Abstract
Homologous recombination safeguards genome integrity, but it can also cause genome instability of important consequences for cell proliferation and organism development. Transcription induces recombination, as shown in prokaryotes and eukaryotes for both spontaneous and developmentally regulated events such as those responsible for immunoglobulin class switching. Deciphering the molecular basis of transcription-associated recombination (TAR) is important in understanding genome instability. Using novel plasmid-borne recombination constructs in Saccharomyces cerevisiae, we show that RNA polymerase II (RNAPII) transcription induces recombination by impairing replication fork progression. RNAPII transcription concomitant to head-on oncoming replication causes a replication fork pause (RFP) that is linked to a significant increase in recombination. However, transcription that is codirectional with replication has little effect on replication fork progression and recombination. Transcription occurring in the absence of replication does not affect either recombination or replication fork progression. The Rrm3 helicase, which is required for replication fork progression through nucleoprotein complexes, facilitates replication through the transcription-dependent RFP site and reduces recombination. Therefore, our work provides evidence that one mechanism responsible for TAR is RNAP-mediated replication impairment.
Keywords: genetic instability, homologous recombination, replication, transcription
Introduction
The maintenance of genome integrity is essential to safeguard genetic information and to prevent the loss of cell fitness that is generally associated with cancer and a number of genetic disorders (Lengauer et al, 1998; Kolodner et al, 2002). In mitosis, homologous recombination is a major mechanism of DNA repair that uses as template an intact homologous DNA sequence. Depending on the template used, whether or not an allelic sequence is in the sister chromatid or in the homologous chromosome, homologous recombination can either safeguard genetic integrity or cause DNA rearrangements.
Transcription has been shown to induce homologous recombination from bacteria to humans, thus affecting genome stability (Aguilera, 2002). This phenomenon, termed transcription-associated recombination (TAR), is also linked to the generation of genetic diversity during developmentally regulated processes such as class switching of immunoglobulin (Ig) genes (Jung et al, 1993; Peters and Storb, 1996). Despite its importance in genome stability and programmed genome rearrangements, the mechanisms by which TAR occurs are poorly understood.
Transcription elongation introduces local changes in DNA topology and chromatin structure that could lead to a transient accumulation of single-stranded DNA (ssDNA) regions (Gangloff et al, 1994). The observation that transcription increases synergistically the hyper-recombinogenic effect of 4-nitroquinoline (4-NQO) and methyl methane-sulphonate (MMS) suggests that transcription makes DNA more accessible to genotoxic agents (Garcia-Rubio et al, 2003), possibly by generating transient ssDNA regions. Studies performed with the hpr1 mutant of the THO complex in Saccharomyces cerevisiae, which shows a strong increase of recombination linked to transcription (Chavez and Aguilera, 1997; Prado et al, 1997), have revealed that TAR in these mutants is mediated by the nascent mRNA. Cotranscriptionally formed DNA:RNA hybrids accumulate in hpr1 mutants and are responsible for TAR (Huertas and Aguilera, 2003), suggesting that an ssDNA sequence might contribute to TAR. Interestingly, DNA:RNA hybrids also accumulate during class switching of Ig genes (Yu et al, 2003).
Homologous recombination is a major DNA repair pathway of breaks occurring during DNA replication (Cox et al, 2000; Rothstein et al, 2000). Indeed, replication seems to be a major source of spontaneous genetic instability, as suggested by the accumulation in yeast to humans of DNA breaks and chromosomal rearrangements in S-phase checkpoint mutants (Myung et al, 2001; Casper et al, 2002; Cha and Kleckner, 2002). The inhibition of replication by physical obstacles (DNA–protein complexes or DNA lesions), chemical inhibitors or mutations may lead to replication fork breakages that require homologous recombination to resume replication (Horiuchi and Fujimura, 1995; Zou and Rothstein, 1997; Seigneur et al, 1998; Ivessa et al, 2000, 2003; McGlynn and Lloyd, 2000; Sogo et al, 2002; Courcelle et al, 2003; Torres et al, 2004a). In this regard, a number of in vitro and in vivo studies suggest that transcription could occasionally inhibit replication fork progression. Bacteriophage T4 and φ29 replication machineries are transiently paused in vitro by collisions with the bacterial RNAP ternary transcription complex (Liu et al, 1993; Liu and Alberts, 1995; Elias-Arnanz and Salas, 1997, 1999). Replication fork progression is inhibited by transcription in vivo in bacteria and yeast (French, 1992; Krasilnikova et al, 1998; Takeuchi et al, 2003), and natural replication fork pauses (RFPs) have been detected at yeast transfer RNA (tRNA) genes that depend on RNA polymerase III (RNAPIII)-mediated transcription (Deshpande and Newlon, 1996).
Here we have tested the possibility that RNAPII-mediated transcription induced recombination by impairing replication fork progression in S. cerevisiae. Using plasmid-borne direct-repeat constructs under the control of regulated promoters, we showed that a head-on and, to a much lesser extent, codirectional encounter between transcription and replication promotes TAR. Besides, TAR was associated with the appearance of an RFP at the recombining region that was partially suppressed by the Rrm3 helicase. These results indicate that TAR is a consequence of the impairment of replication fork progression caused by RNAPII-mediated transcription, and provides a molecular link between TAR and DNA replication.
Results
TAR requires head-on oncoming replication
To determine whether TAR is the result of a conflict between RNAPII-mediated transcription and replication, novel in vivo recombination constructs were designed (Figure 1A). These constructs were made in centromeric plasmids and were based on two direct repeats of a 0.6-kb internal fragment of the LEU2 gene, which generate a selectable wild-type copy of LEU2 by recombination. Transcription through the leu2 repeats is driven by the regulated GAL1 promoter, which is repressed in 2% glucose (GAL-IN (Glu)) and activated in 2% galactose (GAL-IN (Gal)). The leu2 repeats are oriented according to their transcription either inward (IN) or outward (OUT) with respect to the unique ARSH4 replication origin contained in the plasmid, and are replicated by the proximal replication fork (shown later). Northern analysis showed that the level of transcription through the repeats was similar in the GAL-IN and GAL-OUT constructs (Figure 1B). The constructs permit the analysis of recombination caused by the encounter—either head-on or codirectional—between the replication fork and RNAPII-mediated transcription.
Figure 1.
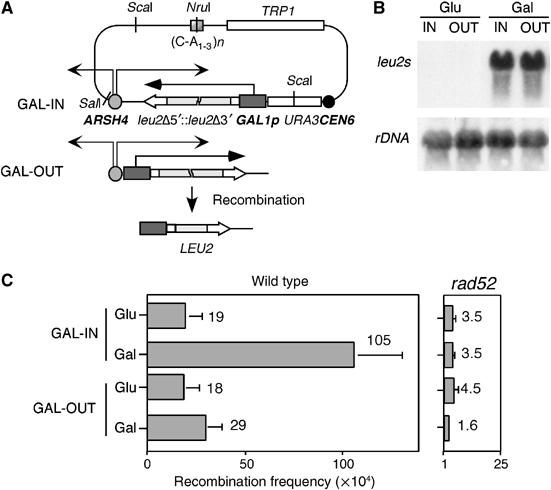
Homologous recombination is induced by RNAPII-mediated transcription if this occurs in the opposite direction to an oncoming replication fork. (A) Schemes of the centromeric plasmids harbouring the recombination constructs GAL-IN and GAL-OUT, and of the LEU2 recombination product. The arrows indicate the progression orientation of RNAPII transcription driven from the GAL1 promoter and of the replication forks initiated at ARSH4. The distance that each fork has to traverse from ARSH4 to the promoter are approximately 2.5 and 6 kb for the rightward- and leftward-advancing forks, respectively. (B) Northern analysis of transcripts emerging from the direct-repeat constructs in wild-type cells grown either in glucose (Glu; transcription OFF) or galactose (Gal; transcription ON). (C) Recombination frequencies in wild-type and rad52Δ strains. The average and standard deviation are indicated.
As shown in Figure 1C, in the absence of transcription, the frequency of recombination is the same regardless of the orientation of the leu2 repeats relative to ARSH4 (GAL-IN versus GAL-OUT in glucose). When transcription was active, the frequency of recombination was dependent on the orientation of the leu2 repeats relative to ARSH4. In GAL-OUT (Gal), the frequency of recombination was 1.6-fold higher than in GAL-OUT (Glu). This indicates that transcription by itself had little effect on recombination if it advanced in the same direction as the replication fork. In contrast, transcription through the leu2 repeats increased the frequency of recombination 5.5-fold in the GAL-IN construct, in which transcription and replication are convergent (Figure 1C, compare GAL-IN (Gal) versus GAL-IN (Glu)). This increase in recombination was dependent on RAD52, indicating that the events occurred by homologous recombination.
The results suggest that an impairment of replication fork progression caused by the oncoming transcription could be responsible for the transcription-mediated increase in recombination. If this were the case, transcription should induce recombination only during the S phase. To test this hypothesis, three new constructs were generated by replacing the GAL1 promoter in the IN construct by cell-cycle-specific promoters whose cell-cycle-specific regulation was known to be maintained in centromeric plasmids: CLN-IN, in which transcription is driven by the promoter of the CLN2 G1-cyclin gene, is expressed in G1 (Wittenberg et al, 1990; Stuart and Wittenberg, 1994; Spellman et al, 1998), and HHF-IN and HHO-IN, in which transcription is driven from the promoters of the H4- and H1-like histone genes HHF2 and HHO1, respectively, are expressed in late G1/S phase (Hereford et al, 1981; Freeman et al, 1992; Spellman et al, 1998) (Figure 2A). Consequently, whereas transcription and replication would take place at nonoverlapping times in the CLN-IN construct and should not affect recombination, they would occur concomitantly in the HHF-IN and HHO-IN constructs and are expected to increase recombination. As can be seen in Figures 1C and 2B, the frequency of recombination in the CLN-IN construct was similar to that observed in GAL-IN (Glu). In contrast, transcription driven from the S-phase-induced promoters in the HHF-IN and HHO-IN constructs increased recombination 5- and 2.5-fold, respectively. To determine whether the increase in recombination caused by S-phase transcription required a head-on encounter of transcription and replication, recombination was determined in the HHF-OUT construct carrying the OUT repeat system under the control of the HHF2 promoter (Figure 2A, HHF-OUT). As can be seen in Figures 1C and 2B, S-phase-induced transcription in the HHF-OUT construct increased recombination two-fold above the GAL-OUT levels in the absence of transcription.
Figure 2.
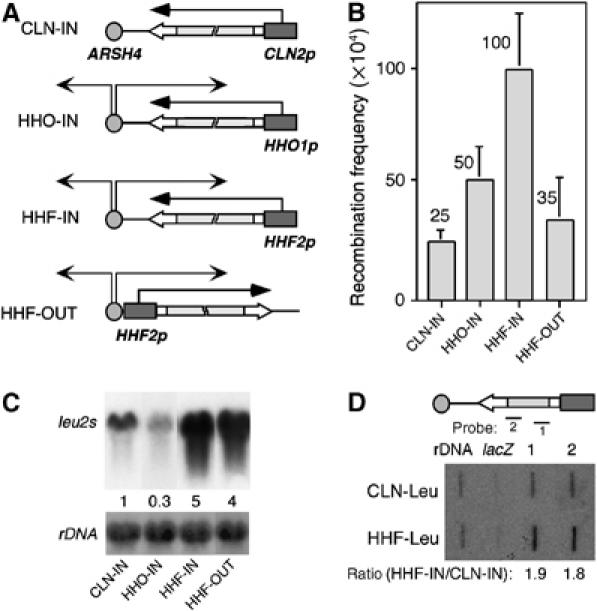
Recombination is induced by transcription only if this is active during S phase. (A) Schemes of the CLN-IN, HHO-IN, HHF-IN and HHF-OUT constructs. The leu2 direct repeats are under the control of the CLN2, HHO and HHF2 promoters, respectively, which are activated in G1 phase in the CLN-IN construct and in late G1/S phase in the HHF-IN and HHO-IN constructs. (B) Recombination frequencies of each construct in the wild-type strain. The average and standard deviation are indicated. (C) Northern analysis of leu2 transcripts emerging from each system (leu2s). RNA levels are normalized with respect to the CLN-IN values, taken as 1. (D) Run-on analysis of leu2 transcripts emerging from the CLN-Leu and HHF-Leu constructs. A scheme of these constructs, which are driven by the CLN2 and HHF2 promoters, respectively, is shown on top. The amount of mRNA bound to each probe is normalized with respect to the CLN-IN values, taken as 1. lacZ was used as negative control.
Since transcription levels have a direct effect on the frequency of recombination in yeast (Thomas and Rothstein, 1989; Saxe et al, 2000; Gonzalez-Barrera et al, 2002), we determined whether our results could be explained as a consequence of different transcription rates. As shown in Figure 2C, the levels of recombination in the HHO-IN and HHF-IN constructs correlated with transcript levels, as determined by Northern. However, transcripts in the non-hyper-recombinant CLN-IN construct accumulated at levels three-fold higher than in HHO-IN, whereas transcription levels were similar in HHF-IN and HHF-OUT (Figure 2C). To confirm that transcript levels determined by Northern correlated with transcription rates, we performed run-on analysis of the HHF-Leu and CLN-Leu constructs (Figure 2D), which were obtained directly from yeast Leu+ recombinants derived from HHF-IN and CLN-IN transformants, respectively. As expected, the rate of transcription in the HHF-Leu construct was higher (two-fold) than in the CLN-Leu construct. Altogether, these results confirm that transcription by itself is not sufficient to induce recombination. Instead, the increase in recombination mediated by transcription requires concomitant replication.
TAR is linked to the appearance of an RFP
To further explore the molecular nature of TAR, we determined whether replication fork progression was negatively affected by RNAPII-mediated transcription in our constructs. For this, replication intermediates were analysed by 2D-gel electrophoresis (Figure 3). Two overlapping restriction fragments covering the leu2 repeats in the IN constructs were analysed: a SalI–ScaI fragment that leads to the formation of an arc of Y-shaped replication intermediates (Figure 3B), and a ScaI fragment in which the internal position of ARSH4 leads to the formation of an additional bubble arc (Figure 3C). The absence of termination arcs—double Y- and X-shaped replication intermediates—in both fragments suggested that the proximal replication fork replicated the leu2 repeats, validating the interpretation concerning orientation of replication relative to transcription.
Figure 3.
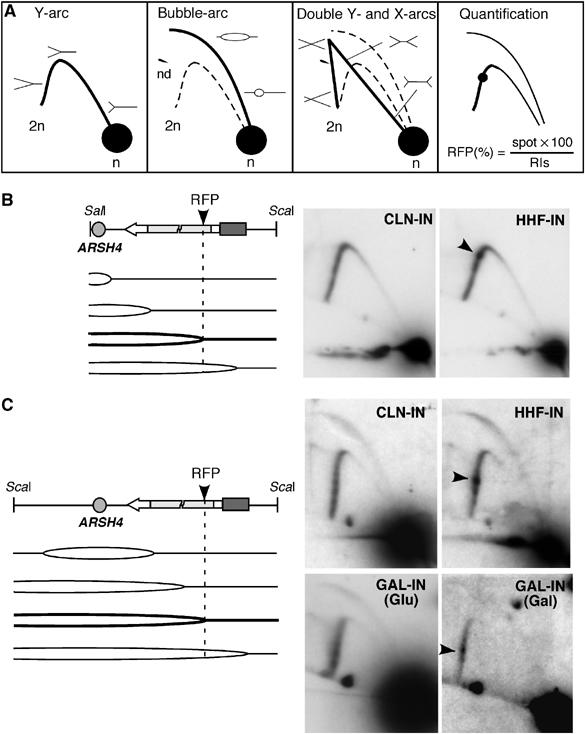
Converging transcription and replication leads to a 2D-gel-detectable RFP. (A) Schematic representation of the migration pattern of the Y-, bubble-, double Y- and X-shaped replication intermediates in 2D-gel electrophoresis. n indicates unreplicated molecules, RIs replication intermediates and nd not determined molecules. (B) 2D-gel analysis of the SalI–ScaI restriction fragment covering the leu2 repeats in the CLN-IN and HHF-IN constructs. Note that bubble molecules are not detected due to the terminal location of ARSH4 in the SalI–ScaI fragment. (C) 2D-gel analysis of the ScaI restriction fragment covering the leu2 repeats in the GAL-IN (either from cells grown in glucose (Glu) or galactose (Gal)), CLN-IN and HHF-IN constructs. The location of ARSH4 in an internal position leads to the detection of a bubble arc in addition to the Y-arc. A scheme of the replication intermediates expected for each restriction fragment and the position of the transcription-dependent RFP in the HHF-IN and GAL-IN constructs (solid arrow) is shown on the left.
As can be seen in Figure 3B and C, a region of intense hybridization in the arc corresponding to the Y-shaped replication intermediates was detected in the HHF-IN construct, which is specifically transcribed during the S phase. The mapping of this hybridization signal in the two overlapping restriction fragments revealed that there was an accumulation of replication forks at the leu2Δ3′ repeat, at approximately 350 bp from the translation initiation site. Despite this accumulation of replication forks, a complete arc of Y-shaped replication intermediates was formed, indicating that most of the replication forks at the leu2Δ3′ repeat were not blocked, but paused or slowed down at a specific RFP site.
To determine whether the RFP at the HHF-IN construct was a consequence of transcription through the leu2 repeats during S phase, we analysed replication intermediates in the CLN-IN, which is specifically transcribed during G1, and the GAL-IN constructs. As can be seen in Figure 3B and C, the RFP was not detected in CLN-IN. In addition, the RFP was not detected in GAL-IN (Glu), while it appeared in GAL-IN (Gal) at the same position in the leu2Δ3′ repeat as in HHF-IN (Figure 3C). Therefore, the formation of this RFP requires the presence of active transcription during the S phase. Quantification of the hybridization signal revealed a significant accumulation of replication intermediates at this RFP in HHF-IN (24±2.5% of replication intermediates) and GAL-IN (Gal) (21±2%). Such intermediates were not observed in CLN-IN (13±1.5%) or in GAL-IN (Glu) (14±1%).
The appearance of an RFP in HHF-IN and GAL-IN (Gal) but not in CLN-IN or GAL-IN (Glu) is consistent with a link of the RFP to TAR. If this were the case, active transcription should not lead to a significant RFP in the GAL-OUT (Gal) or HHF-OUT constructs, as the codirectional progression of transcription and replication hardly affected recombination (Figures 1 and 2). As shown in Figure 4, the GAL-OUT construct did not show detectable RFPs both in glucose and galactose, whereas the HHF-OUT construct seems to show a weak transcription-dependent RFP at the leu2Δ5′ repeat at a similar distance from ARSH4 as the RFP observed in HHF-IN. Altogether, these results indicate that TAR is linked to a transcription-dependent RFP, suggesting that the increase in recombination may result from the impairment of replication fork progression caused by transcription.
Figure 4.
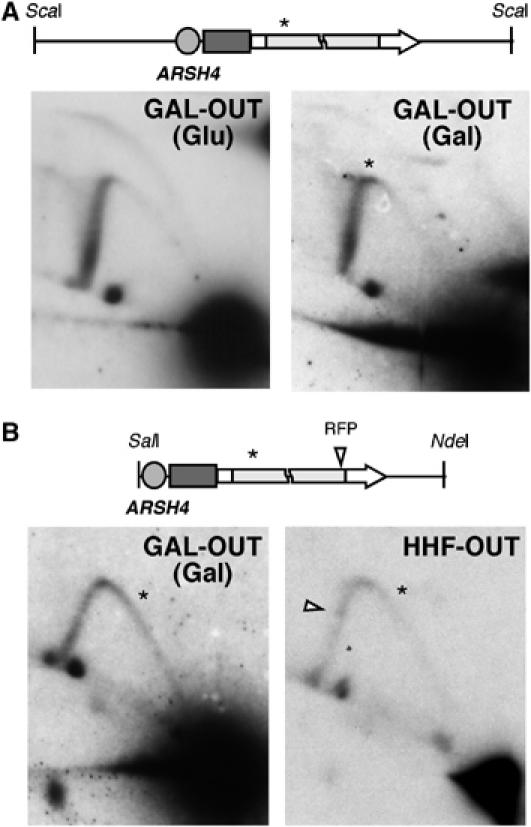
Analysis of the progression of replication forks that are codirectional with transcription. (A) 2D-gel analysis of the ScaI restriction fragment covering the leu2 repeats in the GAL-OUT construct isolated from cells grown in glucose (Glu; transcription OFF) or galactose (Gal; transcription ON). (B) 2D-gel electrophoresis of the SalI–NdeI restriction fragment covering the leu2 repeats in the GAL-OUT and HHF-OUT constructs isolated from cells grown in galactose or glucose, respectively. An open arrow indicates a weak transcription-dependent RFP in the HHF-OUT construct. The asterisks indicate the expected position corresponding for the RFP at the leu2Δ3′ repeat as detected in the GAL-IN and HHF-IN constructs (Figure 3). Note that the signal accumulated in the inflection points of the Y-arcs in the GAL-OUT construct is the consequence of the overlapping left and right arcs, and does not correspond to a true RFP, since it does not change its position in the overlapping fragments.
RFP is independent of the direct repeats
We reported the first evidence of an RFP associated with RNAPII-mediated transcription. We accordingly wondered whether this RFP could be due to the presence of direct repeats in our constructs that lead to pairing intermediates able to pause replication fork progression. Influence of the repeats in the formation of the transcription-dependent RFP was determined by 2D-gel analysis of the HHF-Leu construct, which differs from the HHF-IN construct by the presence of just one repeat unit. As shown in Figure 5, in two overlapping restriction fragments, an RFP of similar intensity and at the same position in the LEU2 gene as in the HHF-IN construct was detected. This indicates that the RFP was independent of the presence or absence of direct repeats.
Figure 5.
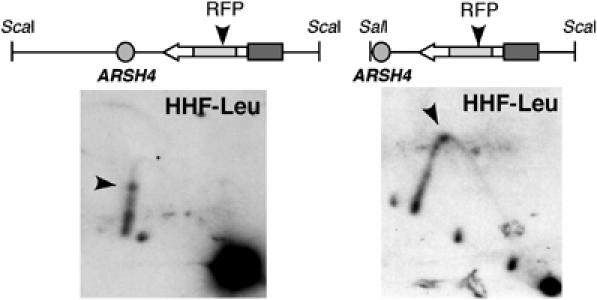
The transcription-dependent RFP is independent of direct repeats. 2D-gel analysis of Leu+ recombinants from the HHF-IN construct containing just one repeat unit (HHF-Leu construct). The overlapping ScaI (left) and SalI–ScaI (right) restriction fragments are shown on top of each panel. A solid arrow indicates the RFP.
The helicase Rrm3 facilitates the progression of the replication fork through the transcription-dependent RFP site
To get further insight into the molecular nature of the transcription-dependent RFP associated with TAR, replication intermediates were analysed by 2D-gel electrophoresis in mutants affected in recombination and/or replication fork progression. Genetic evidence suggests that the rescue of replication forks in yeast requires the coordinated activity of the recombination proteins Rad51 and Rad52, together with at least two 3′- to -5′ DNA helicases, Srs2 and Sgs1, with roles in replication fork progression (Lee et al, 1999; Cobb et al, 2003; Versini et al, 2003) and recombination (Ira et al, 2003; Krejci et al, 2003; Veaute et al, 2003). Srs2 and Sgs1 seem to prevent the accumulation of genotoxic recombination intermediates, as suggested by the observation that the synthetic lethality of the srs2 sgs1 double mutant is rescued by the absence of Rad51 and Rad52 (Gangloff et al, 2000). These helicases are genetically related to Rrm3, a 5′- to -3′ DNA helicase that facilitates both replication fork progression through natural RFPs at centromeres, tRNA genes, inactive replication origins, telomeres and ribosomal DNA (rDNA) (Ivessa et al, 2000, 2002, 2003), and resolution of convergent replication forks at the replication fork barrier (RFB) in the rDNA (Ivessa et al, 2000). As shown for srs2 sgs1, the absence of Rad51 and Rad52 suppresses the synthetic growth defects of the srs2 rrm3 and sgs1 rrm3 double mutants (Ooi et al, 2003; Schmidt and Kolodner, 2004; Torres et al, 2004b). Thus, replication intermediates were analysed in rad51Δ, rad52Δ, srs2Δ, sgs1Δ and rrm3Δ strains.
As shown in Figure 6A, the transcription-dependent RFP at the leu2Δ3′ repeat in the HHF-IN construct (solid arrow) was significantly increased in rrm3Δ cells (37±2% of replication intermediates versus 24±2.5% in the wild type). This increase was accompanied by an accumulation of convergent forks (X-shaped replication intermediates) at the transcription-dependent RFP site, suggesting that the pause lasted long enough as to allow a fraction of the opposite replication forks to reach the RFP site. To determine whether the increase in this RFP detected in rrm3Δ depended on transcription during the S phase, replication intermediates in the GAL-IN and CLN-IN constructs were analysed. The transcription-dependent RFP at the leu2Δ3′ repeat was not detected in rrm3Δ cells in the absence of transcription (GAL-IN (Glu)), when transcription occurred in G1 (CLN-IN) (Figure 6A) or when transcription and replication were codirectional (Figure 6B). However, the transcription-dependent RFP at the leu2Δ5′ repeat in the HHF-OUT construct was also increased in rrm3Δ cells (Figure 6B, open arrow, 20±2% of replication intermediates). These results indicate that the helicase Rrm3 facilitates replication fork progression through an RNAPII transcription-dependent RFP site. Since Rrm3 has been shown to be required for the advance of the replication fork through non-nucleosomal protein–DNA complexes (Ivessa et al, 2003), this suggests that the RNAPII-dependent RFP detected in our constructs is due to the presence of a transcription-dependent nucleoprotein complex. In addition to the transcription-dependent RFP detected in the Y arc, the absence of Rrm3 led to an accumulation of intermediates in the cone formed by the arcs of the X- and double Y-shaped molecules in all constructs (Figure 6A and B), consistent with a general replication impairment. As expected from these results, recombination was increased in all constructs in rrm3Δ (3- to 10-fold above the wild type) (Figure 6C).
Figure 6.
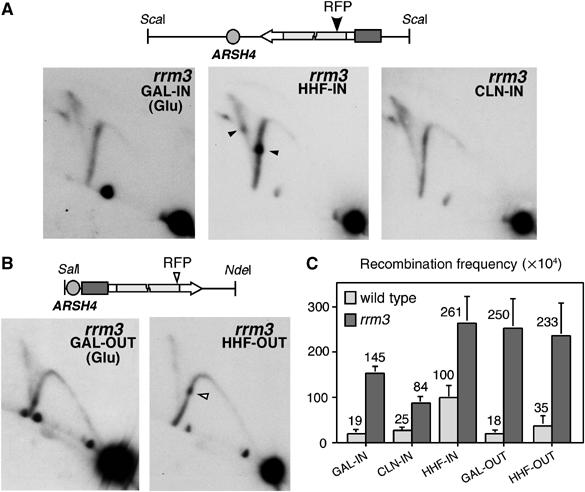
Increase of transcription-dependent RFP and recombination in the absence of the Rrm3 helicase. (A) 2D-gel analysis of the ScaI restriction fragment covering the leu2 repeats in the GAL-IN, HHF-IN and CLN-IN constructs isolated from rrm3Δ cells grown in glucose. Solid arrows indicate the transcription-dependent RFP in the HHF-IN construct. (B) 2D-gel analysis of the SalI–NdeI restriction fragment covering the leu2 repeats in the GAL-OUT and HHF-OUT constructs isolated from rrm3Δ cells grown in glucose. Open arrows indicate the transcription-dependent RFP in the HHF-OUT construct. (C) Recombination frequencies of the GAL-IN, CLN-IN, HHF-IN, GAL-OUT and HHF-OUT constructs in the wild-type and rrm3Δ strains grown in glucose. The average and standard deviation are indicated.
The intensity of the signal at the transcription-dependent RFP site in the HHF-IN construct did not change in rad51Δ, rad52Δ, srs2Δ and sgs1Δ mutants with respect to wild-type cells (Figure 7). In these strains, the proportion of paused replication forks at the transcription-dependent RFP site represented 20–25% of replication intermediates.
Figure 7.
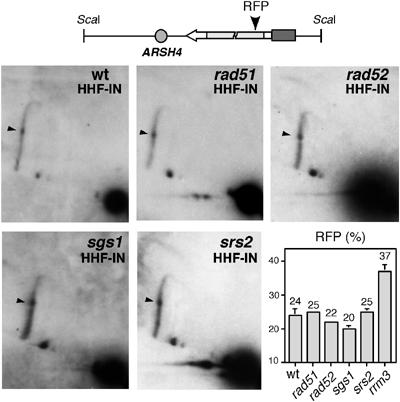
The transcription-dependent RFP in the HHF-IN construct is not affected by rad51Δ, rad52Δ, sgs1Δ and srs2Δ. 2D-gel analysis of the ScaI restriction fragment covering the leu2 repeats in the HHF-IN construct in wild type, rad51Δ, rad52Δ, sgs1Δ and srs2Δ. Solid arrows indicate the transcription-dependent RFP at the leu2Δ3′ repeat. Quantification data of the RFPs relative to the replication intermediates, taken as 100, are shown.
Discussion
Using specifically designed plasmid-borne constructs, we have analysed the effect in yeast that transcription has on recombination, depending on whether or not the transcribed DNA sequence is simultaneously replicated either codirectionally or in a head-on orientation. We show that transcription by itself is not sufficient to induce recombination. TAR requires replication fork progression opposite to transcription and is associated with the appearance of an RFP. The codirectional advance of transcription has little effect on replication fork progression and recombination. The helicase Rrm3 facilitates the advance of the replication fork through the transcription-dependent RFP, suggesting that an RNAPII-dependent nucleoprotein complex participates in the impairment of replication. These results indicate that TAR can be mediated by impairment of replication fork progression.
The effect on genomic integrity of the potential collisions between transcription and replication is poorly understood. Here, we observed that transcription driven from the GAL1 promoter along two direct repeats increased recombination between the repeats if these were simultaneously replicated by an oncoming replication fork. In addition, transcription driven from the HHF2 and HHO1 promoters, which are activated in S phase, but not from the CLN2 promoter, which is activated in G1, increased recombination in the head-on orientation. In contrast, transcription occurring codirectionally with replication had little effect on recombination (Figures 1 and 2). These results indicate that transcription by itself is not sufficient to induce recombination. A head-on and, to a lesser extent, a codirectional encounter of RNAPII-mediated transcription and DNA replication leads to recombinogenic DNA damage. Consistent with this conclusion, transcription of a DNA fragment that is simultaneously replicated by an oncoming replication fork led to the appearance of a transcription-dependent RFP, while transcription occurring codirectionally led to a much weaker transcription-dependent RFP. Transcription occurring in the absence of replication did not affect replication fork progression (Figures 3 and 4).
The appearance of a transcription-dependent RFP indicates that DNA replication is impaired by RNAPII-mediated transcription in our constructs. This impairment is more pronounced when the encounter between transcription and replication is head-on than when it is codirectional. Replication impairment could generate recombinogenic intermediates. In accordance with this possibility, defective DNA replication caused by either replication inhibitors or mutations affecting components of the replication machinery accumulates recombination intermediates in yeast (Zou and Rothstein, 1997; Sogo et al, 2002), and a number of studies have suggested a tight connection between inefficient DNA replication and homologous recombination from bacteria to humans (Rothstein et al, 2000).
A general impairment of replication fork progression along the whole DNA repeat region that would not be detected in the form of RFPs could contribute to TAR. Nevertheless, the transcription-dependent RFP detected in this study could be associated with the initiation of recombination, because it lies in the region of homology and because its appearance correlates with TAR. Indeed, RFPs have been shown to promote recombination. Thus, RFPs generated either by the binding of the Tus protein to the replication termination Ter site in Escherichia coli (Horiuchi et al, 1994; Horiuchi and Fujimura, 1995) or by the absence of the helicase Rrm3 in yeast (Ivessa et al, 2003; Torres et al, 2004a) are associated with an increase in homologous recombination. Also, yeast mutants lacking the RFB-binding protein Fob1, required for RFB activity, display slowed down replication fork progression and repeat instability at the rDNA locus as a consequence of head-on collisions between RNAPI transcription and replication (Takeuchi et al, 2003).
Rrm3 promotes the advance of the replication fork through non-nucleosomal nucleoprotein complexes (Ivessa et al, 2003). Therefore, the observation that the helicase Rrm3 facilitates replication through the transcription-dependent RFP sites (Figure 6) suggests that a transcription-dependent nucleoprotein complex could be responsible for the RFP in our constructs. A transiently arrested RNAPII ternary transcription complex could impair the advance of the replication fork. This is consistent with the observation that natural RFPs found at tRNA genes—which are also polar and increased in the absence of Rrm3 regardless of the transcript length (see below)—require the assembly of the RNAPIII transcription initiation complex (Deshpande and Newlon, 1996; Ivessa et al, 2003). Along the same line, studies using the in vitro-purified bacteriophage T4 and φ29 systems have shown that head-on collisions between the replication fork and the RNAP transcription ternary complex are more disadvantageous than codirectional collisions (Liu et al, 1993; Liu and Alberts, 1995; Elias-Arnanz and Salas, 1997, 1999). In the head-on collision, the φ29 DNA polymerase was blocked by a halted RNAP complex and resumed replication once the RNAP was allowed to move (Elias-Arnanz and Salas, 1999), suggesting that an arrested, in contrast to an elongating, RNAPII ternary transcription complex could impose a physical hindrance to the advance of the replication fork. The more complex replication apparatus of the bacteriophage T4 was shown to pass the RNAP complex, either halted or elongating, after a pause of a few seconds. Interestingly, the T4 replication apparatus required the activity of the gene 41 DNA helicase to solve the head-on, but not the codirectional, collision (Liu et al, 1993; Liu and Alberts, 1995). The DNA helicase Rrm3 might help to detach the RNAPII transcription ternary complex from DNA, as proposed for other nucleoprotein complexes that inhibit replication fork progression (Ivessa et al, 2003). This function is specific of Rrm3 in our constructs, since the transcription-dependent RFP was not affected by the absence of the DNA helicases Sgs1 or Srs2 (Figure 7).
Transcription could also impair replication either by increasing the torsional stress of the DNA or by facilitating the binding of sequence-specific proteins that could act as roadblocks for replication fork progression. The torsional stress is expected to be higher in a head-on than in a codirectional arrangement because the former would accumulate positive supercoiling at the converging region. A head-on, but not a codirectional, collision between transcription and replication has been shown to increase the knotting of the sister chromatids behind the fork in E. coli (Olavarrieta et al, 2002). However, the observations that the transcription-dependent RFP is independent of the transcript length (1.8 kb in the HHF-IN versus 1.2 kb in the HHF-Leu) (Figure 5), and is not spread along the transcribed region, make replication pauses unlikely as a consequence of torsional stress in our constructs. Also unlikely is that a sequence-specific DNA-binding protein pauses replication because the transcription-dependent RFP in the IN constructs was detected at the leu2Δ3′ repeat but not at the same DNA sequence at the leu2Δ5′ repeat. Finally, it is worth noticing that the detection of transcription-dependent RFPs may depend on topological constraints, because such RFPs are not easily detected in linearized plasmids (our preliminary observations with the pARSGLB-IN plasmid, containing GAL-IN, linearized at the telomeric sequences). In this regard, RFPs are not detected downstream of the endogenous GAL1 and GAL10 promoters (Ivessa et al, 2003). Further molecular analyses would be required to determine the importance of supercoiling stress and/or other DNA structural parameters in the formation of transcription-dependent RFPs.
In summary, the link between a transcription-dependent RFP and recombination observed here sheds light on understanding the mechanism by which transcription induces recombination and underlines the relevance of homologous recombination as a DNA repair mechanism connected with DNA replication. Our work raises the question of whether other cases of TAR, such as recombination associated with cotranscriptionally formed RNA:DNA hybrids of yeast THO mutants (Huertas and Aguilera, 2003) or the generation of genetic diversity during class switching of Ig genes (Jung et al, 1993; Peters and Storb, 1996), are also linked to impairment of replication fork progression. Our results provide evidence that head-on collisions between transcription and replication can be a source of genomic instability. This might explain the preferential positioning in the leading strand of essential genes in bacteria (Rocha and Danchin, 2003), and the presence of polar RFBs that prevent head-on collisions between RNAPI transcription and replication at the rDNA locus in eukaryotes (Brewer and Fangman, 1988; Little et al, 1993; Wiesendanger et al, 1994; Gerber et al, 1997). A tight control of transcription during DNA replication may be, therefore, essential for the prevention of genetic instability and for proper control of developmentally programmed chromosomal rearrangements.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study were BY4741 (a his3Δ0 leu2Δ0 ura3Δ0) and its isogenic Y10540 (rad52Δkan), Y16401 (rad51Δkan), Y01331 (srs2Δkan), Y10775 (sgs1Δkan) and Y00994 (rrm3Δkan). Yeast cells were grown in synthetic complete (SC) medium as described (Kaiser et al, 1994). Plasmids pARSGLB-OUT, pARSGLB-IN, pARSHLB-OUT, pARSHLB-IN, pARSCLB-IN and pARSXLB-IN are yeast centromeric plasmids containing the GAL-OUT, GAL-IN, HHF-OUT, HHF-IN, CLN-IN and HHO-IN recombination constructs, respectively. They are based on plasmid pFERNU, which was constructed by cloning ARSH4, URA3, CEN6 and the 83 bp (C-A1-3)n telomeric sequences in pRS304 (Sikorski and Hieter, 1989) lacking the EcoRI site at the polylinker. The PCR-amplified ARSH4 sequence was inserted at the ClaI–SalI site; the URA3 marker obtained by HindIII digestion from YEp24 and made blunt ended was inserted at the SmaI site; and the PCR-amplified CEN6 sequence was inserted at the SacII site. The telomeric sequences were cloned in two steps: (1) two BamHI (made blunt ended)–EcoRI (C-A1-3)n fragments were inserted as inverted repeats at the AatII site (made blunt ended) and (2) an NruI-containing linker was inserted at the resulting EcoRI site. pARSGLB-IN and pARSGLB-OUT were constructed by inserting the SacI–ApaI (made blunt ended) fragment of p314GLB (Piruat and Aguilera, 1998), containing the leu2Δ3′∷leu2Δ5′ direct-repeat recombination system (Prado and Aguilera, 1995) under control of the GAL1 promoter, at the ClaI (made blunt ended) site of pFERNU. The fragment was cloned either inward (pARSGLB-IN) or outward (pARSGLB-OUT) of the ARSH4 sequence with respect to the direction of transcription. pARSHLB-IN and pARSHLB-OUT were cloned in two steps: (1) the SacI–BamHI fragment from pRS314-GLB, containing the GAL1 promoter, was replaced by a PCR fragment that contained the HHF2 promoter (pRS314-HLB) and (2) the SacI–ApaI fragment (made blunt ended) from pRS314-HLB, containing the HHF2pr∷leu2Δ3′∷leu2Δ5′ recombination system, was inserted at the ClaI (made blunt ended) site of pFERNU either in the IN (pARSHLB-IN) or the OUT orientation (pARSHLB-OUT). pARSCLB-IN and pARSXLB-IN were constructed by replacing either the AatII or the NarI–SphI fragment of pARSHLB-IN, containing the HHF2 promoter, with a PCR fragment containing the CLN2 or HHO1 promoter, respectively. PCR amplifications were made with oligos 5′-attcatcgattatatgtaaagtacgctttt-3′ and 5′-tcaggtcgactaataatggtttcttaggac-3′ (ARSH4), 5′-cattaccgcggcttttcatcacgtgctataa- 3′ and 5′-tgaatccgcggttttacatcttcggaaaaca- 3′ (CEN6), 5′-cattagagctcgacgtcgcatgcgttatcacg caaactatgttttgac-3′ and 5′-gcgtaggatccggcgccgacgtctattttatt gtattgattgttgttt-3′ (HHF2 promoter), 5′-cattagacgtcgcatgccgaactaaagcaact atacattg-3′ and 5′-gcgtaggcgccgacgtctgtctgtcgttaaat ttaatgaa-3′ (CLN2 promoter) and 5′-cattagacgtcgcatgctataactgatatgta aactgtgc-3′ and 5′-gcgtaggcgccgacgtcgttgctttagttgta ttataatt-3′ (HHO1 promoter). Plasmids pARSHLB-Leu and pARSCLB-Leu are yeast centromeric plasmids containing the HHF-Leu and CLN-Leu constructs, respectively. They resulted by homologous recombination between the leu2 repeats of the HHF-IN and CLN-IN constructs, respectively.
Recombination analysis
Spontaneous recombination frequencies were obtained as the average value of median frequencies obtained by 6–10 fluctuation tests performed with 2–3 independent transformants. For each fluctuation test, six independent colonies were analysed as previously described (Prado and Aguilera, 1995). Note that the entire endogenous LEU2 gene was completely deleted in all strains used in this study, so that Leu+ recombinants could only arise by recombination between the leu2 repeats of the plasmid-borne recombination constructs.
RNA level analysis
Total RNA from 2–4 independent transformants was extracted and analysed by Northern hybridization as previously published (Chavez and Aguilera, 1997). mRNA was probed with the 598 bp ClaI–EcoRV internal LEU2 fragment, quantified in a Fuji FLA3000 and normalized with respect to the 25S rRNA value. Run-on analysis was performed as previously described (Chavez and Aguilera 1997). As probes we used PCR fragments obtained with primers 5′-gttccacttccagatgaggc-3′ and 5′-ttagcaaattgtggcttgat-3′ (leu2-1), 5′-gttttggcctcttcaagatt-3′ and 5′-gttcttgtctggcaaagagg-3′ (leu2-2), 5′-ttggagagggcaactttgg-3′ and 5′-caggatcggtcgattgtgc-3′ (rDNA) and 5′-tcgttgctgcataaaccg-3′ and 5′-tcgataatttcaccgccg-3′ (lacZ). A 1 μg portion of each DNA probe was immobilized in Hybond-N membranes. Filters were hybridized with in vivo α32P-UTP-labelled total RNA extracted from strains harbouring either the CLN-Leu or HHF-Leu construct. The run-on was performed twice with similar results.
Analysis of replication intermediates
Total DNA from 0.5 l of mid-log-phase cells was isolated according to Allers and Lichten (2000) with the modifications of Wellinger et al (2003) in the absence of hexamine cobalt trichloride (HCC). After digestion with the appropriate restriction enzymes, DNA was enriched in replication intermediates by selective adsorption to BND-cellulose (Huberman et al, 1987). DNA molecules were resolved by neutral/neutral 2-D gel electrophoresis as described previously (Brewer and Fangman, 1987) and probed with the 598 bp ClaI–EcoRV LEU2 fragment. Quantification of the RFP signal was determined relative to the total intensity of the replication intermediates, and the average and standard deviations of two (rad51Δ, rad52Δ, sgs1Δ, srs2Δ) or three (wild type and rrm3Δ) independent quantifications are plotted.
Acknowledgments
We thank M Nieto and J Escalante for technical assistance, RE Wellinger for help on the replication intermediate analysis, F Antequera for critical reading of the manuscript and D Haun for style supervision. The research was funded by the Spanish Ministry of Science and Technology (BMC2000-0439 and SAF2003-00204 grants).
References
- Aguilera A (2002) The connection between transcription and genomic instability. EMBO J 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Lichten M (2000) A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res 28: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 [DOI] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW (2002) ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chavez S, Aguilera A (1997) The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 11: 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT (2003) DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064–1067 [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ Marians KJ (2000) The importance of repairing stalled replication forks. Nature 404: 37–41 [DOI] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS (1996) DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Elias-Arnanz M, Salas M (1997) Bacteriophage phi29 DNA replication arrest caused by codirectional collisions with the transcription machinery. EMBO J 16: 5775–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias-Arnanz M, Salas M (1999) Resolution of head-on collisions between the transcription machinery and bacteriophage phi29 DNA polymerase is dependent on RNA polymerase translocation. EMBO J 18: 5675–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman KB, Karns LR, Lutz KA, Smith MM (1992) Histone H3 transcription in Saccharomyces cerevisiae is controlled by multiple cell cycle activation sites and a constitutive negative regulatory element. Mol Cell Biol 12: 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S (1992) Consequences of replication fork movement through transcription units in vivo. Science 258: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Gangloff S, Lieber MR, Rothstein R (1994) Transcription, topoisomerases and recombination. Experientia 50: 261–269 [DOI] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25: 192–194 [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A (2003) Recombinogenic effects of DNA-damaging agents are synergistically increased by transcription in Saccharomyces cerevisiae. New insights into transcription-associated recombination. Genetics 165: 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber JK, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F (1997) Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90: 559–567 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Barrera S, García-Rubio M, Aguilera A (2002) Transcription and double-strand-breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics 162: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford LM, Osley MA, Ludwig TR II, McLaughlin CS (1981) Cell-cycle regulation of yeast histone mRNA. Cell 24: 367–375 [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y (1995) Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol 177: 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M (1994) The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol 176: 4656–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman JA, Spotila LD, Nawotka KA, el-Assouli SM, Davis LR (1987) The in vivo replication origin of the yeast 2 microns plasmid. Cell 51: 473–481 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA (2002) Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev 16: 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Zakian VA (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489 [DOI] [PubMed] [Google Scholar]
- Jung S, Rajewsky K, Radbruch A (1993) Shutdown of class switch recombination by deletion of a switch region control element. Science 259: 984–987 [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis M, Mitchell A (1994) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Kolodner RD, Putnam CD, Myung K (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM (1998) Transcription through a simple DNA repeat blocks replication elongation. EMBO J 17: 5095–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286: 2339–2342 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B (1998) Genetic instabilities in human cancers. Nature 396: 643–649 [DOI] [PubMed] [Google Scholar]
- Little RD, Platt TH, Schildkraut CL (1993) Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol 13: 6600–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Alberts BM (1995) Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267: 1131–1137 [DOI] [PubMed] [Google Scholar]
- Liu B, Wong ML, Tinker RL, Geiduschek EP, Alberts BM (1993) The DNA replication fork can pass RNA polymerase without displacing the nascent transcript. Nature 366: 33–39 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG (2000) Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101: 35–45 [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD (2001) Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408 [DOI] [PubMed] [Google Scholar]
- Olavarrieta L, Hernandez P, Krimer DB, Schvartzman JB (2002) DNA knotting caused by head-on collision of transcription and replication. J Mol Biol 322: 1–6 [DOI] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD (2003) DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet 35: 277–286 [DOI] [PubMed] [Google Scholar]
- Peters A, Storb U (1996) Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4: 57–65 [DOI] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A (1998) A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 17: 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (1995) Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics 139: 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Piruat JI, Aguilera A (1997) Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J 16: 2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A (2003) Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet 34: 377–378 [DOI] [PubMed] [Google Scholar]
- Rothstein R, Michel B, Gangloff S (2000) Replication fork pausing and recombination or ‘gimme a break'. Genes Dev 14: 1–10 [PubMed] [Google Scholar]
- Saxe D, Datta A, Jinks-Robertson S (2000) Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol Cell Biol 20: 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD (2004) Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol 24: 3213–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B (1998) RuvAB acts at arrested replication forks. Cell 95: 419–430 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C (1994) Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol 14: 4788–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Torres JZ, Bessler JB, Zakian VA (2004a) Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev 18: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Schnakenberg SL, Zakian VA (2004b) Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol Cell Biol 24: 3198–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P (2003) The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J 22: 1939–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Schar P, Sogo JM (2003) Rad52-independent accumulation of joint circular minichromosomes during S phase in Saccharomyces cerevisiae. Mol Cell Biol 23: 6363–6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesendanger B, Lucchini R, Koller T, Sogo JM (1994) Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res 22: 5038–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Sugimoto K, Reed SI (1990) G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell 62: 225–237 [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR (2003) R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4: 442–451 [DOI] [PubMed] [Google Scholar]
- Zou H, Rothstein R (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90: 87–96 [DOI] [PubMed] [Google Scholar]


