, human mitochondrial DNA, human Alu sequence) [254,255,259] to determine chimerism for NHSM [41], t2iL [176], 5i/L/A [107], FAC [60,254], and LCDM [255] human naïve or intermediate culture methods.
Using a mitochondrial DNA PCR detection assay, Theunissen et al. reported rare and sporadic chimerism in less than 1% of E10.5 embryos following injection of NHSM, t2iL, or variants of 5i/L/A human naïve cells into murine morulae and blastocysts [259]. A similar approach was employed by Yang et al. for the LCDM culture system and the authors similarly measured limited (≤1%) human chimeric integration in E10.5 embryos, but with significantly higher frequencies of human cell contribution within the murine trophectoderm [255].
Interestingly, the authors of the LCDM method recommend the addition of the tankyrase inhibitor IWR-1-endo to their chemical cocktail when culturing human cells [255], and as such, their results further corroborate our own studies establishing that tankyrase inhibition stabilizes a human naïve-like state with improved functional pluripotency [119].
Belmonte's group similarly reported extremely low levels of human–animal chimerism using a PCR-based genomic assay of human-specific Alu sequences that detected human cells in the chimeric offspring of human–pig blastocyst injection experiments. Collectively, these experiments demonstrated that robust levels of interspecies chimerism with human naïve PSCs have not yet been achieved [254].
However, it remains unclear whether limited chimerism of human naïve PSCs in mouse blastocysts [41,107,257,259] represents a poorly obtained human naïve functional pluripotency or alternatively reflects obstacles of genetic distance, phenotypic differences, and/or developmental divergences of human and mouse embryos (eg, early postimplantation epiblast morphogenesis and ontogenesis variations in size, shape, and speed). Efforts to improve host compatibility in chimera assays have included allogeneic (monkey-to-monkey) transfer [258] and injection of human naïve PSCs into ungulate (ie, pig and cattle) blastocysts [254]. Unfortunately, both strategies have still yielded particularly low levels of chimerism that did not match the successes of rat–mouse interspecies [254,260,261] or large animal (ie, pig) allogeneic [262] blastocyst complementation.
Strategies that provide a selective advantage for donor PSCs may augment the incorporation of human cells into host embryos. Future studies will test candidate naïve PSCs with such improved methodologies for assaying the functional pluripotency of putative bona fide naïve human PSCs.
Several reports have now demonstrated impaired functional pluripotency from selected human naïve-reverted pluripotent states [263,264], including the NHSM [41,263], 5i/L/A [259,264,265], reverse toggle [178,263], and naïve conversion medium (NCM) [182,263] systems. Moreover, the 5i/L/A method produced hPSCs in a naïve-like state that either required prolonged repriming for proper differentiation [259,265] or displayed pronounced neuroectodermal bias and impaired terminal differentiation [264]. Other human naïve reversion methods similarly displayed limited capacity to undergo terminal differentiation toward functional phenotypes when compared with primed isogenic cultures [263].
Notwithstanding the hypothesis that these states may embrace a more primitive and paradoxically less competent pluripotent state [266], the impaired functional pluripotency that has been detected using these human naïve states is in clear contradiction with results obtained from mouse [9,35,267,268] and rabbit [269,270] PSCs, especially in regard to neural differentiation. Primed mEpiSC lines display disparate lineage priming and differentiation capacity [9,54], while naïve cultures not only exhibit restored or augmented neuroectodermal capacity [9,35,267] but also generate terminally differentiated neural populations that resemble more closely the mouse adult brain [267].
Unlike the aforementioned variable human naïve-like states described above, chemical naïve reversion that supplements classical MEK/ERK and GSK3β inhibition with a tankyrase inhibiter (LIF-3i) is the only method described thus far that improves functional pluripotency across germ layers of a large repertoire of hESC and hiPSC lines [119]. Importantly, karyotypic and epigenomic imprinting aberrations were not detected in LIF-3i-reverted naïve-like hPSCs [119].
In contrast, the impaired functional pluripotency in other naïve reversion methods described thus far may derive from either the reported chromosomal instability or aberrant erasure of genomic imprints following chemical manipulation from these systems. For example, abnormal karyotypes were reported in 5i/L/A [259,264,265], NHSM [263], and reverse toggle [263] methods, while aberrant parental imprinting was inherent to the 5i/L/A [259,271] and t2iL [271] methods. The further optimization of tankyrase inhibitor-utilizing LIF-3i methods in defined feeder-free, xeno-free culture conditions may allow efficient and clinically useful generation of functional and engraftable adult-like cell types for therapeutic use, including for hemato-vascular regeneration [272].
Acknowledgments
This work was supported by grants from NIH/NHLBI [U01HL099775 (E.T.Z.), NIH/NEI R01EY023962 (E.T.Z.)], NIH/NICHD R01HD082098 (E.T.Z.), RPB Stein Innovation Award (E.T.Z.), The Maryland Stem Cell Research Fund 2013-MSCRF-III-114936 (L.Z.), 2013-MSCRFII-0032-00 (E.T.Z.), 2014-MSCRFE-118153 (T.S.P.), and Novo Nordisk Science Forum Award (E.T.Z.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Driesch H. (1894). Analytische Theorie der Organischen Entwicklung. W. Engelmann, Leipzig [Google Scholar]
- 2.Condic ML. (2014). Totipotency: what it is and what it is not. Stem Cells Dev 23:796–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholas JS. and Hall B. (1942). Experiments on developing rats. II. The development of isolated blastomeres and fused eggs. J Exp Zool 90:441–459 [Google Scholar]
- 4.Sheng G. (2015). Epiblast morphogenesis before gastrulation. Dev Biol 401:17–24 [DOI] [PubMed] [Google Scholar]
- 5.Suwinska A, Czolowska R, Ozdzenski W. and Tarkowski AK. (2008). Blastomeres of the mouse embryo lose totipotency after the fifth cleavage division: expression of Cdx2 and Oct4 and developmental potential of inner and outer blastomeres of 16- and 32-cell embryos. Dev Biol 322:133–144 [DOI] [PubMed] [Google Scholar]
- 6.Maehle AH. (2011). Ambiguous cells: the emergence of the stem cell concept in the nineteenth and twentieth centuries. Notes Rec R Soc Lond 65:359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Häecker V. (1914). Über Gedächtnis, Vererbung und Pluripotenz: August Weismann zum achtzigten Geburtstage gewidmet. Mit 14 Abbildungen im Text. G. Fischer, Jena [Google Scholar]
- 8.Nichols J. and Smith A. (2012). Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol 4:a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernemann C, Greber B, Ko K, Sterneckert J, Han DW, Arauzo-Bravo MJ. and Scholer HR. (2011). Distinct developmental ground states of epiblast stem cell lines determine different pluripotency features. Stem Cells 29:1496–1503 [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, Osorno R, Tsakiridis A. and Wilson V. (2012). In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep 2:1571–1578 [DOI] [PubMed] [Google Scholar]
- 11.Kleinsmith LJ. and Pierce GB., Jr. (1964). Multipotentiality of single embryonal carcinoma cells. Cancer Res 24:1544–1551 [PubMed] [Google Scholar]
- 12.Papaioannou VE, McBurney MW, Gardner RL. and Evans MJ. (1975). Fate of teratocarcinoma cells injected into early mouse embryos. Nature 258:70–73 [DOI] [PubMed] [Google Scholar]
- 13.Gardner RL. (1968). Mouse chimeras obtained by the injection of cells into the blastocyst. Nature 220:596–597 [DOI] [PubMed] [Google Scholar]
- 14.Gardner RL. and Johnson MH. (1973). Investigation of early mammalian development using interspecific chimaeras between rat and mouse. Nat New Biol 246:86–89 [DOI] [PubMed] [Google Scholar]
- 15.Rossant J, Gardner RL. and Alexandre HL. (1978). Investigation of the potency of cells from the postimplantation mouse embryo by blastocyst injection: a preliminary report. J Embryol Exp Morphol 48:239–247 [PubMed] [Google Scholar]
- 16.Nichols J. and Smith A. (2009). Naive and primed pluripotent states. Cell Stem Cell 4:487–492 [DOI] [PubMed] [Google Scholar]
- 17.Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NM, Campbell A, Devito LG, et al. (2016). Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol 18:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED. and Brivanlou AH. (2016). Self-organization of the in vitro attached human embryo. Nature 533:251–254 [DOI] [PubMed] [Google Scholar]
- 19.Tachibana M, Sparman M, Ramsey C, Ma H, Lee HS, Penedo MC. and Mitalipov S. (2012). Generation of chimeric rhesus monkeys. Cell 148:285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans MJ. and Kaufman MH. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156 [DOI] [PubMed] [Google Scholar]
- 21.Martin GR. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 78:7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K, Shoyab M, Taga T. and Kishimoto T. (1994). Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech Dev 45:163–171 [DOI] [PubMed] [Google Scholar]
- 23.Niwa H, Burdon T, Chambers I. and Smith A. (1998). Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12:2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA. and Gough NM. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336:684–687 [DOI] [PubMed] [Google Scholar]
- 25.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M. and Rogers D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336:688–690 [DOI] [PubMed] [Google Scholar]
- 26.Sato N, Meijer L, Skaltsounis L, Greengard P. and Brivanlou AH. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med 10:55–63 [DOI] [PubMed] [Google Scholar]
- 27.Ying QL, Nichols J, Chambers I. and Smith A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115:281–292 [DOI] [PubMed] [Google Scholar]
- 28.Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P. and Scholer HR. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6:215–226 [DOI] [PubMed] [Google Scholar]
- 29.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL. and McKay RD. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448:196–199 [DOI] [PubMed] [Google Scholar]
- 30.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA. and Vallier L. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448:191–195 [DOI] [PubMed] [Google Scholar]
- 31.Kim H, Wu J, Ye S, Tai CI, Zhou X, Yan H, Li P, Pera M. and Ying QL. (2013). Modulation of beta-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat Commun 4:2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi K, Lopes SM, Tang F. and Surani MA. (2008). Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453:519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, et al. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol 20:311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A. and Stunnenberg HG. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149:590–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boroviak T, Loos R, Lombard P, Okahara J, Behr R, Sasaki E, Nichols J, Smith A. and Bertone P. (2015). Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev Cell 35:366–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boroviak T, Loos R, Bertone P, Smith A. and Nichols J. (2014). The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol 16:516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X. and Levasseur DN. (2008). The transcriptional network controlling pluripotency in ES cells. Cold Spring Harb Symp Quant Biol 73:195–202 [DOI] [PubMed] [Google Scholar]
- 39.Festuccia N, Osorno R, Wilson V. and Chambers I. (2013). The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr Opin Genet Dev 23:504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veillard AC, Marks H, Bernardo AS, Jouneau L, Laloe D, Boulanger L, Kaan A, Brochard V, Tosolini M, et al. (2014). Stable methylation at promoters distinguishes epiblast stem cells from embryonic stem cells and the in vivo epiblasts. Stem Cells Dev 23:2014–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504:282–286 [DOI] [PubMed] [Google Scholar]
- 42.Rugg-Gunn PJ, Cox BJ, Ralston A. and Rossant J. (2010). Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci U S A 107:10783–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T. and Dent SY. (2014). Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15:93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T. and Wysocka J. (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14:838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Factor DC, Corradin O, Zentner GE, Saiakhova A, Song L, Chenoweth JG, McKay RD, Crawford GE, Scacheri PC. and Tesar PJ. (2014). Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell 14:854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandolfini L, Luzi E, Bressan D, Ucciferri N, Bertacchi M, Brandi R, Rocchiccioli S, D'Onofrio M. and Cremisi F. (2016). RISC-mediated control of selected chromatin regulators stabilizes ground state pluripotency of mouse embryonic stem cells. Genome Biol 17:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roper SJ, Chrysanthou S, Senner CE, Sienerth A, Gnan S, Murray A, Masutani M, Latos P. and Hemberger M. (2014). ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res 42:8914–8927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin H, Hejna M, Liu Y, Percharde M, Wossidlo M, Blouin L, Durruthy-Durruthy J, Wong P, Qi Z, et al. (2016). YAP induces human naive pluripotency. Cell Rep 14:2301–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24:1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, et al. (2014). YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 158:157–170 [DOI] [PubMed] [Google Scholar]
- 51.Schoorlemmer J, Perez-Palacios R, Climent M, Guallar D. and Muniesa P. (2014). Regulation of mouse retroelement MuERV-L/MERVL expression by REX1 and epigenetic control of stem cell potency. Front Oncol 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter M, Teissandier A, Perez-Palacios R. and Bourc'his D. (2016). An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. Elife 5:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera-Perez JA. and Magnuson T. (2005). Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol 288:363–371 [DOI] [PubMed] [Google Scholar]
- 54.Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DA, Jones V, Hor A, de Alencastro G, et al. (2014). The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 14:107–120 [DOI] [PubMed] [Google Scholar]
- 55.Chou YF, Chen HH, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, Lu J, McKay RD. and Geijsen N. (2008). The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell 135:449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexandrova S, Kalkan T, Humphreys P, Riddell A, Scognamiglio R, Trumpp A. and Nichols J. (2016). Selection and dynamics of embryonic stem cell integration into early mouse embryos. Development 143:24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joo JY, Choi HW, Kim MJ, Zaehres H, Tapia N, Stehling M, Jung KS, Do JT. and Scholer HR. (2014). Establishment of a primed pluripotent epiblast stem cell in FGF4-based conditions. Sci Rep 4:7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han DW, Tapia N, Joo JY, Greber B, Arauzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT. and Scholer HR. (2010). Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell 143:617–627 [DOI] [PubMed] [Google Scholar]
- 59.Ozawa M, Kawakami E, Sakamoto R, Shibasaki T, Goto A. and Yoshida N. (2014). Development of FGF2-dependent pluripotent stem cells showing naive state characteristics from murine preimplantation inner cell mass. Stem Cell Res 13:75–87 [DOI] [PubMed] [Google Scholar]
- 60.Tsukiyama T. and Ohinata Y. (2014). A modified EpiSC culture condition containing a GSK3 inhibitor can support germline-competent pluripotency in mice. PLoS One 9:e95329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC. and Smith A. (2010). Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 7:319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murayama H, Masaki H, Sato H, Hayama T, Yamaguchi T. and Nakauchi H. (2015). Successful reprogramming of epiblast stem cells by blocking nuclear localization of beta-catenin. Stem Cell Reports 4:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR. and Chambers I. (2012). Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11:477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gillich A, Bao S, Grabole N, Hayashi K, Trotter MW, Pasque V, Magnusdottir E. and Surani MA. (2012). Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell 10:425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W. and Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136:1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, et al. (2009). Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4:513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeon H, Waku T, Azami T, Khoa le TP, Yanagisawa J, Takahashi S. and Ema M. (2016). Comprehensive identification of Kruppel-like factor family members contributing to the self-renewal of mouse embryonic stem cells and cellular reprogramming. PLoS One 11:e0150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, et al. (2013). Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502:65–70 [DOI] [PubMed] [Google Scholar]
- 69.Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I. and Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138:722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo G. and Smith A. (2010). A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137:3185–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K. and Surani MA. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461:1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Onishi K, Tonge PD, Nagy A. and Zandstra PW. (2012). Microenvironment-mediated reversion of epiblast stem cells by reactivation of repressed JAK-STAT signaling. Integr Biol (Camb) 4:1367–1376 [DOI] [PubMed] [Google Scholar]
- 73.Zhang H, Gayen S, Xiong J, Zhou B, Shanmugam AK, Sun Y, Karatas H, Liu L, Rao RC, et al. (2016). MLL1 inhibition reprograms epiblast stem cells to naive pluripotency. Cell Stem Cell 18:481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ware CB, Wang L, Mecham BH, Shen L, Nelson AM, Bar M, Lamba DA, Dauphin DS, Buckingham B, et al. (2009). Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell 4:359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou H, Li W, Zhu S, Joo JY, Do JT, Xiong W, Kim JB, Zhang K, Scholer HR. and Ding S. (2010). Conversion of mouse epiblast stem cells to an earlier pluripotency state by small molecules. J Biol Chem 285:29676–29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niwa H, Ogawa K, Shimosato D. and Adachi K. (2009). A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460:118–122 [DOI] [PubMed] [Google Scholar]
- 77.Martello G, Bertone P. and Smith A. (2013). Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J 32:2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takao Y, Yokota T. and Koide H. (2007). Beta-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem Biophys Res Commun 353:699–705 [DOI] [PubMed] [Google Scholar]
- 79.Li J, Li J. and Chen B. (2012). Oct4 was a novel target of Wnt signaling pathway. Mol Cell Biochem 362:233–240 [DOI] [PubMed] [Google Scholar]
- 80.Hawkins K, Mohamet L, Ritson S, Merry CL. and Ward CM. (2012). E-cadherin and, in its absence, N-cadherin promotes Nanog expression in mouse embryonic stem cells via STAT3 phosphorylation. Stem Cells 30:1842–1851 [DOI] [PubMed] [Google Scholar]
- 81.Kelly KF, Ng DY, Jayakumaran G, Wood GA, Koide H. and Doble BW. (2011). beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 8:214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi X, Li TG, Hao J, Hu J, Wang J, Simmons H, Miura S, Mishina Y. and Zhao GQ. (2004). BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A 101:6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morikawa M, Koinuma D, Mizutani A, Kawasaki N, Holmborn K, Sundqvist A, Tsutsumi S, Watabe T, Aburatani H, Heldin CH. and Miyazono K. (2016). BMP sustains embryonic stem cell self-renewal through distinct functions of different Kruppel-like factors. Stem Cell Reports 6:64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133:1106–1117 [DOI] [PubMed] [Google Scholar]
- 85.Zylicz JJ, Dietmann S, Gunesdogan U, Hackett JA, Cougot D, Lee C. and Surani MA. (2015). Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife 4:e09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ. and Gomez-Skarmeta JL. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22:2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Galonska C, Ziller MJ, Karnik R. and Meissner A. (2015). Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell 17:462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K. and Scholer HR. (1996). Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122:881–894 [DOI] [PubMed] [Google Scholar]
- 89.Leese HJ. and Barton AM. (1984). Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J Reprod Fertil 72:9–13 [DOI] [PubMed] [Google Scholar]
- 90.Houghton FD, Thompson JG, Kennedy CJ. and Leese HJ. (1996). Oxygen consumption and energy metabolism of the early mouse embryo. Mol Reprod Dev 44:476–485 [DOI] [PubMed] [Google Scholar]
- 91.Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C. and Ruohola-Baker H. (2012). HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J 31:2103–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banuelos CA, Banath JP, MacPhail SH, Zhao J, Eaves CA, O'Connor MD, Lansdorp PM. and Olive PL. (2008). Mouse but not human embryonic stem cells are deficient in rejoining of ionizing radiation-induced DNA double-strand breaks. DNA Repair (Amst) 7:1471–1483 [DOI] [PubMed] [Google Scholar]
- 93.Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, Babcock GF. and Stambrook PJ. (2010). Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev 19:1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, et al. (2007). Telomere lengthening early in development. Nat Cell Biol 9:1436–1441 [DOI] [PubMed] [Google Scholar]
- 95.Kalmbach K, Robinson LG, Jr., Wang F, Liu L. and Keefe D. (2014). Telomere length reprogramming in embryos and stem cells. Biomed Res Int 2014:925121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chiruvella KK, Sebastian R, Sharma S, Karande AA, Choudhary B. and Raghavan SC. (2012). Time-dependent predominance of nonhomologous DNA end-joining pathways during embryonic development in mice. J Mol Biol 417:197–211 [DOI] [PubMed] [Google Scholar]
- 97.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. and Jones JM. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147 [DOI] [PubMed] [Google Scholar]
- 98.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP. and Thomson JA. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20:1261–1264 [DOI] [PubMed] [Google Scholar]
- 99.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J. and Thomson JA. (2000). Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 227:271–278 [DOI] [PubMed] [Google Scholar]
- 100.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT. and Wu JC. (2009). Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A 106:15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, et al. (2011). Chemically defined conditions for human iPSC derivation and culture. Nat Methods 8:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J. and Rao MS. (2004). Differences between human and mouse embryonic stem cells. Dev Biol 269:360–380 [DOI] [PubMed] [Google Scholar]
- 103.Schnerch A, Cerdan C. and Bhatia M. (2010). Distinguishing between mouse and human pluripotent stem cell regulation: the best laid plans of mice and men. Stem Cells 28:419–430 [DOI] [PubMed] [Google Scholar]
- 104.Koestenbauer S, Zech NH, Juch H, Vanderzwalmen P, Schoonjans L. and Dohr G. (2006). Embryonic stem cells: similarities and differences between human and murine embryonic stem cells. Am J Reprod Immunol 55:169–180 [DOI] [PubMed] [Google Scholar]
- 105.Rossant J. (2008). Stem cells and early lineage development. Cell 132:527–531 [DOI] [PubMed] [Google Scholar]
- 106.Simerly C, McFarland D, Castro C, Lin CC, Redinger C, Jacoby E, Mich-Basso J, Orwig K, Mills P, et al. (2011). Interspecies chimera between primate embryonic stem cells and mouse embryos: monkey ESCs engraft into mouse embryos, but not post-implantation fetuses. Stem Cell Res 7:28–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, et al. (2014). Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15:471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mascetti VL. and Pedersen RA. (2016). Human-mouse chimerism validates human stem cell pluripotency. Cell Stem Cell 18:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Allegrucci C. and Young LE. (2007). Differences between human embryonic stem cell lines. Hum Reprod Update 13:103–120 [DOI] [PubMed] [Google Scholar]
- 110.Nazareth EJ, Ostblom JE, Lucker PB, Shukla S, Alvarez MM, Oh SK, Yin T. and Zandstra PW. (2013). High-throughput fingerprinting of human pluripotent stem cell fate responses and lineage bias. Nat Methods 10:1225–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mehta A, Mathew S, Viswanathan C. and Sen Majumdar A. (2010). Intrinsic properties and external factors determine the differentiation bias of human embryonic stem cell lines. Cell Biol Int 34:1021–1031 [DOI] [PubMed] [Google Scholar]
- 112.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR. and Melton DA. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol 26:313–315 [DOI] [PubMed] [Google Scholar]
- 113.Mikkola M, Olsson C, Palgi J, Ustinov J, Palomaki T, Horelli-Kuitunen N, Knuutila S, Lundin K, Otonkoski T. and Tuuri T. (2006). Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Dev Biol 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chang KH, Nelson AM, Fields PA, Hesson JL, Ulyanova T, Cao H, Nakamoto B, Ware CB. and Papayannopoulou T. (2008). Diverse hematopoietic potentials of five human embryonic stem cell lines. Exp Cell Res 314:2930–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burridge PW, Anderson D, Priddle H, Barbadillo Munoz MD, Chamberlain S, Allegrucci C, Young LE. and Denning C. (2007). Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells 25:929–938 [DOI] [PubMed] [Google Scholar]
- 116.Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC. and Bhatia M. (2006). Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods 3:807–815 [DOI] [PubMed] [Google Scholar]
- 117.Hong SH, Rampalli S, Lee JB, McNicol J, Collins T, Draper JS. and Bhatia M. (2011). Cell fate potential of human pluripotent stem cells is encoded by histone modifications. Cell Stem Cell 9:24–36 [DOI] [PubMed] [Google Scholar]
- 118.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, et al. (2011). Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144:439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zimmerlin L, Park TS, Huo JS, Verma K, Pather SR, Talbot CC, Agarwal J, Steppan D, Zhang YW, et al. (2016). Tankyrase inhibition promotes a stable human naïve pluripotent state with improved functionality. Development 143:4368–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, et al. (2015). A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 33:1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, et al. (2010). Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141:872–883 [DOI] [PubMed] [Google Scholar]
- 122.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 123.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 124.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 125.Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS. and Lanza R. (2010). Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28:704–712 [DOI] [PubMed] [Google Scholar]
- 126.Choi KD, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, Thomson J. and Slukvin I. (2009). Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells 27:559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, et al. (2011). A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol 29:279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA. and Zhang SC. (2010). Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A 107:4335–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, et al. (2009). Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Salomonis N, Dexheimer PJ, Omberg L, Schroll R, Bush S, Huo J, Schriml L, Ho Sui S, Keddache M, et al. (2016). Integrated genomic analysis of diverse induced pluripotent stem cells from the progenitor cell biology consortium. Stem Cell Reports 7:110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park TS, Zimmerlin L. and Zambidis ET. (2013). Efficient and simultaneous generation of hematopoietic and vascular progenitors from human induced pluripotent stem cells. Cytometry A 83:114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L. and Zambidis ET. (2011). A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One 6:e18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O'Sullivan JF, Grainger SJ, Kapp FG, Sun L, et al. (2015). Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol 17:994–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weng Z, Kong CW, Ren L, Karakikes I, Geng L, He J, Chow MZ, Mok CF, Keung W, et al. (2014). A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells. Stem Cells Dev 23:1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Toyoda T, Mae S, Tanaka H, Kondo Y, Funato M, Hosokawa Y, Sudo T, Kawaguchi Y. and Osafune K. (2015). Cell aggregation optimizes the differentiation of human ESCs and iPSCs into pancreatic bud-like progenitor cells. Stem Cell Res 14:185–197 [DOI] [PubMed] [Google Scholar]
- 136.Rostovskaya M, Bredenkamp N. and Smith A. (2015). Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philos Trans R Soc Lond B Biol Sci 370:20140365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koehler KR, Tropel P, Theile JW, Kondo T, Cummins TR, Viville S. and Hashino E. (2011). Extended passaging increases the efficiency of neural differentiation from induced pluripotent stem cells. BMC Neurosci 12:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, et al. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471:68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H. and Umezawa A. (2011). DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet 7:e1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ruiz S, Diep D, Gore A, Panopoulos AD, Montserrat N, Plongthongkum N, Kumar S, Fung HL, Giorgetti A, et al. (2012). Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci U S A 109:16196–16201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, et al. (2014). Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, et al. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28:848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nukaya D, Minami K, Hoshikawa R, Yokoi N. and Seino S. (2015). Preferential gene expression and epigenetic memory of induced pluripotent stem cells derived from mouse pancreas. Genes Cells 20:367–381 [DOI] [PubMed] [Google Scholar]
- 145.Quattrocelli M, Palazzolo G, Floris G, Schoffski P, Anastasia L, Orlacchio A, Vandendriessche T, Chuah MK, Cossu G, Verfaillie C. and Sampaolesi M. (2011). Intrinsic cell memory reinforces myogenic commitment of pericyte-derived iPSCs. J Pathol 223:593–603 [DOI] [PubMed] [Google Scholar]
- 146.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, et al. (2011). Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 29:1117–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bar-Nur O, Russ HA, Efrat S. and Benvenisty N. (2011). Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 9:17–23 [DOI] [PubMed] [Google Scholar]
- 148.Sanchez-Freire V, Lee AS, Hu S, Abilez OJ, Liang P, Lan F, Huber BC, Ong SG, Hong WX, Huang M. and Wu JC. (2014). Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J Am Coll Cardiol 64:436–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu S, Zhao MT, Jahanbani F, Shao NY, Lee WH, Chen H, Snyder MP. and Wu JC. (2016). Effects of cellular origin on differentiation of human induced pluripotent stem cell-derived endothelial cells. JCI Insight 1:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Phetfong J, Supokawej A, Wattanapanitch M, Kheolamai P, U-Pratya Y. and Issaragrisil S. (2016). Cell type of origin influences iPSC generation and differentiation to cells of the hematoendothelial lineage. Cell Tissue Res 365:101–112 [DOI] [PubMed] [Google Scholar]
- 151.Quattrocelli M, Swinnen M, Giacomazzi G, Camps J, Barthelemy I, Ceccarelli G, Caluwe E, Grosemans H, Thorrez L, et al. (2015). Mesodermal iPSC-derived progenitor cells functionally regenerate cardiac and skeletal muscle. J Clin Invest 125:4463–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kang X, Yu Q, Huang Y, Song B, Chen Y, Gao X, He W, Sun X. and Fan Y. (2015). Effects of integrating and non-integrating reprogramming methods on copy number variation and genomic stability of human induced pluripotent stem cells. PLoS One 10:e0131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sobol M, Raykova D, Cavelier L, Khalfallah A, Schuster J. and Dahl N. (2015). Methods of reprogramming to induced pluripotent stem cell associated with chromosomal integrity and delineation of a chromosome 5q candidate region for growth advantage. Stem Cells Dev 24:2032–2040 [DOI] [PubMed] [Google Scholar]
- 154.Congras A, Barasc H, Canale-Tabet K, Plisson-Petit F, Delcros C, Feraud O, Oudrhiri N, Hadadi E, Griscelli F, et al. (2016). Non integrative strategy decreases chromosome instability and improves endogenous pluripotency genes reactivation in porcine induced pluripotent-like stem cells. Sci Rep 6:27059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, Teramoto I, Narita M, Sato Y, et al. (2013). Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci U S A 110:20569–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Galat V, Galat Y, Perepitchka M, Jennings LJ, Iannaccone PM. and Hendrix MJ. (2016). Transgene reactivation in induced pluripotent stem cell derivatives and reversion to pluripotency of induced pluripotent stem cell-derived mesenchymal stem cells. Stem Cells Dev 25:1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Okita K, Ichisaka T. and Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317 [DOI] [PubMed] [Google Scholar]
- 158.Nori S, Okada Y, Nishimura S, Sasaki T, Itakura G, Kobayashi Y, Renault-Mihara F, Shimizu A, Koya I, et al. (2015). Long-term safety issues of iPSC-based cell therapy in a spinal cord injury model: oncogenic transformation with epithelial-mesenchymal transition. Stem Cell Reports 4:360–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, et al. (2015). A comparison of non-integrating reprogramming methods. Nat Biotechnol 33:58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Park TS, Huo JS, Peters A, Talbot CC, Jr., Verma K, Zimmerlin L, Kaplan IM. and Zambidis ET. (2012). Growth factor-activated stem cell circuits and stromal signals cooperatively accelerate non-integrated iPSC reprogramming of human myeloid progenitors. PLoS One 7:e42838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Park TS, Donnenberg VS, Donnenberg AD, Zambidis ET. and Zimmerlin L. (2014). Dynamic interactions between cancer stem cells and their stromal partners. Curr Pathobiol Rep 2:41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zimmerlin L, Park TS, Zambidis ET, Donnenberg VS. and Donnenberg AD. (2013). Mesenchymal stem cell secretome and regenerative therapy after cancer. Biochimie 95:2235–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E. and Cooke JP. (2012). Activation of innate immunity is required for efficient nuclear reprogramming. Cell 151:547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Park TS, Bhutto I, Zimmerlin L, Huo JS, Nagaria P, Miller D, Rufaihah AJ, Talbot C, Aguilar J, et al. (2014). Vascular progenitors from cord blood-derived induced pluripotent stem cells possess augmented capacity for regenerating ischemic retinal vasculature. Circulation 129:359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, et al. (2014). Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Park TS, Zambidis ET, Lucitti JL, Logar A, Keller BB. and Peault B. (2009). Human embryonic stem cell-derived hematoendothelial progenitors engraft chicken embryos. Exp Hematol 37:31–41 [DOI] [PubMed] [Google Scholar]
- 167.Panicker LM, Miller D, Awad O, Bose V, Lun Y, Park TS, Zambidis ET, Sgambato JA. and Feldman RA. (2014). Gaucher iPSC-derived macrophages produce elevated levels of inflammatory mediators and serve as a new platform for therapeutic development. Stem Cells 32:2338–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Thompson SA, Burridge PW, Lipke EA, Shamblott M, Zambidis ET. and Tung L. (2012). Engraftment of human embryonic stem cell derived cardiomyocytes improves conduction in an arrhythmogenic in vitro model. J Mol Cell Cardiol 53:15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Burridge PW. and Zambidis ET. (2013). Highly efficient directed differentiation of human induced pluripotent stem cells into cardiomyocytes. Methods Mol Biol 997:149–161 [DOI] [PubMed] [Google Scholar]
- 170.Kyttala A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, et al. (2016). Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Reports 6:200–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, Endo H, Eto K, Toguchida J, Uemoto S. and Yamanaka S. (2012). Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci U S A 109:12538–12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L. and Gaffney D. (2014). Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet 10:e1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Burrows CK, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK. and Gilad Y. (2016). Genetic variation, not cell type of origin, underlies the majority of identifiable regulatory differences in iPSCs. PLoS Genet 12:e1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Feraud O, Valogne Y, Melkus MW, Zhang Y, Oudrhiri N, Haddad R, Daury A, Rocher C, Larbi A, et al. (2016). Donor dependent variations in hematopoietic differentiation among embryonic and induced pluripotent stem cell lines. PLoS One 11:e0149291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW. and Jaenisch R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A 107:9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, et al. (2014). Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158:1254–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS. and Ng HH. (2013). Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13:663–675 [DOI] [PubMed] [Google Scholar]
- 178.Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, et al. (2014). Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A 111:4484–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K. and Geijsen N. (2010). A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell 6:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Valamehr B, Robinson M, Abujarour R, Rezner B, Vranceanu F, Le T, Medcalf A, Lee TT, Fitch M, Robbins D. and Flynn P. (2014). Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Reports 2:366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen H, Aksoy I, Gonnot F, Osteil P, Aubry M, Hamela C, Rognard C, Hochard A, Voisin S, et al. (2015). Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat Commun 6:7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Duggal G, Warrier S, Ghimire S, Broekaert D, Van der Jeught M, Lierman S, Deroo T, Peelman L, Van Soom A, et al. (2015). Alternative routes to induce naive pluripotency in human embryonic stem cells. Stem Cells 33:2686–2698 [DOI] [PubMed] [Google Scholar]
- 183.Carter MG, Smagghe BJ, Stewart AK, Rapley JA, Lynch E, Bernier KJ, Keating KW, Hatziioannou VM, Hartman EJ. and Bamdad CC. (2016). A primitive growth factor, NME7AB, is sufficient to induce stable naive state human pluripotency; reprogramming in this novel growth factor confers superior differentiation. Stem Cells 34:847–859 [DOI] [PubMed] [Google Scholar]
- 184.Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, Smith A. and Nichols J. (2016). Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports 6:437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Onishi K, Tonge PD, Nagy A. and Zandstra PW. (2014). Local BMP-SMAD1 signaling increases LIF receptor-dependent STAT3 responsiveness and primed-to-naive mouse pluripotent stem cell conversion frequency. Stem Cell Reports 3:156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Illich DJ, Zhang M, Ursu A, Osorno R, Kim KP, Yoon J, Arauzo-Bravo MJ, Wu G, Esch D, et al. (2016). Distinct signaling requirements for the establishment of ESC pluripotency in late-stage EpiSCs. Cell Rep 15:787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sumi T, Oki S, Kitajima K. and Meno C. (2013). Epiblast ground state is controlled by canonical Wnt/beta-catenin signaling in the postimplantation mouse embryo and epiblast stem cells. PLoS One 8:e63378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sugimoto M, Kondo M, Koga Y, Shiura H, Ikeda R, Hirose M, Ogura A, Murakami A, Yoshiki A, Chuva de Sousa Lopes SM. and Abe K. (2015). A simple and robust method for establishing homogeneous mouse epiblast stem cell lines by wnt inhibition. Stem Cell Reports 4:744–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang J, Wang W, Ooi J, Campos LS, Lu L. and Liu P. (2015). Signalling through retinoic acid receptors is required for reprogramming of both mouse embryonic fibroblast cells and epiblast stem cells to induced pluripotent stem cells. Stem Cells 33:1390–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gomes Fernandes M, Dries R, Roost MS, Semrau S, de Melo Bernardo A, Davis RP, Ramakrishnan R, Szuhai K, Maas E, et al. (2016). BMP-SMAD signaling regulates lineage priming, but is dispensable for self-renewal in mouse embryonic stem cells. Stem Cell Reports 6:85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kurek D, Neagu A, Tastemel M, Tuysuz N, Lehmann J, van de Werken HJ, Philipsen S, van der Linden R, Maas A, et al. (2015). Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Reports 4:114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R. and Smith A. (2011). Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13:838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S. and Smith A. (2007). FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134:2895–2902 [DOI] [PubMed] [Google Scholar]
- 194.ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, Eroglu E, Siu RK. and Nusse R. (2011). Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol 13:1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Waghray A, Saiz N, Jayaprakash AD, Freire AG, Papatsenko D, Pereira CF, Lee DF, Brosh R, Chang B, et al. (2015). Tbx3 controls Dppa3 levels and exit from pluripotency toward mesoderm. Stem Cell Reports 5:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Russell R, Ilg M, Lin Q, Wu G, Lechel A, Bergmann W, Eiseler T, Linta L, Kumar PP, et al. (2015). A dynamic role of TBX3 in the pluripotency circuitry. Stem Cell Reports 5:1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ye S, Zhang D, Cheng F, Wilson D, Mackay J, He K, Ban Q, Lv F, Huang S, Liu D. and Ying QL. (2016). Wnt/beta-catenin and LIF-Stat3 signaling pathways converge on Sp5 to promote mouse embryonic stem cell self-renewal. J Cell Sci 129:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ye S, Li P, Tong C. and Ying QL. (2013). Embryonic stem cell self-renewal pathways converge on the transcription factor Tfcp2l1. EMBO J 32:2548–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Stuart HT, van Oosten AL, Radzisheuskaya A, Martello G, Miller A, Dietmann S, Nichols J. and Silva JC. (2014). NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr Biol 24:340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bourillot PY, Aksoy I, Schreiber V, Wianny F, Schulz H, Hummel O, Hubner N. and Savatier P. (2009). Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells 27:1760–1771 [DOI] [PubMed] [Google Scholar]
- 201.Hamilton WB. and Brickman JM. (2014). Erk signaling suppresses embryonic stem cell self-renewal to specify endoderm. Cell Rep 9:2056–2070 [DOI] [PubMed] [Google Scholar]
- 202.Illingworth RS, Holzenspies JJ, Roske FV, Bickmore WA. and Brickman JM. (2016). Polycomb enables primitive endoderm lineage priming in embryonic stem cells. Elife 5:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nichols J, Silva J, Roode M. and Smith A. (2009). Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136:3215–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li J, Wang G, Wang C, Zhao Y, Zhang H, Tan Z, Song Z, Ding M. and Deng H. (2007). MEK/ERK signaling contributes to the maintenance of human embryonic stem cell self-renewal. Differentiation 75:299–307 [DOI] [PubMed] [Google Scholar]
- 205.Hao J, Li TG, Qi X, Zhao DF. and Zhao GQ. (2006). WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev Biol 290:81–91 [DOI] [PubMed] [Google Scholar]
- 206.Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C. and Kahn M. (2007). Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A 104:5668–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D. and Niwa H. (2006). Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun 343:159–166 [DOI] [PubMed] [Google Scholar]
- 208.Davidson KC, Adams AM, Goodson JM, McDonald CE, Potter JC, Berndt JD, Biechele TL, Taylor RJ. and Moon RT. (2012). Wnt/beta-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc Natl Acad Sci U S A 109:4485–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bone HK, Nelson AS, Goldring CE, Tosh D. and Welham MJ. (2011). A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci 124:1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Price FD, Yin H, Jones A, van Ijcken W, Grosveld F. and Rudnicki MA. (2013). Canonical Wnt signaling induces a primitive endoderm metastable state in mouse embryonic stem cells. Stem Cells 31:752–764 [DOI] [PubMed] [Google Scholar]
- 211.Chatterjee SS, Saj A, Gocha T, Murphy M, Gonsalves FC, Zhang X, Hayward P, Akgol Oksuz B, Shen SS, et al. (2015). Inhibition of beta-catenin-TCF1 interaction delays differentiation of mouse embryonic stem cells. J Cell Biol 211:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kim SH, Kim MO, Cho YY, Yao K, Kim DJ, Jeong CH, Yu DH, Bae KB, Cho EJ, et al. (2014). ERK1 phosphorylates Nanog to regulate protein stability and stem cell self-renewal. Stem Cell Res 13:1–11 [DOI] [PubMed] [Google Scholar]
- 213.Yeo JC, Jiang J, Tan ZY, Yim GR, Ng JH, Goke J, Kraus P, Liang H, Gonzales KA, et al. (2014). Klf2 is an essential factor that sustains ground state pluripotency. Cell Stem Cell 14:864–872 [DOI] [PubMed] [Google Scholar]
- 214.Kim MO, Kim SH, Cho YY, Nadas J, Jeong CH, Yao K, Kim DJ, Yu DH, Keum YS, et al. (2012). ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat Struct Mol Biol 19:283–290 [DOI] [PubMed] [Google Scholar]
- 215.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S. and Ng HH. (2008). A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10:353–360 [DOI] [PubMed] [Google Scholar]
- 216.Zhang P, Andrianakos R, Yang Y, Liu C. and Lu W. (2010). Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J Biol Chem 285:9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, et al. (2013). FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13:351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K. and Saitou M. (2013). PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell 12:368–382 [DOI] [PubMed] [Google Scholar]
- 219.Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, Hashimoto S, Nakamura T, Sugasawa K, et al. (2014). PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development 141:269–280 [DOI] [PubMed] [Google Scholar]
- 220.von Meyenn F, Iurlaro M, Habibi E, Liu NQ, Salehzadeh-Yazdi A, Santos F, Petrini E, Milagre I, Yu M, et al. (2016). Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol Cell 62:983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tee WW, Shen SS, Oksuz O, Narendra V. and Reinberg D. (2014). Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 156:678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Qiu D, Ye S, Ruiz B, Zhou X, Liu D, Zhang Q. and Ying QL. (2015). Klf2 and Tfcp2l1, two Wnt/beta-catenin targets, act synergistically to induce and maintain naive pluripotency. Stem Cell Reports 5:314–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H, Mahmoudi T, Decraene C, Calin GA, Merrill BJ. and Fodde R. (2013). Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet 9:e1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, et al. (2013). Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497:122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hino S, Kishida S, Michiue T, Fukui A, Sakamoto I, Takada S, Asashima M. and Kikuchi A. (2001). Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol Cell Biol 21:330–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wu J, Okamura D, Li M, Suzuki K, Luo C, Ma L, He Y, Li Z, Benner C, et al. (2015). An alternative pluripotent state confers interspecies chimaeric competency. Nature 521:316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cong F. and Varmus H. (2004). Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc Natl Acad Sci U S A 101:2882–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wiechens N, Heinle K, Englmeier L, Schohl A. and Fagotto F. (2004). Nucleo-cytoplasmic shuttling of Axin, a negative regulator of the Wnt-beta-catenin Pathway. J Biol Chem 279:5263–5267 [DOI] [PubMed] [Google Scholar]
- 229.Henderson BR. (2000). Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat Cell Biol 2:653–660 [DOI] [PubMed] [Google Scholar]
- 230.Schmitz Y, Rateitschak K. and Wolkenhauer O. (2013). Analysing the impact of nucleo-cytoplasmic shuttling of beta-catenin and its antagonists APC, Axin and GSK3 on Wnt/beta-catenin signalling. Cell Signal 25:2210–2221 [DOI] [PubMed] [Google Scholar]
- 231.Krieghoff E, Behrens J. and Mayr B. (2006). Nucleo-cytoplasmic distribution of beta-catenin is regulated by retention. J Cell Sci 119:1453–1463 [DOI] [PubMed] [Google Scholar]
- 232.Faunes F, Hayward P, Descalzo SM, Chatterjee SS, Balayo T, Trott J, Christoforou A, Ferrer-Vaquer A, Hadjantonakis AK, Dasgupta R. and Arias AM. (2013). A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 140:1171–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Marucci L, Pedone E, Di Vicino U, Sanuy-Escribano B, Isalan M. and Cosma MP. (2014). Beta-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep 8:1686–1696 [DOI] [PubMed] [Google Scholar]
- 234.Abu-Remaileh M, Gerson A, Farago M, Nathan G, Alkalay I, Zins Rousso S, Gur M, Fainsod A. and Bergman Y. (2010). Oct-3/4 regulates stem cell identity and cell fate decisions by modulating Wnt/beta-catenin signalling. EMBO J 29:3236–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Munoz Descalzo S, Rue P, Faunes F, Hayward P, Jakt LM, Balayo T, Garcia-Ojalvo J. and Martinez Arias A. (2013). A competitive protein interaction network buffers Oct4-mediated differentiation to promote pluripotency in embryonic stem cells. Mol Syst Biol 9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Christoforou A, Mulvey CM, Breckels LM, Geladaki A, Hurrell T, Hayward PC, Naake T, Gatto L, Viner R, Martinez Arias A. and Lilley KS. (2016). A draft map of the mouse pluripotent stem cell spatial proteome. Nat Commun 7:8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ. and Smith A. (2013). Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153:335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620 [DOI] [PubMed] [Google Scholar]
- 239.Ye S, Tan L, Yang R, Fang B, Qu S, Schulze EN, Song H, Ying Q. and Li P. (2012). Pleiotropy of glycogen synthase kinase-3 inhibition by CHIR99021 promotes self-renewal of embryonic stem cells from refractory mouse strains. PLoS One 7:e35892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Umehara H, Kimura T, Ohtsuka S, Nakamura T, Kitajima K, Ikawa M, Okabe M, Niwa H. and Nakano T. (2007). Efficient derivation of embryonic stem cells by inhibition of glycogen synthase kinase-3. Stem Cells 25:2705–2711 [DOI] [PubMed] [Google Scholar]
- 241.Tsakiridis A, Huang Y, Blin G, Skylaki S, Wymeersch F, Osorno R, Economou C, Karagianni E, Zhao S, Lowell S. and Wilson V. (2014). Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development 141:1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Blauwkamp TA, Nigam S, Ardehali R, Weissman IL. and Nusse R. (2012). Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun 3:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Meek S, Wei J, Sutherland L, Nilges B, Buehr M, Tomlinson SR, Thomson AJ. and Burdon T. (2013). Tuning of beta-catenin activity is required to stabilize self-renewal of rat embryonic stem cells. Stem Cells 31:2104–2115 [DOI] [PubMed] [Google Scholar]
- 244.Chen Y, Blair K. and Smith A. (2013). Robust self-renewal of rat embryonic stem cells requires fine-tuning of glycogen synthase kinase-3 inhibition. Stem Cell Reports 1:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sineva GS. and Pospelov VA. (2014). beta-Catenin in pluripotency: adhering to self-renewal or Wnting to differentiate? Int Rev Cell Mol Biol 312:53–78 [DOI] [PubMed] [Google Scholar]
- 246.Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H. and Lim B. (2008). T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells 26:2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Evans PM, Chen X, Zhang W. and Liu C. (2010). KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol 30:372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Heuberger J. and Birchmeier W. (2010). Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol 2:a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D. and Knowles BB. (2004). Maternal beta-catenin and E-cadherin in mouse development. Development 131:4435–4445 [DOI] [PubMed] [Google Scholar]
- 250.Gottardi CJ, Wong E. and Gumbiner BM. (2001). E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol 153:1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.del Valle I, Rudloff S, Carles A, Li Y, Liszewska E, Vogt R. and Kemler R. (2013). E-cadherin is required for the proper activation of the Lifr/Gp130 signaling pathway in mouse embryonic stem cells. Development 140:1684–1692 [DOI] [PubMed] [Google Scholar]
- 252.Lyashenko N, Winter M, Migliorini D, Biechele T, Moon RT. and Hartmann C. (2011). Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol 13:753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ohtsuka S, Nishikawa-Torikai S. and Niwa H. (2012). E-cadherin promotes incorporation of mouse epiblast stem cells into normal development. PLoS One 7:e45220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Wu J, Platero-Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, Suzuki K, Bogliotti YS, Cuello C, et al. (2017). Interspecies chimerism with mammalian pluripotent stem cells. Cell 168:473–486 e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, et al. (2017). Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169:243–257 e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.James D, Noggle SA, Swigut T. and Brivanlou AH. (2006). Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol 295:90–102 [DOI] [PubMed] [Google Scholar]
- 257.Fang R, Liu K, Zhao Y, Li H, Zhu D, Du Y, Xiang C, Li X, Liu H, et al. (2014). Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell 15:488–496 [DOI] [PubMed] [Google Scholar]
- 258.Chen Y, Niu Y, Li Y, Ai Z, Kang Y, Shi H, Xiang Z, Yang Z, Tan T, et al. (2015). Generation of cynomolgus monkey chimeric fetuses using embryonic stem cells. Cell Stem Cell 17:116–124 [DOI] [PubMed] [Google Scholar]
- 259.Theunissen TW, Friedli M, He Y, Planet E, O'Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, et al. (2016). Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19:502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yamaguchi T, Sato H, Kato-Itoh M, Goto T, Hara H, Sanbo M, Mizuno N, Kobayashi T, Yanagida A, et al. (2017). Interspecies organogenesis generates autologous functional islets. Nature 542:191–196 [DOI] [PubMed] [Google Scholar]
- 261.Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee YS, Usui J, et al. (2010). Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142:787–799 [DOI] [PubMed] [Google Scholar]
- 262.Matsunari H, Nagashima H, Watanabe M, Umeyama K, Nakano K, Nagaya M, Kobayashi T, Yamaguchi T, Sumazaki R, Herzenberg LA. and Nakauchi H. (2013). Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc Natl Acad Sci U S A 110:4557–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Warrier S, Van der Jeught M, Duggal G, Tilleman L, Sutherland E, Taelman J, Popovic M, Lierman S, Chuva De Sousa Lopes S, et al. (2017). Direct comparison of distinct naive pluripotent states in human embryonic stem cells. Nat Commun 8:15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lee JH, Laronde S, Collins TJ, Shapovalova Z, Tanasijevic B, McNicol JD, Fiebig-Comyn A, Benoit YD, Lee JB, Mitchell RR. and Bhatia M. (2017). Lineage-specific differentiation is influenced by state of human pluripotency. Cell Rep 19:20–35 [DOI] [PubMed] [Google Scholar]
- 265.Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R. and Plath K. (2017). Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20:87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Smith A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development 144:365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Jang HJ, Kim JS, Choi HW, Jeon I, Choi S, Kim MJ, Song J. and Do JT. (2014). Neural stem cells derived from epiblast stem cells display distinctive properties. Stem Cell Res 12:506–516 [DOI] [PubMed] [Google Scholar]
- 268.Hirose H, Kato H, Kikuchi-Taura A, Soma T. and Taguchi A. (2012). Mouse ES cells maintained in different pluripotency-promoting conditions differ in their neural differentiation propensity. In Vitro Cell Dev Biol Anim 48:143–148 [DOI] [PubMed] [Google Scholar]
- 269.Honda A, Hatori M, Hirose M, Honda C, Izu H, Inoue K, Hirasawa R, Matoba S, Togayachi S, Miyoshi H. and Ogura A. (2013). Naive-like conversion overcomes the limited differentiation capacity of induced pluripotent stem cells. J Biol Chem 288:26157–26166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Honsho K, Hirose M, Hatori M, Yasmin L, Izu H, Matoba S, Togayachi S, Miyoshi H, Sankai T, Ogura A. and Honda A. (2015). Naive-like conversion enhances the difference in innate in vitro differentiation capacity between rabbit ES cells and iPS cells. J Reprod Dev 61:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE. and Clark AT. (2016). Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18:323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Peters A, Burridge PW, Pryzhkova MV, Levine MA, Park TS, Roxbury C, Yuan X, Peault B. and Zambidis ET. (2010). Challenges and strategies for generating therapeutic patient-specific hemangioblasts and hematopoietic stem cells from human pluripotent stem cells. Int J Dev Biol 54:965–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Stevens LC. (1970). The development of transplantable teratocarcinomas from intratesticular grafts of pre- and postimplantation mouse embryos. Dev Biol 21:364–382 [DOI] [PubMed] [Google Scholar]
- 274.Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH. and Surani MA. (2015). SOX17 is a critical specifier of human primordial germ cell fate. Cell 160:253–268 [DOI] [PMC free article] [PubMed] [Google Scholar]