Abstract
Tick-borne encephalitis (TBE) is the most common tick-borne viral infection in Eurasia; thousands of human cases are annually reported from several European countries. Several tick species are vectors of the tick-borne encephalitis virus (TBEV), while TBE appears to be spreading from the Eurasian continent westward to Europe. Fifteen study sites were chosen from five territories of southern Ukraine, including Odessa, Mykolaiv, Kherson Oblast, the Autonomous Republic of Crimea, and Sevastopol. Tick collection was performed in spring season of three consecutive years (1988–1990) using either flagging technique or direct collection of specimens feeding on cattle. A total of 15,243 tick imagoes and nymphs were collected from nine species, including Dermacentor marginatus, D. reticulatus, Haemaphysalis parva, H. punctata, Hyalomma marginatum, Ixodes ricinus, Rhipicephalus bursa, R. rossicus, and R. sanguineus, pooled in 282 monospecific samples. Supernatant of grinded pool was used for inoculation to suckling mice for virus isolation. Eight TBEV isolates were identified from ticks among six study sites. Ticks showed a minimum infection rate from 0.11% to 0.81%. Phylogenetic analysis of the envelope (E) protein gene of seven isolates, assigned all to the European subtype (TBEV-Eu) showing a maximum identity of 97.17% to the “Pan” TBEV-Eu reference strain. Compared to 104 TBEV-Eu isolates they clustered within the same clade as the Pan reference strain and distinguished from other TBEV-Eu isolates. Amino acid sequence analysis of the South Ukrainian TBEV-Eu isolates revealed the presence of four amino acid substitutions 67 (N), 266 (R), 306 (V), and 407 (R), in the ectodomains II and III and in the stem-anchor region of the E protein gene. This study confirmed TBEV-Eu subtype distribution in the southern region of Ukraine, which eventually overlaps with TBEV-FE (Far Eastern subtype) and TBEV-Sib (Siberian subtype) domains, showing the heterogeneity of TBEV circulating in Ukraine.
Keywords: : envelope protein, nucleotide sequencing, phylogenetic analysis, tick-borne encephalitis virus, Ukraine
Introduction
Tick-borne encephalitis virus (TBEV) is a human-pathogenic arbovirus transmitted by ixodid ticks and a member of the genus Flavivirus within the family Flaviviridae (Simmonds et al. 2011). Tick-borne encephalitis (TBE), caused by the flavivirus, is endemic in most of the European countries, Russia, northern parts of China, and Japan. During the past few decades endemic areas have expanded and within many endemic areas the number of reported cases increased (Bogovic and Strle 2015). TBE has an important impact on public health—the incidence of clinical cases is reported to be between 10,000 and 15,000 per year worldwide, although it is very probably underestimated, because notification of the disease is not mandatory in all countries (Amicizia et al. 2013).
The established method of TBEV genotyping has been based on envelope (E) protein gene structure (Ecker et al. 1999), while subtype-specific sites are also located in all virus genes (Pogodina et al. 2007, Zlobin et al. 2007, Verkhozina et al. 2008). Now three TBEV subtypes are distinguished—the European/Western subtype (TBEV-Eu), the Far Eastern (TBEV-FE), and the Siberian subtype (TBEV-Sib) (Ecker et al. 1999, Zlobin et al. 2007, Simmonds et al. 2011). In addition, two other potential subtypes have been identified in a few instances. The fourth subtype represents only one isolate (178-79 TBE isolate), originated from Irkutsk region, Russia, and preliminary typed as TBEV-FE. The fifth subtype includes 10 isolates, originated from Buryatia, Chita, and Irkutsk regions, Russia (886-84 isolate preliminary typed as TBEV-Sib) (Verkhozina et al. 2008, Demina et al. 2010).
A clear association of TBE pathogenesis has been demonstrated with TBEV subtype, as well as geographical location (Charrel et al. 2004). Indeed, subtypes are associated with a particular clinical presentation, including variable case fatality rate, chronic forms, or neurological involvement (Dumpis et al. 1999, Gratz 2005). In addition, ticks harbor epidemiological pattern of TBE, which appears closely related to the ecology and biology of the main tick vector species, including Ixodes persulcatus (TBEV-Sib; TBEV-FE), I. ricinus (TBEV-Eu) with respect to their distribution, seasonality, and tick feeding habit (Dumpis et al. 1999, Ecker et al. 1999, Randolph et al. 2000). Ultimately, each of three subtypes predominates within a particular area, where concurrently other subtypes can cocirculate (Süss et al. 2002, Zlobin et al. 2007).
To date, from the limited virus isolates done in Ukraine, there is evidence that all TBEV subtypes—TBEV-FE (Ecker et al. 1999), TBEV-Eu (Iurchenko et al. 2012), and TBEV-Sib (Kozlova et al., personal communication; GenBank data) are present on the Crimean peninsula, and TBEV-Sib can be found in the northwestern region of the country (Volyn Oblast) (Adel'shin et al. 2006).
The aim of the present work was to investigate TBEV genetic variability in southern Ukraine and to identify any genetically determined changes within the E protein of virus isolates. In this article, for a better understanding of TBEV ecology and epidemiology in the country, we will present the characterization of the TBEV isolates from the study sites and a comparison to all TBEV E protein gene sequences available from Ukraine.
Methods
Tick collection
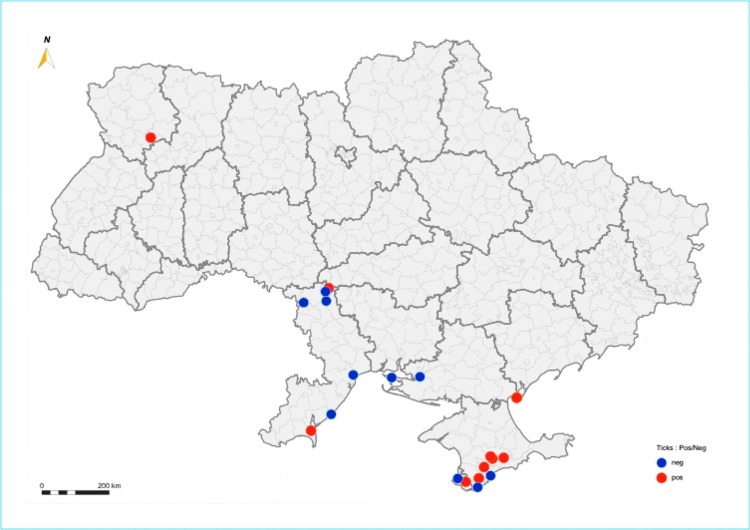
Tick collection was done across 15 sites among 5 administrative territories of southern Ukraine, including Odessa, Mykolaiv, Kherson Oblast, the Autonomous Republic of Crimea, and Sevastopol, and performed predominantly in a spring season (May–September 1988; March–June 1989; March–June, September 1990) from three consecutive years 1988 to 1990 using either flagging technique (free ticks) or direct tick collection from cattle (feeding stage) (Fig. 1, blue and purple dots). Ticks were identified by morphotaxonomy, transported to the laboratory, and stored alive in humid chambers or in tubes immersed into liquid nitrogen before testing.
FIG. 1.
Identified sites where tick-borne encephalitis virus has been isolated in Ukraine. See Supplementary Table S2 for virus strain characteristics. Credit: Dr. Marc Souris, Laos and Cambodia representative, Institute of Research for Development (IRD-UMR 190).
The sites sampled for ticks, and identified sites where TBEV has been isolated, were mapped using the SavGIS 9.07.007 software (SavGIS, 2015).
Virus isolation and identification
Virus isolation was conducted in the same year the ticks were collected and identified. For virus isolation ticks were pooled approximately till 50 imagoes and 200 nymphs of one species were collected in one site at the same time. Pooled ticks were washed once with ethanol and then two to three times with Hanks' balanced salt solution pH 7.2. Then ticks were ground with quartz sand by a pestle in a porcelain mortar in a sterile environment and added 2–4 mL of medium 199 pH 7.2 with 10% fetal calf serum and antibiotics (100 units per 1 mL). The obtained suspensions were centrifuged at 1500 g at 4°C for 15 min, and the supernatants were used for the intracerebral inoculation of 1- to 3-day-old (newborn) white mice (0.01–0.02 mL per mouse) as described earlier (Gould and Clegg 1985). The mice were observed for 3 weeks. With the development of symptoms (anorexia, inactivity, paralysis, and paresis) sick mice were euthanized and their brains were used for subsequent passaging. The 10% brain homogenates on the medium 199 pH 7.2 were prepared, centrifuged at 1500 g at 4°C for 15 min, filtered through 220 nm pore diameter syringe filters (Millipore) to eliminate the bacteria, and used for intracerebral inoculation of newborn mice. TBEV isolates were stored frozen with the periodical passaging through newborn mice brain.
Virus identification was done in complement fixation test (CFT) according to the standard procedure (Casey 1965). For CFT viral sucrose-acetone antigens (Clarke and Casals 1958) and mouse hyperimmune ascitic fluids (Tikasingh et al. 1966) were produced. The reference antigens and immune sera were provided by D.I. Ivanovsky Institute of Virology.
TBEV prevalence calculation
Calculation of TBEV prevalence in ticks was done using the minimum infection rate (MIR) as a method for estimating infection rates from pooled sampling when the prevalence is extremely low (<0.1%) as described elsewhere (Cowling et al. 1999) and eventually applied for TBEV prevalence by others (Andreassen et al. 2012). MIR calculation was done using the following formula with the assumption that each positive pool is infected by a single individual:
 |
where:
k = pool size,
m = the number of pools tested,
x = the number of positive pools.
The MIR values are expressed as percentage of potentially infected ticks in a given population at a given time.
The confidence intervals were estimated by calculating the corresponding binomial confidence limits (P) using the formula, which is applicable when the estimated prevalence ≤25% and ≥75% and expressed as percentage (Lakin 1990):
 |
where symbols are corresponded to the MIR formula.
Sequencing
Sanger technique was used for the nucleotide sequencing (Sanger et al. 1977). TBEV RNA was extracted from mice brain tissue containing virus. The isolate history varied from 7 to 13 passages. Isolation of total RNA was performed using a set of TRIzol® Plus RNA Purification Kit Reagent (Invitrogen, USA). Complementary DNA (cDNA) was synthesized with High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, USA) and random primers. Amplification of three overlapping fragments of the E protein gene was performed in the reaction mixture Platinum PCR SuperMix (Invitrogen) with the oligonucleotide primers developed with the Primer3 software (Rozen et al. 2000), available on the website http://frodo.wi.mit.edu/primer3/and published earlier (Iurchenko et al. 2012). The PCR products were purified with NucleoSpin Extract II Kit (Macherey-Nagel, Germany), amplified in a reaction mixture with terminators (BigDye Terminator v3.1 Cycle Sequencing Kit; Applied Biosystems) with the same primers (Iurchenko et al. 2012). After purification on NucleoSEQ columns (Macherey-Nagel) the products of PCR with terminators were analyzed by the capillary electrophoresis in automatic Applied Biosystems 3130 Genetic Analyzer. All stages of the sequencing were conducted in accordance with reagent's manufacturer instructions.
Sequencing alignment and assembly of the consensus sequences of E protein gene were done using the BioEdit version 7.0.5.3 software (Hall 1999).
Phylogenetic analysis
Phylogenetic analysis was carried out using the software MEGA7 (Kumar et al. 2016). To determine evolutionary distance and genetic relationships of TBEV isolates, the complete E protein gene analysis was performed and applied to 111 TBEV strain nucleotide sequences available in GenBank database and seven TBEV isolate sequences, including the four present and three others already published (Iurchenko et al. 2012) (Supplementary Tables S2 and S3). Spanish sheep encephalitis (X77470.1), louping ill (M94957.1), Negishi (M94956.1), Greek goat encephalitis (X77732.1), Turkish sheep encephalitis (DQ235151.1), Omsk hemorrhagic fever (X66694.1), Langat (DQ845244.1), and Kyasanur forest disease (X74111.1) viruses of mammalian tick-borne virus group were used as the out-group.
To illustrate that all three TBEV subtypes are circulating in Ukraine, the phylogenetic tree was generated and applied to all available 12 nucleotide sequences (complete and partial) of the E protein gene of TBEV isolated in Ukraine and three reference isolates belonging to three different subtypes (Supplementary Tables S2 and S3).
The reliability of the phylogenetic trees was tested by bootstrap resampling (1000 replications).
TBEV E protein analysis
Jalview 2.9.0b2 software was used to align TBEV E protein amino acid sequences and discover marker amino acid substitutions (Waterhouse et al. 2009).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its supplementary files.
Results
TBEV prevalence and isolate characteristics
Collection sites were originally selected within the All-Union Program on Arbovirus Surveillance led by the Ivanovsky Institute of Virology (Moscow, Russia). In addition, TBEV strains from sporadic registered cases in Crimea were used. A total of 15,243 tick imagoes and nymphs were collected from nine tick species, including Dermacentor marginatus, D. reticulatus, Haemaphysalis parva, H. punctata, I. ricinus, Hyalomma marginatum, Rhipicephalus bursa, R. rossicus, and R. sanguineus, pooled in 282 monospecific samples for virus isolation by intracerebral inoculation of newborn mice.
Eight TBEV isolates were from ticks collected at 6 of the 15 sites of three administrative territories (Fig. 1). In one site (Byriuchyj Island, Heniches'k District, Kherson Oblast) TBEV was detected for two consecutive years showing the temporal persistence of the pathogen (Table 1; Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/vbz). An estimated TBEV pooled tick prevalence using the MIR was recorded from 0.11% to 0.81% among positive sites and 95% upper confidence limit did not exceed 4.49% (Table 1).
Table 1.
Tick-Borne Encephalitis Virus Estimated Pooled Prevalence as Minimum Infection Rate and 95% Confidence Interval Among Ixodid Ticks in the Positive Sites of Southern Ukraine, 1988–1990
| District | Year | Positive pools/pools tested | Pool average size | MIR (95% CI), % |
|---|---|---|---|---|
| Kiliia | 1988 | 1/3 | 43 | 0.77 (0.13–4.28) |
| Savran | 1989 | 1/6 | 44 | 0.38 (0.07–2.13) |
| Bakhchysarai | 1989 | 1/6 | 95 | 0.17 (0.03–0.98) |
| Simferopol | 1989 | 1/4 | 31 | 0.81 (0.14–4.49) |
| Henichesk | 1989 | 2/10 | 37 | 0.54 (0.15–1.95) |
| 1990 | 1/5 | 48 | 0.41 (0.07–2.33) | |
| Bilohirsk | 1990 | 1/10 | 90 | 0.11 (0.02–0.62) |
CI, confidence interval; MIR, minimum infection rate.
Phylogenetic analysis
Nucleotide sequences used for analysis included the one produced by the present study, as well as the sequences obtained from GenBank or other sources of the literature. All sequence origins are listed in detail in the Supplementary Table S2. The complete E protein gene nucleotide sequences of seven of the eight TBEV isolates from ixodid ticks in Autonomous Republic of Crimea, Odessa, and Kherson Oblasts from 1988 to 1990 were used for a phylogenetic analysis with 111 TBEV strains available from GenBank (Supplementary Tables S2 and S3). Comparative analysis revealed a 100% identity of the E protein gene sequence among all isolates studied (isolates 120, 150, 70, 80, 85, 290, and Savran 160). Calculation of genetic or evolutionary distances showed a high level of identity of the study isolates with TBEV-Eu, including a nucleotide sequence identity from 93.38% with strain ZH RuetiZH (I. ricinus ticks, Switzerland, 2009) (Gäumann et al. 2010) to 97.17% with strain Pan (human blood, Russia, 1957) (Ecker et al. 1999), compared to a nucleotide sequence identity with TBEV-Sib and TBEV-FE subtypes ranging from 81.72% (isolate Crimea, I. ricinus ticks, Ukraine, 1987) to 82.59% (Vasilchenko isolate from human blood, Russia, 1969). The study isolates showed 80.32% of identity with a fourth subtype (i.e., isolate 178-79, I. persulcatus ticks, Russia, 1986) and 83.08% with a fifth subtype (i.e., isolate 886-84, Myodes rufocanus rodent, Russia, 1984) (Demina et al. 2010). These findings confirmed that all seven isolates from ticks of southern Ukraine belong to the European subtype showing a maximum identity of 97.17% to the “Pan TBEV-Eu” reference strain and clustering together in the same clade, distinguishing itself from other TBEV-Eu strains (Fig. 2; Supplementary Fig. S1).
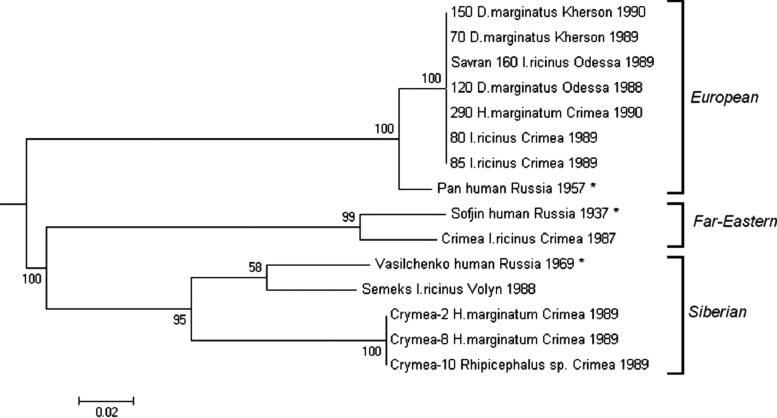
FIG. 2.
Phylogenetic tree construction (MEGA 7 software) based on the nucleotide sequence of the E protein gene illustrating the presence of all three TBEV subtypes in Ukraine. E gene nucleotide sequences were aligned with Muscle and analyzed by Maximum Likelihood method. The reliability of the tree was tested by bootstrap resampling (1000 replications). In addition, comparison of the Ukrainian strains with other TBEV-Eu strains isolated in Europe and Asia is presented in the Supplementary Figure S1A and B. *Reference strain subtype. E, envelope; TBEV, tick-borne encephalitis virus.
Phylogenetic analysis of all available E protein gene nucleotide sequences (complete and partial) of the 12 TBEV isolates in Ukraine, as well as three reference strains belonging to different subtypes (Supplementary Tables S2 and S3), indicated that all three TBEV subtypes were present in Ukraine (Fig. 2). Ultimately, the current study confirmed the presence of TBEV-Eu in southern Ukraine that overlapped a same territory as TBEV-FE and TBEV-Sib showing the heterogeneity of TBEV cocirculating on the Crimean peninsula (Fig. 2).
E protein characterization
Altogether, TBEV isolates 120, 150, 70, 80, 85, 290 and Savran 160 were clearly identified to be of TBEV-Eu subtype with respect to their dedicated amino acid signature—47(A), 88(G), 115(A), 178(E), 206(V), 267(A), 277(E), 317(A), 426(A), 431(S), 433(I), and 437(V), which appears entirely contained among all of the seven isolates and specific to TBEV-Eu type as previously described (Ecker et al. 1999).
Amino acid sequence analysis of all TBEV-Eu South Ukrainian isolates showed consistent presence of four marker amino acid substitutions, including 67(N), 266(R), 306(V), and 407(R) (Table 2). The 67(N) appears unique in South Ukrainian isolates, while all other known TBEV isolates, regardless of the subtype, have aspartic acid (D) at position 67. Arginine (R) at position 266 was previously identified only in two isolates from I. ricinus ticks in Switzerland in 2009—ZH Langnau a.A.1 and ZH Langnau a.A.2 (Gäumann et al. 2011), while all other TBEV strains have lysine (K) at position 266. Besides South Ukrainian isolates, valine (V) at position 306 was found only in five isolates: TBEV-Eu Pan, ZZ9 (I. ricinus ticks, Austria, 1985) (Ecker et al. 1999), VD Cudrefin (I. ricinus ticks, Switzerland, 2009) (Gäumann et al. 2010), and TBEV-FE strains Oshima 5-10 isolates (human, Japan, 1993) (Takashima et al. 1997) and 178-79 isolates. All other known isolates have methionine (M) at the position 306. Arginine (R) at position 407 detected in South Ukrainian isolates is typical for the TBEV-Sib and TBEV-FE (including Crimea isolate) subtypes and was revealed only in two TBEV-Eu strains—Pan and Stara Ves (Croatia) (Ecker et al. 1999).
Table 2.
Marker Amino Acid Substitutions in the Envelope Protein of Tick-Borne Encephalitis Virus Strains Isolated in Southern Ukraine Compared with the Prototype Strains
| Amino acid substitutions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ectodomain | ||||||||||
| Domain I | Domain II | Domain III | Stem-anchor region | |||||||
| TBEV subtype | Strain name | 39a | 189 | 67 | 100 | 246 | 266 | 306 | 407 | 487 |
| European | 120b | P | A | N | G | A | R | V | R | V |
| ZH Langnau a.A.1c | P | A | D | G | A | R | M | K | V | |
| ZZ9d | P | A | D | G | A | K | V | K | V | |
| Pane | P | A | D | G | A | K | V | R | V | |
| Neudoerfl | P | A | D | G | A | K | M | K | V | |
| Siberian | Crimea-2f | – | – | – | – | – | – | – | – | I |
| Semeks | – | P | – | S | – | – | – | – | – | |
| Zausaev | P | A | D | G | A | K | M | R | V | |
| 886-84 | P | A | D | G | A | K | M | R | V | |
| Buzuuchuk | P | A | D | G | A | K | M | R | V | |
| Vasilchenko | P | A | D | G | A | K | M | R | V | |
| Far Eastern | Crimea | L | A | D | G | V | K | M | R | V |
| Sofjin | P | A | D | G | A | K | M | R | V | |
| Oshima 5-10g | P | A | D | G | A | K | V | R | V | |
Position numbers are counted from the first amino acid of Envelope protein. The names of the strains isolated in Ukraine are highlighted in bold. The unique amino acid substitutions highlighted in black, the rare ones in TBEV in gray, and the rare ones in this subtype and typical for others framed with black line square. In the table one of the strains represents a group with identical amino acid substitutions in the selected positions: bstrains 120, 150, 70, 80, 85, 290 and Savran 160; cstrains ZH Langnau a.A.1 and ZH Langnau a.A.2; dstrains ZZ9 and VD Cudrefin; estrains Pan and Stara Ves; fstrains Crimea-2, Crimea-8 and Crimea-10; gstrains Oshima 5–10 and 178–79.
TBEV, tick-borne encephalitis virus.
Two of the amino acid substitutions revealed in the South Ukrainian TBEV isolates, 67(N) and 266(R), are located in the domain II (amino acid residues 52-136 and 190-284), the 306(V) in the domain III (amino acid residues 303-395) (Rey et al. 1995), and 407(R) in the stem-anchor region (residues 401-496) (Allison et al., 1999) (Table 2).
Furthermore, the unique amino acid substitutions were found in other TBEV isolates in Ukraine. TBEV-FE strain Crimea has amino acid substitutions 39(L) in the domain I (residues 1-51, 137-189, and 285-302) (Rey et al. 1995) and 246(V) in the domain II. Both substitutions are not found in TBEV, but valine (V) at position 246 is a typical for 2 other flaviviruses—Omsk hemorrhagic fever and Negishi (results not showed). Crimea-2, Crimea-8 and Crimea-10 isolates are distinguished from all other TBEV isolates by the presence of isoleucine (I) at position 487 in the stem-anchor region, while all other TBEV isolates have valine (V) at the position. Volhynian isolate Semeks has unique amino acid substitutions 189(P) and 100(S) located in the domain I and the domain II correspondingly (Table 2).
Discussion
With respect to the TBEV activity in Ukraine and its vectors, the virological testing revealed the presence of TBEV among ixodid ticks in southern Ukraine, in 1988–1990. TBEV estimated pooled prevalence of ticks in the region varied from 0.11% to 0.81%. These findings are consistent with the results of the virological examination of ixodid ticks on the Crimean peninsula in 1985–1990, when 63 isolates were recovered from 35,170 ticks, when the average MIR of ticks with TBEV was 0.18% (Markeshin 1994) [where the infection prevalence of TBEV in unfed host-questing ticks is typically 0.1–5.0%, with some exceptions in Far East Russia (Randolph 2001)], is also consistent with the current study's findings.
TBEV was detected in I. ricinus tick—the main vector of TBEV-Eu (Dumpis et al. 1999, Ecker et al. 1999, Randolph et al. 2000) and also among ticks of three other species indicating a consistent virus presence in the study area through the identified vector hosts.
Phylogenetic analysis of seven of the eight TBEV isolates from southern Ukraine showed 100% identity of the E protein gene nucleotide sequences in all isolates studied and their belonging to European/Western subtype. Moreover, these isolates form a separate phylogenetic cluster inside TBEV-Eu subtype with Pan strain. Ultimately, one hypothesis is that TBEV-Eu strains isolated in southern Ukraine were evolving independently of the rest of TBEV-Eu strains and diverged from a common ancestor with the Pan strain after differentiation of TBEV-Eu strains into two clusters (Supplementary Fig. S1).
We observed several original structural protein gene mutations (i.e., amino acid substitutions) that eventually provide more insight for the understanding of their significance in terms of functionality, as well as TBEV circulation in Ukraine. Indeed, analysis of dedicated amino acid sequences of E protein revealed the presence of four amino acid substitutions that distinguished TBEV-Eu strains isolated in the current study region (120, 150, 70, 80, 85, 290, and Savran 160) from the other known isolates. Furthermore, unique amino acid substitutions in the E protein were revealed in TBEV-FE (Crimea isolate) and TBEV-Sib (isolates Semeks, Crimea-2, Crimea-8 and Crimea-10 isolates) in Ukraine by other researchers (Ecker et al. 1999, Adel'shin et al. 2006, Kozlova et al., personal communication; GenBank data). The biological role of amino acid substitutions so far has not been totally elucidated; however, it is known that all domains of the ectodomain possess antigenic activity and can determine the virulence for different isolates. Furthermore, the domain II is presumably participating in the virus fusion with a cell membrane, and the domain III is a potential binding site of a cellular receptor (Rey et al. 1995). The stem-anchor region of the E protein is important for a number of functions, including trimerization of soluble protein E, interactions with the precursor of membrane protein M during viral assembly, particle formation, and low-pH-induced structural changes associated with membrane fusion (Allison et al. 1999). Ultimately, the presence of the unique and rare amino acid substitutions in the Ukrainian TBEV isolates distinguished them from the isolates circulating in other territories. Further molecular-genetic studies are necessary to conclude if these substitutions serve as markers of the regional TBEV population in Ukraine.
However, the E protein gene sequence analyzed is associated with specific TBEV isolates recovered from different origins and could eventually be subject to genetic variations introduced by host-virus interactions during repeated passaging through newborn mice brain cells. In addition, cross-contamination cannot be excluded in the process of long-term maintenance. Although E protein gene data allowed defining a subtype of TBEV isolated in southern Ukraine from 1988 to 1990, whole genome sequencing could provide new knowledge about the genetic variability of TBEV population. Such an approach is indeed of great importance for the understanding of the origin and evolution of the virus, as well as the formation and maintenance of TBE natural foci (Kovalev and Mukhacheva 2014). In addition, genotyping circulating virus strains should be useful for epidemiological surveillance, as well as improving diagnostic and prophylaxis of the disease in Ukraine.
Conclusion
This study confirmed the occurrence of European subtype of TBEV in southern Ukraine. A comparative phylogenetic analysis of the E protein gene showed the homogeneity of European subtype TBEV population in Ukraine, while the seven isolates from southern Ukraine are most related to Pan strain and form with it a separate phylogenetic cluster within TBEV-Eu subtype. At the same time, concurrent circulation of two other subtypes, including TBEV-Sib and TBEV-FE in Crimean peninsula, is showing the heterogeneity of TBEV population in the region. Because it has been nearly 30 years since the sampling, we referred to, was carried out, the epidemiology could have changed, and further studies are needed to evaluate the risk of TBEV expansion and therefore identify driving forces for TBEV and ticks' geographical range.
Supplementary Material
Acknowledgments
The authors express sincere thanks to Vladimir Partyga (SB “I.I. Mechnikov Ukrainian Anti-Plague Research Institute of the Ministry of Health of Ukraine,” Odessa, Ukraine) for tick collection and identification, to D.K. Lvov (D.I. Ivanovsky Institute of Virology) for his assistance in confirming TBEV isolate identification, to Irina Kozlova, Natalya Gusarova (FSSFE Scientific Centre of Family Health and Human Reproduction Problems SB RAMS, Irkutsk, Russia), Vladimir Zlobin (Institute of Biomedical Technology, Irkutsk State Medical University of Russian Ministry of Heath, Irkutsk, Russia), and Sergey Tkachev (Institute of Chemical Biology and Fundamental Medicine SB RAS, Novosibirsk, Russia) for providing the geographical coordinates of TBEV isolates of Crimea. The authors acknowledge the United States Department of Defense, Defense Threat Reduction Agency (DTRA), Cooperative Biological Engagement Program (CBEP) for their support to develop this article. While DTRA/CBEP did not support the research described in this publication, the program supported the presentation of this research in an international forum and supported grantsmanship training related to the development of this article. The contents of this publication are the responsibility of the authors and do not necessarily reflect the views of DTRA or the United States Government. A special thanks to Marc Souris (Institute of Research for Development, Marseille, France) for his support in generating the map of the tick collection sites.
Authors' Contributions
O.O.Y. made substantial contributions to conception, design, and coordination of the genetic study, carried out the molecular genetic studies, phylogenetic analysis, statistical analysis of the pooled prevalence, and drafted the article. D.O.D. made substantial contributions to conception, design, and coordination of virological studies, took part in the tick collection, processed all laboratory samples, performed live virus isolation, identification, and maintenance, and drafted article. N.O.V. participated in the design of the study, drafting the article, and revising it critically for important intellectual content. O.O.Y. and J.P.G. reanalyzed all data, developed the final versions of figures and tables, and produced the final version of the article to be published. All authors read and approved the final article.
Author Disclosure Statement
No competing financial interests exist.
References
- Adel'shin RV, Zlobin VI, Belikov SI, Dzhioev YuP, et al. Molecular epidemiology of tick-borne encephalitis in the European part of Russia and in some countries of Baltic, Eastern and South-Eastern of Europe (in Russian). Epidemiol Vaktsinoprofil 2006; 27:27–34 [Google Scholar]
- Allison SL, Stiasny K, Stadler K, Mandl CW, et al. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol 1999; 73:5605–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amicizia D, Domnich A, Panatto D, Lai PL, et al. Epidemiology of tick-borne encephalitis (TBE) in Europe and its prevention by available vaccines. Hum Vaccin Immunother 2013; 9:1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen A, Jore S, Cuber P, Dudman S, et al. Prevalence of tick borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Parasit Vectors 2012; 5:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogovic P, Strle F. Tick-borne encephalitis: A review of epidemiology, clinical characteristics, and management. World J Clin Cases 2015; 3:430–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey HL. Standardized diagnostic complement fixation method and adaptation to micro test. I. Laboratory branch complement fixation method. By laboratory branch task force. II. Adaptation of LBCF method to micro technique. Public Health Monogr 1965; 74:1–34 [PubMed] [Google Scholar]
- Charrel RN, Attoui H, Butenko AM, Clegg JC, et al. Tick-borne virus diseases of human interest in Europe. Clin Microbiol Infect 2004; 10:1040–1055 [DOI] [PubMed] [Google Scholar]
- Clarke D, Casals J. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg 1958; 7:561–573 [DOI] [PubMed] [Google Scholar]
- Cowling DW, Gardner IA, Johnson WO. Comparison of methods for estimation of individual-level prevalence based on pooled samples. Prev Vet Med 1999; 39:211–225 [DOI] [PubMed] [Google Scholar]
- Demina TV, Dzhioev YP, Verkhozina MM, Kozlova IV, et al. Genotyping and characterization of the geographical distribution of tick-borne encephalitis virus variants with a set of molecular probes. J Med Virol 2010; 82:965–976 [DOI] [PubMed] [Google Scholar]
- Dumpis U, Crook D, Oksi J. Tick-borne encephalitis. Clin Infect Dis 1999; 28:882–890 [DOI] [PubMed] [Google Scholar]
- Ecker M, Allison SL, Meixner T, Heinz FX. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J Gen Virol 1999; 80:179–185 [DOI] [PubMed] [Google Scholar]
- Gäumann R, Muhlemann K, Strasser M, Beuret CM. High-throughput procedure for tick surveys of tick-borne encephalitis virus and its application in a national surveillance study in Switzerland. Appl Environ Microbiol 2010; 76:4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäumann R, Růžek D, Mühlemann K, Strasser M, et al. Phylogenetic and virulence analysis of tick-borne encephalitis virus field isolates from Switzerland. J Med Virol 2011; 83:853–863 [DOI] [PubMed] [Google Scholar]
- Gould EA, Clegg JCS. Growth, titration and purification of alphaviruses and flaviviruses. In: Mahy BWJ, ed. Virology—A Practical Approach. Oxford: IRL, 1985:43–78 [Google Scholar]
- Gratz N. Transmissible Infectious Diseases in Europe. Their Distribution and Public Health Impact (in Russian). WHO Regional Office in Europe, 2005:158 [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999; 41:95–98 [Google Scholar]
- Iurchenko OA, Vynograd NA, Dubina DA. Molecular genetic characteristics of tick-borne encephalitis virus in the Crimea (in Russian). Vopr Virusol 2012; 57:40–43 [PubMed] [Google Scholar]
- Kovalev SY, Mukhacheva TA. Clusterons as a tool for monitoring populations of tick-borne encephalitis virus. J Med Virol 2014; 86:283–289 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin GF. Biometrics (in Russian). 4th ed., revised. Moscow: Vysshaya Shkola, 1990:352 [Google Scholar]
- Markeshin SY. The Study of Natural Foci of Tick-Borne Encephalitis, Hemorrhagic Fever with Renal Syndrome and the Crimean-Congo Hemorrhagic Fever in Crimea (in Russian). Abstract of the thesis for the degree of candidate of medical sciences: 03.00.06. Moscow, 1994:24 [Google Scholar]
- Pogodina VV, Karan LS, Kolyasnikova NM, Levina LS, et al. Evolution of tick-borne encephalitis and a problem of evolution of its causative agent (in Russian). Vopr Virusol 2007; 52:16–21 [PubMed] [Google Scholar]
- Randolph SE. The shifting landscape of tick-borne zoonoses: Tick-borne encephalitis and Lyme borreliosis in Europe. Phil Trans R Soc Lond B 2001; 356:1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph SE, Green RM, Peacey MF, Rogers DJ. Seasonal synchrony: The key to tick-borne encephalitis foci identified by satellite data. Parasitology 2000; 121:15–23 [DOI] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, et al. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 1995; 375:291–298 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S, eds. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press, 2000:365–386 [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci (USA) 1977; 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SavGIS. Système d'information géographique pour Windows. 2015. Available at www.savgis.org/SavGIS/Telechargement/Logiciel.html
- Simmonds P, Becher P, Collett MS, Gould EA, et al. Family Flaviviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, eds. Virus Taxonomy: The Classification and Nomenclature of Viruses. The Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Academic Press, 2011:1003–1020 [Google Scholar]
- Süss J, Schrader C, Abel U, Bormane A, et al. Characterization of tick-borne encephalitis (TBE) foci in Germany and Latvia (1997–2000). Int J Med Microbiol 2002; 291:34–42 [DOI] [PubMed] [Google Scholar]
- Takashima I, Morita K, Chiba M, Hayasaka D, et al. A case of tick-borne encephalitis in Japan and isolation of the virus. J Clin Microbiol 1997; 35:1943–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikasingh ES, Spence L, Downs WG. The use of adjuvant and sarcoma 180 cells in the production of mouse hyperimmune ascitic fluids to arboviruses. Am J Trop Med Hyg 1966; 15:219–226 [DOI] [PubMed] [Google Scholar]
- Verkhozina MM, Zlobin VS, Kozlova IV, Demina TV, et al. Ecology-epidemiological and molecular-genetic analysis of the population of tick-borne encephalitis virus on the territory of Irkutsk region (in Russian). Epidemiol Vaktsinoprofil 2008; 38:12–18 [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, et al. Jalview version 2: A multiple sequence alignment and analysis workbench. Bioinformatics 2009; 25:1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlobin VI, Verkhozina MM, Demina TV, Demina TV, et al. Molecular epidemiology of tick-borne encephalitis (in Russian). Vopr Virusol 2007; 52:4–13 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its supplementary files.